Molecular-genetic comparisons and manipulations of regulators of stomatal development raise the possibility of a single origin for stomata early in land plant evolution.
Abstract
The fossil record suggests stomata-like pores were present on the surfaces of land plants over 400 million years ago. Whether stomata arose once or whether they arose independently across newly evolving land plant lineages has long been a matter of debate. In Arabidopsis, a genetic toolbox has been identified that tightly controls stomatal development and patterning. This includes the basic helix-loop-helix (bHLH) transcription factors SPEECHLESS (SPCH), MUTE, FAMA, and ICE/SCREAMs (SCRMs), which promote stomatal formation. These factors are regulated via a signaling cascade, which includes mobile EPIDERMAL PATTERNING FACTOR (EPF) peptides to enforce stomatal spacing. Mosses and hornworts, the most ancient extant lineages to possess stomata, possess orthologs of these Arabidopsis (Arabidopsis thaliana) stomatal toolbox genes, and manipulation in the model bryophyte Physcomitrella patens has shown that the bHLH and EPF components are also required for moss stomatal development and patterning. This supports an ancient and tightly conserved genetic origin of stomata. Here, we review recent discoveries and, by interrogating newly available plant genomes, we advance the story of stomatal development and patterning across land plant evolution. Furthermore, we identify potential orthologs of the key toolbox genes in a hornwort, further supporting a single ancient genetic origin of stomata in the ancestor to all stomatous land plants.
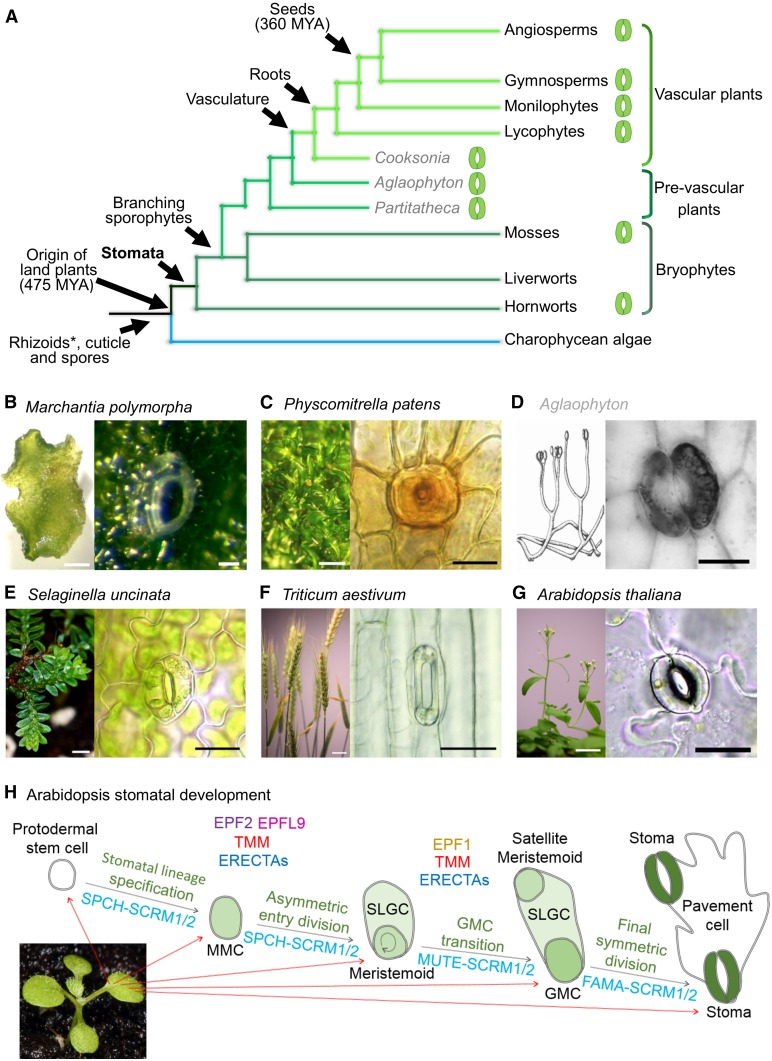
Stomata, microscopic turgor-driven valves formed by guard cells, are present on the aerial surfaces of most land plants (Fig. 1, A–G). The regulation of stomatal apertures controls plant water loss, promotes the uptake of carbon dioxide, and in many cases assists in regulating internal temperatures (Zeiger et al., 1987; Mustilli et al., 2002; Xu et al., 2016). Stomata are also a major site of pathogen entry and plant defense (Gudesblat et al., 2009). Despite their central role in so many processes, their origins and evolutionary history have long been a matter of considerable debate (Payne, 1979; Chater et al., 2011; Pressel et al., 2014; Franks and Britton-Harper, 2016; McAdam and Brodribb, 2016). Along with root-like structures, a waxy cuticle, and vasculature, stomata were a key innovation that enabled plants to conquer the land (Fig. 1A; Berry et al., 2010). The presence of stoma-like structures on very ancient land plant fossils, the absence of stomata in liverworts, the apparent secondary losses of stomata from several basal and highly derived clades, as well as developmental, morphological, and physiological variation have presented plant biologists with many quandaries when interpreting how and when stomata have evolved (Haig, 2013; Rudall et al., 2013; Pressel et al., 2014). Their presence and absence across the land plant phylogeny (Fig. 1A) presents difficulties in understanding major transitions in plant evolution. Owing to the apparent conflicting evidence, the fundamental question remains as to whether stomata are monophyletic in origin. We are now in an exciting era where tractable genetic plant systems and corresponding sequenced genomes are plentiful, so the definitive answer to this question is close. In this work, we discuss the recent literature relating to the evolution of the signaling components that regulate stomatal development and propose what future research might be needed to shed more light on the origin and role of stomata in aiding in the terrestrialization of life on Earth.
Figure 1.
The evolution and origin of stomata in land plants. A, Recently proposed land plant phylogeny including extinct early land plant representatives (labeled gray) based on Wickett et al. (2014), Edwards et al. (2014), and Chen et al. (2017). Lineages that have stoma-bearing representatives are marked with an adjacent stomatal image. Rhizoids are marked with an asterisk, as the evolution of these structures is still debated (Tam et al., 2015). B to G, Representatives of nonvascular and vascular land plant species with images of pore (B) or stomata (C–G). In extant plants, stomata are found on the sporophyte. Left image in D reproduced with permission of the Linnean Society of London. Right image in D is reproduced from Edwards and Kerp, Stomata in early land plants: An anatomical and ecophysiological approach, 1998, 49, 255–278, with permission of Oxford University Press. G, Diagram to illustrate the control of stomatal developmental transitions in the angiosperm Arabidopsis (seedling image). A subset of protodermal cells enters the stomatal lineage and take on MMC identity. MMCs undergo an asymmetric cell division producing a smaller meristemoid and a larger SLGC, through the actions of the bHLH transcription factors SPCH and ICE1/SCRM or SCRM/2. EPF2 and EPFL9 compete for the binding of a number of ERECTA plasma membrane receptors. These interactions are modulated via the membrane protein TMM. After the asymmetric division, the larger SLGC either exits the stomatal lineage and takes on a pavement cell identity or undergoes a further division to form a satellite meristemoid (not shown). Meristemoids differentiate in to a GMC via the activity of heterodimeric bHLH MUTE and SCRM/2. EPF1 peptide signals extracellularly via ERECTAs (preferentially ER-like1), again modulated by TMM, to restrict GMC formation. GMCs undergo a symmetric division induced by FAMA and SCRM/2 activity to form a pair of guard cells. Scale bars: B, left and right 100 µm; C, left 200 µm, right 20 µm; D, right 50 µm; E, left 200 µm, right 20 µm; F, left 20 mm, right 50 µm; G, left 20 mm, 20 µm; H, 250 µm.
This update focuses on the origins and evolution of the molecular and genetic machinery involved in stomatal production on the plant epidermis. Although we discuss the origins of stomatal function in the context of these new discoveries, the evolution of guard cell signaling and stomatal behavior has recently been reviewed (Assmann and Jegla, 2016; Chen et al., 2017; Xu et al., 2016). The complex cellular processes underpinning stomatal development, also the subject of several recent reviews (Torii, 2015; Han and Torii, 2016; Simmons and Bergmann, 2016), will be outlined briefly to provide the background to the evo-devo context.
SUPERFICIAL SIMILARITIES, SUPERFICIAL DIFFERENCES: LESSONS FROM ACROSS THE CLADES
The strikingly similar morphologies of stomata across evolutionary time and across extant land plants (Fig. 1, B–G) arguably belie the often stark variation that has arisen from natural selection. This variation includes differences in ontogenetic decision making, environmental control of patterning, and final stomatal size and shape. For example, the mature stomata of equisetum and some extinct fossil lineages possess silicified radiating ribs not seen in other taxa (Cullen and Rudall, 2016), but silicification has arisen in stomata of diverse lineages (Trembath-Reichert et al., 2015). We therefore have to carefully untangle those shared phenotypes that have come about from convergent processes and those that have a genuinely shared ancestry and shared genetic module. A clear example of this issue is the evolution of epidermal cell files and stomatal rows, as can be observed in monocots such as lilies and grasses, but also in older groups such as conifers and far more ancient groups such as equisetum. By studying the similarities and differences in stomatal development and patterning between these disparate groups, we can more clearly see the pitfalls of assigning homology (or lack of homology) based on morphology and other visible/observable characteristics alone (Rudall et al., 2013; Rudall and Knowles, 2013; Cullen and Rudall, 2016). The wealth of genomic and transcriptomic data becoming available for more species across the land plant phylogeny may now allow us to probe how deep in time such similarities reach and where novel adaptations have arisen along the way. By experimentally probing the conservation of protein function and the gene networks involved in stomatal development and patterning, we can more definitively assign where homology is present.
THE DICOTYLEDONOUS ANGIOSPERM ARABIDOPSIS: THE “ARCHETYPAL” STOMATAL MODEL
Much of what we know regarding the molecular genetic control of stomatal development comes from studies involving the genetic model species Arabidopsis (Fig. 1, G and H). Arabidopsis was the original workbench used for studying stomatal genetics and continues to provide much insight into how stomata develop and function (Yang and Sack, 1995; Chater et al., 2015; Han and Torii, 2016; Qi et al., 2017). Such advances have identified many of the key genetic players responsible for permitting entry into the stomatal lineage, the formation of the meristemoid, and the subsequent divisions and transitions that lead to the formation of stomata (Zhao and Sack, 1999; Ohashi-Ito and Bergmann, 2006; Hara et al., 2007; MacAlister et al., 2007; Pillitteri et al., 2007; Kanaoka et al., 2008; Hunt et al., 2010; Sugano et al., 2010). The activity of the Arabidopsis meristemoid in particular has been shown to be intricately regulated by a multitude of endogenous signaling pathways and environmental cues thereby enabling control over stomatal density and spacing during development (Chater et al., 2014; Lau et al., 2014). Owing to an extensive knowledge base, recent studies in stomatal evolutionary development and physiology invariably call on Arabidopsis to compare and contrast systems when making evolutionary interpretations (Chater et al., 2011; MacAlister and Bergmann, 2011; Caine et al., 2016). Our thinking is inevitably pigeon-holed, however, because Arabidopsis is a dicot angiosperm of the Brassicaceae family, and the caveat remains that apparent “deviations” from what we observe in Arabidopsis stomata may turn out to be more appropriate models for land plants as a whole. Nevertheless, several recent stomatal evolution studies strongly support Arabidopsis’s continuing role in informing our thinking (Caine et al., 2016; Chater et al., 2016; Raissig et al., 2016, 2017)
ARABIDOPSIS STOMATAL DEVELOPMENT: STOMATAL ONTOGENY SPELLED OUT IN GENES
Like most other land plants, stomata in Arabidopsis are comprised of a pair of guard cells that surround a central pore (Fig. 1G). A regulated series of cellular divisions ensure that once mature, each stoma is typically spaced by at least one pavement cell (Fig. 1H; Zhao and Sack, 1999; Geisler et al., 2000; Hara et al., 2007). The development of Arabidopsis stomata begins when epidermal (protodermal) stem cells are specified via group Ia bHLH transcription factor SPCH in a heterodimeric association with its group IIIb bHLH partners, SCRM or SCRM2 (also known as INDUCER OF CBF EXPRESSION1 and 2 in some studies; MacAlister et al., 2007; Kanaoka et al., 2008). Once specified, protodermal cells transition to meristemoid mother cells (MMCs) that then asymmetrically divide, again promoted via SPCH-SCRM/SCRM2 activity, to yield a smaller meristemoid and a larger stomatal lineage ground cell (SLGC). The meristemoid can undergo a number of self-renewing amplifying divisions via continued functioning of SPCH-SCRM/SCRM2 or can transition further into the stomatal lineage to become a guard mother cell (GMC) via the actions of MUTE (a group Ia bHLH related to SPCH) again in combination with SCRM/SCRM2 (Pillitteri et al., 2007, 2008; Kanaoka et al., 2008). For a pair of guard cells to form, a GMC must undergo a final symmetric division, which is facilitated by FAMA (a third group Ia bHLH related to SPCH and MUTE) in partnership with either of the broadly functioning SCRMs (Fig. 1H; Ohashi-Ito and Bergmann, 2006; Kanaoka et al., 2008). Concurrently, SLGCs formed by asymmetric divisions can undergo a further asymmetric spacing division to produce a satellite meristemoid, which itself can advance in the stomatal lineage to yield an additional stoma, spaced by a pavement cell (Zhao and Sack, 1999).
It has become clear in Arabidopsis that for stomatal development to be correctly integrated into other aspects of development and to prevent stomata from forming adjacent to one another, a number of extracellular and plasma membrane-bound proteins are essential to coordinate signals between developing stomatal and epidermal pavement cells (Yang and Sack, 1995; Shpak et al., 2005; Rychel et al., 2010; Meng et al., 2015). Some of the key players include the EPF and EPF-like signaling peptides, the Leu-rich repeat ERECTA family of membrane receptor kinases (ERECTA [ER], ERECTA-LIKE1 [ERL1], and ERL2) and the Leu-rich repeat membrane protein TOO MANY MOUTHS (TMM; Fig. 1H). Of importance during early stomatal development are the negatively acting EPF2 and positively acting EPFL9 (also known as STOMAGEN) peptides that compete antagonistically for binding to ERECTA family proteins (most specifically ER), an interaction modulated by TMM (Fig. 1H; Hara et al., 2009; Hunt and Gray, 2009; Hunt et al., 2010; Kondo et al., 2010; Sugano et al., 2010; Lee et al., 2012, 2015). Later in the stomatal lineage, EPF1 interacts with ERECTAs (primarily ERL1), again possibly overseen by TMM, to prevent GMC transitioning (Hara et al., 2007; Lee et al., 2012; Jewaria et al., 2013; Qi et al., 2017). This prevents neighboring cells from becoming stomata and promotes appropriate stomatal patterning and spacing. The signals transduced via EPF2 peptides are relayed via a MAPK signaling cascade, resulting in phosphorylation and inactivation of the nuclear residing SPCH (Wang et al., 2007; Lampard et al., 2008, 2009). It is still unclear as to whether MUTE and FAMA, which act later in the lineage, are also regulated via a MAPK pathway. The development and patterning modules outlined above and in Figure 1H involve probably hundreds, if not thousands, of up- and downstream components for the proper development and maturation of stomata and their neighboring cells and are modulated further by environmental signals and feedback from other hormone pathways (Casson et al., 2009; Chater et al., 2014, 2015; Engineer et al., 2014; Lau et al., 2014). Nevertheless, the available molecular evidence strongly indicates that the increasingly complex picture we are uncovering of Arabidopsis stomatal development relies on a core module of genes that was first recruited in some of the earliest land plants, well over 400 million years ago (Fig. 1A; Peterson et al., 2010; MacAlister and Bergmann, 2011; Villagarcia et al., 2012; Chater et al., 2013; Takata et al., 2013).
ANGIOSPERM DIVERGENCE IN STOMATAL EVOLUTION: MONOCOTS VERSUS DICOTS
A topical example of the extent to which a core genetic module has been tweaked and rewired over more recent evolutionary time is in the comparison between monocot and dicot stomatal development (Raissig et al., 2016, 2017). At first sight, monocot and dicot stomata appear distinct, but to what extent do these differences in gross morphology reflect molecular divergence? The divergence of angiosperms into monocots, with parallel leaf vasculature and rows of stomata with dumb-bell-shaped guard cells, and dicots, with reticulated venation and irregularly positioned stomata with kidney-shaped guard cells, has long been a point of botanical interest (Zeiger et al., 1987; Rudall et al., 2013). The recent explosion in genomic resources available for grasses and the focus on monocot model species as well as grain crop genetics has enriched our understanding of the evolution of stomatal development pathways in monocots and provided a timely contrast with the model dicot Arabidopsis (Chen et al., 2017). These studies show that the partnership between the ICE/SCRM bHLHs and the SPCH, MUTE, and FAMA-like bHLHs (referred to here as SMFs) is essential for stomatal initiation and maturation in monocots, but that their protein function and regulation differ from Arabidopsis in fundamental ways (Liu et al., 2009; Raissig et al., 2016, 2017). For example, in the grass Brachypodium distachyon, there is specialization of ICE1 and SCRM2 functions, whereas these proteins appear to be redundant in Arabidopsis (Kanaoka et al., 2008; Raissig et al., 2016). Similarly, a novel SPCH duplication and neofunctionalization has occurred in Brachypodium, which suggests that ancestral grass stomatal development as a whole may have come under novel evolutionary pressures (Chen et al., 2016, and references therein). Indeed, SPCH gene duplication appears to be a common theme among monocots (Liu et al., 2009; Chater et al., 2016), but the extent to which this represents a divergence in gene function requires further study. Recent data from the analysis of BdMUTE has revealed how the acquisition of protein mobility has allowed this transcription factor to acquire a function in subsidiary cell patterning in grasses, providing insight into a novel evolutionary mechanism in stomatal evolution (Raissig et al., 2017).
One-cell spacing is tightly controlled across land plants (Hara et al., 2007; Rudall et al., 2013; Caine et al., 2016), superficially appearing even more rigidly imposed in the strict cell files of the monocots. Although to date, few studies have been published that focus on the extracellular signals involved in stomatal patterning in the grasses, it appears that EPF, TMM, and ERECTA orthologs are present within the monocots (Caine et al., 2016). As with dicots such as Arabidopsis, the monocot EPF/L peptide family is diverse, and its members probably partake in both stomatal and nonstomatal processes. The presence of putative grass orthologs of Arabidopsis EPF1, EPF2, and EPFL9 (Caine et al., 2016) suggests that they too act on the SPCH-MUTE-FAMA-mediated transitions that optimize stomatal spacing. However, the functions of EPF/Ls may be subtly divergent between dicots and monocots, in line with distinct differences in their stomatal developmental ontogeny. For example, in Arabidopsis, the negatively acting EPF2 regulates asymmetric entry divisions and subsequent meristemoid activity, thereby inhibiting amplifying divisions (Hara et al., 2009; Hunt and Gray, 2009; Caine et al., 2016). Conversely, in grasses, no such amplifying divisions are apparent, as the asymmetric entry division leads directly to a GMC (and a SLGC; Liu et al., 2009; Luo et al., 2012; Raissig et al., 2016). Moreover, the function of EPF1-like peptides also appears divergent between Arabidopsis and grasses, as Arabidopsis EPF1 predominantly regulates the transition from meristemoid to GMC (Hunt and Gray, 2009; Han and Torii, 2016; Qi et al., 2017), another ontogenetic step not seen in grasses (Liu et al., 2009; Luo et al., 2012). Clearly, understanding how EPF/Ls regulate stomatal development in grasses will not only expand our understanding of stomatal developmental ontogeny, but might also provide crop researchers with invaluable new stomatal phenotypes with which to study biotic and abiotic stresses in socioeconomically important species.
EVIDENCE AND COUNTEREVIDENCE FOR MULTIPLE INDEPENDENT ORIGINS OF STOMATA
Raven (2002) proposed the idea of a “monophyly” of stomata, and the idea has been subsequently expanded and also repeatedly put into question as molecular phylogenies and relationships between bryophytes and other basal clades have been revised (see Fig. 1A for one example; Qiu et al., 2006; Haig, 2013; Pressel et al., 2014; Ruhfel et al., 2014; Wickett et al., 2014; Chen et al., 2017). There are several possible scenarios of stomatal origins, as proposed by Haig (2013), Pressel et al. (2014), and others. These scenarios can be reconsidered in the light of recent revisions to our understanding of the land plant phylogeny (Fig. 1A). One previous consensus view of land plant evolution considers liverworts as the basal lineage followed by the evolution of the mosses, then the hornworts and then the tracheophytes (Qiu et al., 2006; Bowman, 2011). The scenarios proposed are (1) a single origin of stomata in the ancestor of all extant land plants, but with total loss in the ancestor of the stomataless liverwort clade (Chen et al., 2016); (2) a single origin of stomata in the ancestor of mosses, hornworts, and the vascular plants, as supported by evidence of conserved guard cell signaling and function (Chater et al., 2011; Ruszala et al., 2011; Haig, 2013; Franks and Britton-Harper, 2016); and (3) independent origins of stomata in the ancestor of peristomate mosses, the ancestor of the hornworts and the ancestor of modern-day tracheophytes, based on morphological and functional differences between the stomata of different lineages (Pressel et al., 2014). This latter scenario implies multiple independent origins across land plants whereby the stomata of peristomate mosses, hornworts, and vascular plants evolved convergently (Pressel et al., 2014).
One problem with respect to the single origin scenarios is the absence of stomata in the basal mosses Takakia and Andreaea, as well as the presence of so-called psuedostomata in Sphagnum (Duckett et al., 2009). The secondary “losses” of stomata in these clades, however, could be seen to parallel the loss of stomata and stoma-associated gene networks in aquatic and semiaquatic vascular plants, such as Isoetes (Yang and Liu, 2015) or the seagrass Zostera marina (Olsen et al., 2016). Furthermore, such losses appear to be a common occurrence within more derived, typically stomatous moss lineages (Egunyomi, 1982). Similarly, as Chater et al. (2016) show, the genetic ablation of stomata from the moss P. patens results in only apparently minor fitness consequences, suggesting that under certain environmental conditions stomata might be lost.
Further potentially confusing issues that have given rise to unnecessary contention and controversy in the stomatal evo-devo literature depend on interpretations of conservation and homologous form and function. For example, it has recently been stated that there is no evidence of homology between hornwort stomata and those of peristomate mosses and vascular plants, and, instead, these structures are likely to have evolved in parallel (Pressel et al., 2014). These conclusions, based on ontogenetic differences and ultrastructural and cytological considerations such as plastid development, are perhaps a little premature in the absence of molecular studies. What is clear is that when considered in the context of their development, form, and function, the stomata of hornworts and indeed mosses appear to have differences compared with those found in vascular land plants (Merced and Renzaglia, 2013, 2016; Rudall et al., 2013; Pressel et al., 2014; Chater et al., 2016). Such differences in the mosses and hornworts include an absence of asymmetric entry divisions and self-renewing amplifying divisions during development and the presence in these species of initially liquid-filled substomatal cavities, a trait not observed in vascular land plants (Pressel et al., 2014; Merced and Renzaglia, 2016). The loss of this fluid from the substomatal cavities of hornworts and perhaps mosses coincides with sporophyte maturation, perhaps aiding dehydration, dehiscence (lysis), and subsequent spore dispersal.
SINGING FROM THE SAME HYMN SHEET: FUNCTIONAL ORTHOLOGY OF STOMATAL DEVELOPMENTAL GENES BETWEEN LAND PLANTS
The strength of molecular evo-devo and phylogenetic approaches to understanding land plant morphological evolution has been demonstrated in studies of root development (Menand et al., 2007; Jones and Dolan, 2012; Tam et al., 2015). The production of rhizoids on moss gametophytes and the production of root hairs on the sporophytes of both monocot and dicot angiosperms have been shown to be governed by deeply conserved bHLH orthologs despite millions of years of evolutionary divergence. However, unlike with rhizoids and root hairs where deeply conserved homologous genes have been co-opted from gametophyte to sporophyte in extant land plants, stomata only feature on sporophytes.
Two recent studies indicate that there could be strong conservation in the fundamental mechanisms by which all land plants form stomata. Caine et al. (2016) and Chater et al. (2016) show that in the moss P. patens (Fig. 1C), which belongs to one of the most anciently diverging land plant lineages possessing stomata (Fig. 1A), the core molecular machinery required to instigate and pattern stomata is derived from the same common ancestor as Arabidopsis. Specifically, for moss stomata to form, orthologs of a FAMA-like gene, PpSMF1, and an ICE/SCRM like gene, PpSCRM1, must be present; mirroring the key regulatory steps in Arabidopsis stomatal development (Chater et al., 2016). Strikingly, when either PpSMF1 or PpSCRM1 genes are knocked out, moss plants fail to produce stomata. Moreover, and again similar to Arabidopsis, for moss stomata to be correctly spaced and develop properly a functioning EPF-ERECTA-TMM patterning module must be in operation (Caine et al., 2016). This molecular evidence demonstrates the conservation of a stomatal developmental toolkit between taxa separated by over 400 million years of evolution and implies a possible universality in stomata across land plants. As with rhizoids and root hairs (Jones and Dolan, 2012), the conservation of core stomatal development and patterning modules across the land plant phylogeny does not imply the absence of selective pressures during the course of evolution.
The stomatal evolution model of bHLH gene duplication and specialization proposed by MacAlister and Bergmann (2011) and evidenced by Davies and Bergmann (2014), neatly describes the ways a relatively basic form of stomatal development can give rise to the variation and complexity observed in different extant land plant lineages. This simple model, informed by the stomatal development work in P. patens (MacAlister and Bergmann, 2011; Caine et al., 2016; Chater et al., 2016), is invaluable for our interpretation of the divergence of stomatal form and physiology in land plants. Moreover, the confirmation of gene function in P. patens stomatal development gives us confidence in predicting the presence or absence of genes in as-yet-unstudied lineages of plants that have stomata (Caine et al., 2016; Chater et al., 2016). While we now know that P. patens uses orthologous development and patterning genes to set out stomata on its epidermis, the exact mechanisms that enable this to happen remain elusive. For example, we know that PpSMF1 and PpSCRM1 are required for stomatal formation, but how are these genes regulated and at what developmental stage does this occur? Do PpEPF1, PpTMM, and PpERECTAs contribute to bHLH regulation using a MAPK pathway akin to vascular land plant regulation of SPCH, and does this regulation occur on stomatal lineage cells prior to and/or after the formation of GMC cells? Perhaps once these questions are answered, we may truly begin to understand how the described genes enable stomatal development to occur in moss.
DOES STOMATAL PATTERNING ASSIST STOMATAL FUNCTION IN MOSSES?
In Arabidopsis, the control of stomatal patterning has been shown to directly influence plant gas exchange, photosynthetic function, and productivity (Dow and Bergmann, 2014; Dow et al., 2014; Franks and Casson, 2014; Franks et al., 2015; Lehmann and Or, 2015; Papanatsiou et al., 2016). In particular, correct spacing via alterations to stomatal size and density ensures optimal guard cell pore control and faster responses to environmental cues (Dow et al., 2014). In bryophytes, stomatal spacing appears to be controlled by a less refined system involving fewer regulatory checkpoints than in vascular plants, and stomatal clustering is often observed (Paton and Pearce, 1957; Pressel et al., 2014; Merced and Renzaglia, 2016). Nonetheless, the conservation of the one-cell-spacing mechanism and associated EPF signaling system in mosses demonstrates a requirement for stomatal spacing, although the evolutionary drivers for a spacing mechanism are unknown. The position of moss stomata above spongy photosynthetic tissue and active stomatal aperture control suggests that moss stomatal patterning might be governed by the same evolutionary pressures as those in angiosperms, i.e. efficient gas exchange and regulation of water loss (Garner and Paolillo, 1973; Chater et al., 2011; Merced and Renzaglia, 2014). Alternatively (but not exclusively), the correct spacing of stomata around the moss sporophyte base may be important in making sporophyte capsules less vulnerable to invasion by pathogens, or in enabling efficient dehiscence (Paton and Pearce, 1957; Pressel et al., 2014; Caine et al., 2016). The function(s) of moss stomata remain largely untested because of the technical difficulties associated with the small size of the stomatal-bearing spore capsules. However, recently evidence to support a role for stomata in dehiscence has emerged from experiments that produced knockout ppsmf1 or ppscrm1 sporophyte capsules in P. patens (Chater et al., 2016). The resulting spore capsules lacking these key regulatory bHLHs failed to produce stomata and show delayed spore dehiscence.
Arabidopsis adjusts stomatal density in response to subambient or elevated CO2, bymodulation of EPF2 peptide levels (Engineer et al., 2014). Fossilized plant cuticles indicate that early land plants could probably respond to changes in atmospheric CO2 concentration by altering stomatal size and density, suggesting that developmental responses to environmental cues such as CO2 are ancient (McElwain and Chaloner, 1995; Franks and Beerling, 2009). The work by Caine et al. (2016) has highlighted that the orthologous EPF gene: PpEPF1 is present in P. patens but it is unclear as to whether this gene might also enable a CO2 density response like Arabidopsis EPF2. Such PpEPF1 functionality seems unlikely, however, owing to the observations that several moss species do not alter stomatal density (or size) in response to CO2 (Baars and Edwards, 2008; Field et al., 2015). Moreover, PpEPF1 cannot restore stomatal spacing when expressed in Arabidopsis epf2 (Caine et al., 2016), suggesting the two proteins have undergone substantial functional divergence. Based on phylogenetic observations, it seems that the EPF gene family underwent a duplication in vascular land plants, which enabled a more sophisticated and improved regulation of stomatal spacing (Caine et al., 2016), which may also have permitted active genetic control over the altering of stomatal density (and perhaps size) in response to growth at different CO2 concentrations.
ANCIENT STOMATA AND ASSOCIATED PORES
Extant plants provide extensive examples of variation in stomatal form and function, whereas the fossil record is more limited with regard to stomatal evolution. This is especially true of the bryophytes and their stomata, which are absent from the ancient land plant fossil record, although ancient bryophyte-like plants with branching sporophytes and stomata have been recently been identified (Edwards et al., 2014). The oldest fossilized plants discovered with stomata belong to the early vascular plant Cooksonia (Edwards et al., 1992), which diverged sometime after the ancestors of the bryophytes diverged from the common land plant lineage (Fig. 1A). Intriguingly, there is fossil evidence of early land plant gametophyte stomata that may, by the authors’ own interpretation, have predated the emergence of extant bryophyte lineages (Remy et al., 1993). Such findings imply that stomata may have first evolved on the gametophyte and subsequently been co-opted by the sporophyte in a similar manner by which root hairs evolved from rhizoids (Jones and Dolan, 2012). However, the interpretation of Remy et al. (1993) is one of a number proposed and requires the characterization of further fossils to support.
While stomata are absent from extant bryophyte gametophytes, there are similar structures present on the gametophytes of extant hornworts and liverworts. These include mucilage clefts and air pores (Fig. 1B), which have at times been suggested to share homology to stomata (Zeiger et al., 1987; Villarreal A and Renzaglia, 2006; Rudall et al., 2013; Villarreal and Renzaglia, 2015; Shimamura, 2016). While nothing is known about the genes underpinning hornwort mucilage clefts, recent work shows that Marchantia liverwort pore development is controlled by genes not previously linked with stomatal differentiation (Ishizaki et al., 2013; Jones and Dolan, 2017). These include NOPPERABO1, encoding a Plant U-box E3 ubiquitin ligase, which is required for pore formation, and MpWIP, which encodes a zinc finger protein that regulates nascent pore morphogenesis. Neither of these genes appears orthologous to those involved in stomatal development, which further supports the view that air pores and stomata are not homologous structures (Rudall et al., 2013). To date, it is unclear whether the canonical genes associated with stomatal development are present in liverworts and hornworts. Clearly, before a definitive theory can be proposed relating to the origins of stomata in land plants, improved molecular data for basal plant taxa as well as further fossil evidence are required.
NEW PHYLOGENIES RELATING TO STOMATAL DEVELOPMENT GENES SUPPORT A CONSERVATION OF A CORE GENETIC MODULE IN STOMATOUS LAND PLANTS
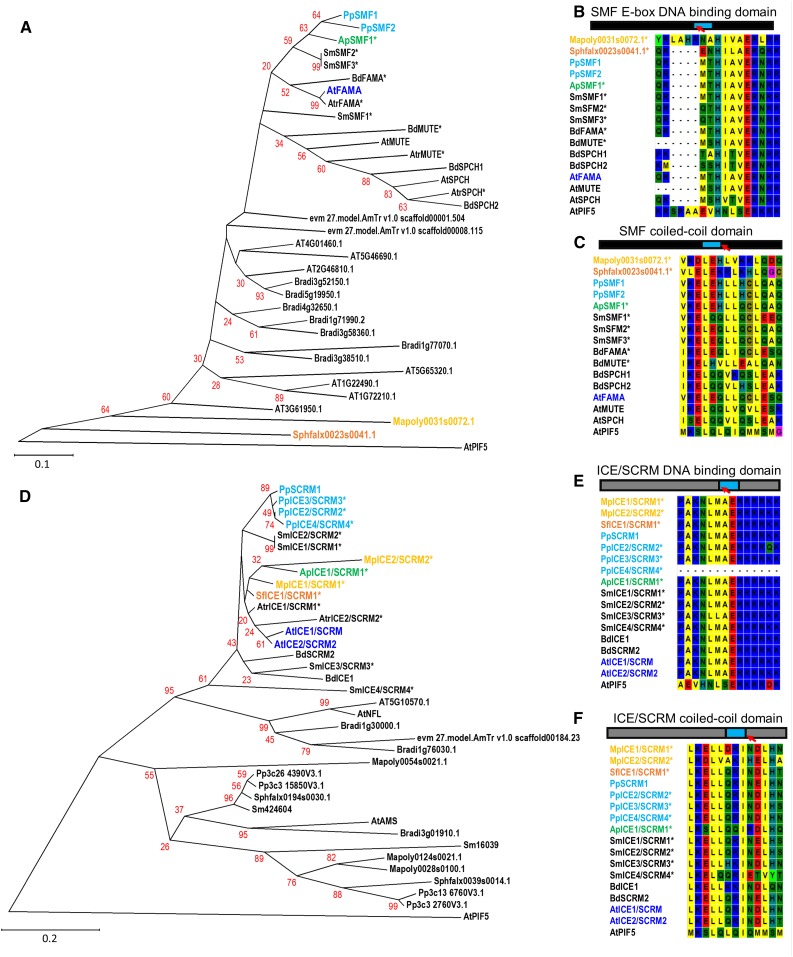
In light of the recent findings in Physcomitrella (Caine et al., 2016; Chater et al., 2016) and following on from MacAlister and Bergmann (2011) and Ran et al. (2013), we can now trace the ancestry of genes involved in the core stomatal developmental bHLH module across the plant kingdom (Fig. 2).
Figure 2.
Phylogenies and domain sequence alignments of the bHLH transcription factors implicated in stomatal development for both stomata- and non-stomata-bearing land plants. A, Phylogeny of SPCH, MUTE, and FAMA orthologs and closest related SMF peptide sequences encoded by genes from the liverwort M. polymorpha (yellow), the mosses S. fallax (orange) and P. patens (light blue), the hornwort A. punctatus (green), and the angiosperm Arabidopsis (dark blue). Color-marked identifiers illustrate in which early taxa bHLHs associated stomatal development may be present. The developmental role of starred peptides and their associated genes (*) has not yet been experimentally confirmed. B, Conservation of SMF E-box DNA-binding domain, and C, coiled-coil domains in SPCH, MUTE, and FAMA sequences of stomata-bearing land plants based on alignments performed for A. D, Phylogeny representing ICE/SCRM and ICE2/SCRM2 orthologs and related genes utilizing peptide sequences from equivalent species to A. Color coding and nomenclature follows A. E, Conservation of ICE/SCRM DNA binding domain, and F, coiled-coil domains based on alignments performed for D. Sequences used to construct phylogenies in A and D were obtained by BLAST comparisons of the sequences of PpSMF1 and PpSCRM1 against the genomes of Mapoly, M. polymorpha; Sphfalx, S. fallax, Pp, P. patens; Sm: Selaginella moellendorffii; Atr, Amborella trichopoda (accession identifier evm); Bradi, Brachypodium distachyon; and AT: Arabidopsis using Phytozome V11 (Goodstein et al., 2012). Retrieved blast sequences of equal to or higher than 80 (PpSMF1 analysis) or 95 (PpSCRM1) were used for sequence alignments. The A. punctatus sequences (ApSMF1 and ApSCRM1) are partial sequences based on the recently publication of this species genome (Szövényi et al., 2015). The SmSMF3 peptide sequence was obtained based on previous analysis by MacAlister and Bergmann (2011). For SmSMF1, SmSMF3, and SmSCRM1-4 gene models were predicted using http://www.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind (Solovyev et al., 2006). For both analyses, identified peptide sequences were aligned using the MUSCLE algorithm (Edgar, 2004) and evolutionary history inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. Positions of gaps and missing data were removed. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). See Supplemental Table S1 for protein accession IDs and the sequences used for A. punctatos and S. moellendorffii. The Arabidopsis bHLH PIF5 was included as an outgroup for rooting the trees.
Using the hornwort Anthoceros punctatus and pseudostomate Sphagnum fallax genomes (Szövényi et al., 2015; Shaw et al., 2016) and the prerelease of the liverwort Marchantia polymorpha genome on Phytozome V11 (Goodstein et al., 2012), we can begin to identify whether genes required for stomatal development are present in unexplored taxa and plant groups that lack stomata. Strikingly, our analyses indicate that the stomatous hornwort A. punctatus possesses genes closely related to both PpSMF1 and PpSCRM1 (Fig. 2, A and D; PpSMF2 is a P. patens in-paralog and has no discernible function during stomatal development; Chater et al., 2016). Observations of key amino acid residues in the bHLH binding domains and coiled-coil domains of the putative A. punctatus SMF1 and SCRM1 reaffirms that the sequences of these peptides share a very high degree of homology with both moss and other land plant orthologs (Fig. 2, B, C, E, and F). This is particularly evident in the DNA binding domains, with ApSMF1 and ApSCRM1 sharing identical residues to almost all FAMA and SCRM/2 sequences identified in the other species analyzed (Fig. 2, B and E).
Assessment of putative stomatal associated bHLH orthologs in M. polymorpha and S. fallax revealed only genes sister to SMF, although orthologs of SCRM genes may be present. These sister SMF genes show clear divergence in their bHLH regions, strongly suggesting that they do not play a role in stomatal development in these species (Fig. 2, B and C). The presence of air pores in M. polymorpha and pseudostomata in S. fallax invites us to speculate that these sister bHLHs may have evolved from genes that once initiated stomata in the ancestors of liverworts and sphagnum, respectively. Sequencing of more liverwort and basal moss taxa, combined with gene-function studies, could shed further light on the molecular evolution of these stoma-like structures, as currently only a limited amount is known relating to the genetics underpinning air pores (Ishizaki et al., 2013; Jones and Dolan, 2017) and nothing is known about the genes underpinning pseudostomata development. Furthermore, phylogenetic studies of genes involved in guard cell function might provide further clues as to the level of homology between gametophyte pores, pseudostomata, and stomata themselves.
ASSESSING SMF GENE FAMILY FUNCTION IN NONVASCULAR AND VASCULAR LAND PLANT REPRESENTATIVES
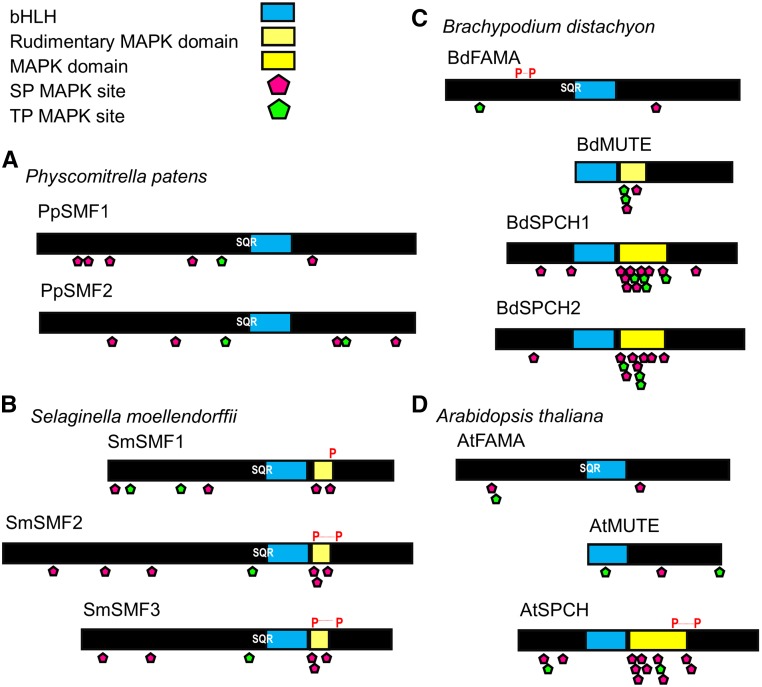
MacAlister and Bergmann (2011) and Davies and Bergmann (2014) have neatly set out a framework by which vascular land plants might have increased the complexity of their stomatal developmental modules over evolutionary time. It is hypothesized that an ancestral FAMA-like bHLH-governed GMC formation (with a role akin to that of MUTE in Arabidopsis) as well as the subsequent production of guard cells (akin to FAMA) in early land plants. Subsequently, this multifunctional bHLH underwent a gene duplication resulting in a MUTE-like gene product and specialization of the two distinct functions. A subsequent duplication event occurred in the ancestral angiosperms, which led to a third SMF gene, SPCH, and further specialization (Fig. 2A; MacAlister and Bergmann, 2011; Ran et al., 2013). In grasses, an additional duplication resulted in two SPCHs, further partitioning the stomatal developmental program (Fig. 2A; Liu et al., 2009; Ran et al., 2013; Raissig et al., 2016). This neofunctionalization of the SMFs and the subsequent divergence of stomatal ontogenetic control can be seen in the comparison of moss, lycophyte, grass, and dicot SMF protein domain structures (Fig. 3; MacAlister and Bergmann, 2011; Davies and Bergmann, 2014; Raissig et al., 2016).
Figure 3.
Comparison of SMF bHLH protein domains and motifs from four representative land plant species. A, Schematic of moss P. patens PpSMF1 and PpSMF2 proteins which contain bHLH domains (light blue) with limited evidence for a downstream MAPK phosphorylation domain. Putative SP (pink diamond) and TP (green diamond) MAPK phosphorylation sites are present, mostly to the N-terminal side of the bHLH region, which could serve as points of regulation. Both PpSMF1 and 2 contain an SQR motif, a potential protein kinase C phosphorylation site. B, Lycophyte S. moellendorffii SmSMF1, SmSMF2, and SmSMF3 sequences contain bHLH and potential MAPK target domains (light yellow) and several SP motifs. Similarly to PpSMF1/2 also contain proximal SP/TP MAPK target sites. An SQR motif is conserved in all three SmSMF sequences immediately upstream of the bHLH domain. Potential PEST domains are present in these sequences (marked with P). C, Grass B. distachyon BdFAMA, BdMUTE, and BdSPCH1, and BdSPCH2 proteins are shorter relative to P. patens and S. moellendorffii proteins, due to a reduced N-terminal region. The MAPK target domain in both BdSPCH1 and BdSPCH2 contains several SP and TP motifs. BdMUTE contains SP and TP motifs within a putative MAPK domain. The SQR motif is present only in BdFAMA. PEST domain identity is weak and therefore excluded omitted from the diagram. D, Dicot Arabidopsis AtFAMA, AtMUTE, and AtSPCH proteins. As with BdSPCH1 and BdSPCH2, AtSPCH has a well-conserved MAPK target domain and additionally contains a potential PEST domain. Protein kinase C sites and PEST domains were predicted using the tools available at http://myhits.isb-sib.ch/cgi-bin/motif_scan with http://emboss.bioinformatics.nl/cgibin/emboss/epestfind.
Arabidopsis SMF bHLHs are becoming well characterized, with key domains and motifs linked directly to protein function (Lampard et al., 2009; Davies and Bergmann, 2014; Yang et al., 2015). As expected for a transcription factor, DNA binding is critical to FAMA’s role in guard cell formation. A bHLH DNA binding domain can be observed across moss, lycophyte, grass, and dicot FAMA variants (Fig. 3, A–D). An adjacent SQR motif may function as a phosphorylation site for a protein kinase C and could represent regulatory point shared across all FAMA orthologs. The analysis of the domain structure of these bHLHs provides some evidence for an ancestral multifunctional bHLH (Fig. 3). New gene models suggest that P. patens and S. moellendorffii possess FAMA-like orthologs and reveal the presence of extensive N-terminal regions that are absent from vascular land plant FAMAs (compare Fig. 3, A and B, with Fig. 3, C and D).
The Arabidopsis SPCH MAPK target domain is C-terminal to the bHLH region. Mutations of residues within this domain lead to incorrect regulation of stomatal entry divisions (Lampard et al., 2009; Yang et al., 2015). In P. patens, there is sparse evidence for this MAPK domain, although one SP motif is present (Fig. 3A). S. moellendorffii contains Ser-Pro/Thr-Pro (SP/TP) motifs in all three SmSMFs, although their lower number compared to angiosperms suggests a more restricted domain with perhaps less regulatory control (Fig. 3, B–D). Interestingly, the presence of SP/TP motifs in BdMUTE may underlie the novel functionality in the grass MUTEs compared to the dicot Arabidopsis (Fig. 3, C and D; Raissig et al., 2017) and may offer insights into potential SPCH-like capabilities that have been proposed for rice (Oryza sativa) OsMUTE (Liu et al., 2009).
In addition to MAPK regulation, PEST domains involved in protein degradation are important for SPCH (and possibly SCRM) regulation in Arabidopsis (Fig. 3D; Raissig et al., 2016). Although Brachypodium SPCH proteins possess only weak conservation of PEST target sites, their presence in earlier diverging homologs suggests a regulatory mechanism that had evolved prior to the lycophytes splitting from the ancestral lineage (Fig. 3B). The S. moellendorffii SmSMFs could be seen as evolutionary intermediates, with putative PEST domains and MAPK target sites suggesting SPCH-like functionality, in combination with bHLH and DNA binding domains reminiscent of FAMA (Fig. 2A). In the moss PpSMF1, SPCH-like signature S/T-P motifs are very limited, and no clear PEST domains are clearly apparent, yet there is clear conservation of the SQR motif and E-box DNA binding domains, suggesting that this protein is more like FAMA than SPCH. Whether PpSMF1 has MUTE-like function based on amino acid sequence comparisons is difficult to discern based on the present evidence but cannot be discounted. Clearly, functional analyses of additional nonvascular and vascular plant bHLHs are required to further understand the evolution of the SMFs and stomatal developmental ontogeny during land plant evolution.
FURTHER EVIDENCE FOR THE CONSERVATION OF STOMATA VIA ANALYSIS OF STOMATAL PATTERNING GENES
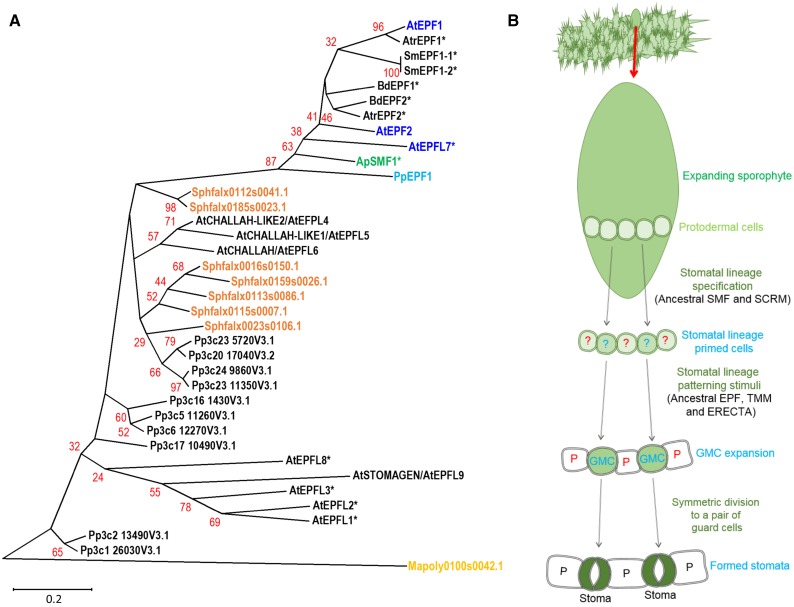
Intercellular signaling components that regulate the SMF/SCRM transcriptional control module, namely EPF, TMM, and ERECTA, are also deeply conserved and, in the case of the EPF/Ls, have undergone considerable expansion across land plant evolution (Takata et al., 2013; Caine et al., 2016). Analysis of stomatal patterning-associated EPF peptide sequences can further inform our understanding of the origins of stomata (Fig. 4A). For example, the hornwort A. punctatus ApEPF1 is closely related to PpEPF1 and other stomatal acting EPFs from later diverging lineages. In contrast, the astomatous M. polymorpha appears to possess only a single more distantly related gene, and the pseudostomatous S. fallax only the EPFL4/5/6-like subgroup of the EPF peptide family. A likely interpretation of these results is that stoma-associated EPFs have been lost in the liverwort and pseudostomatous moss lineages, but conserved in hornworts, mosses, and vascular plants. Taken together with the SMF/SCRM analysis set out in Figure 2, these observations suggest that while the complexity of stomatal development mechanisms has exploded in vascular plants, a more limited basic module has been retained by stomatous nonvascular land plants (Caine et al., 2016; Chater et al., 2016).
Figure 4.
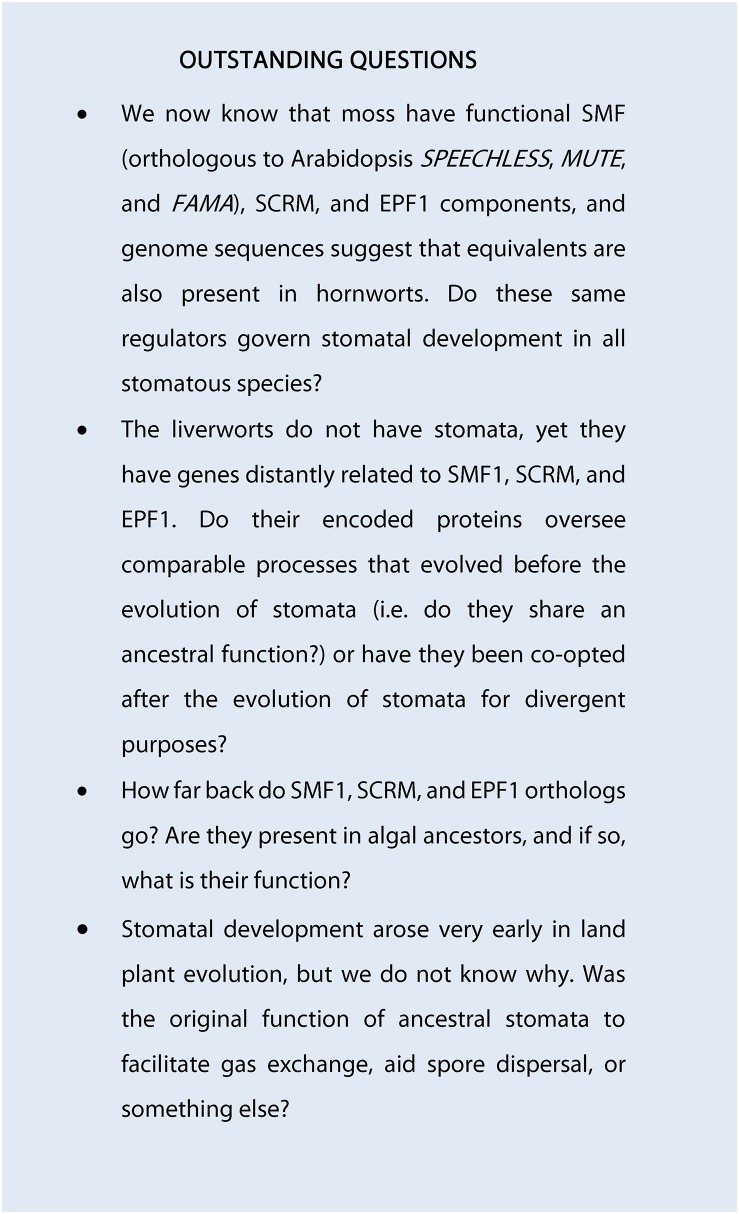
EPF phylogeny of stomata- and non-stomata-bearing land plants and a model for stomatal development in the ancestor of stomata-bearing land plants. A, Phylogeny of EPF peptide sequences across the land plant kingdom. Stomatal and putative stomatal genes or closest equivalents in the liverwort M. polymorpha (yellow), the mosses S. fallax (orange) and P. patens (light blue), the hornwort A. punctatus (green), and the angiosperm Arabidopsis (dark blue) are marked to illustrate in which of these taxa the EPF stomatal patterning gene may be present. Starred genes (*) have not had their functions experimentally determined. The majority of sequences used to construct the phylogeny were obtained by BLAST comparison of the PpEPF1 amino acid sequence against the genomes of Mapoly, M. polymorpha; Sphfalx, S. fallax; Pp, P. patens; Sm, S. moellendorffii; Atr, Amborella trichopoda; Bradi, Brachypodium distachyon; and AT, Arabidopsis. Additional P. patens genes with more limited homology were added based on Caine et al. (2016). For simplicity, non-stomata-associated EPFs from vascular land plants have been omitted from the tree. The A. punctatus ApEPF1 partial sequence is based on Szövényi et al. (2015). Alignment and phylogenetic trees were prepared as described for Figure 2. Arabidopsis EPFL1-6 and 8-9 are included to highlight sequence relationships identified in (Caine et al., 2016). B, A model for stomatal development on the sporophyte of early evolving plants. Stomata precursors may have been specified via activity of an ancestral SMF-SCRM heterodimer. To ensure appropriate spacing of GMCs, cell signaling occurred via an ancestral patterning module (EPF, TMM, and ERECTA), followed by a final symmetric division leading to the formation of mature stomata.
INTEGRATING EMPIRICAL AND PHYLOGENETIC DATA TO PREDICT A MODEL FOR STOMATAL DEVELOPMENT IN THE EARLIEST LAND PLANTS
The recent studies of stomatal development in P. patens (Caine et al., 2016; Chater et al., 2016) combined with newly available genomic data in other early diverging lineages (Figs. 2 and 4) provide a window into the very earliest mechanisms that may have been used by the extinct common ancestor of modern plants to build stomata (Fig. 4B). The production of stomata on the sporophytes of mosses and hornworts appears to require much simpler cellular processes than that of dicots (Pressel et al., 2014; Merced and Renzaglia, 2016). For example, there is no evidence for asymmetric cell divisions in either stomatal lineage. It is probable that the earliest evolving stomatal development mechanisms were also relatively uncomplicated and did not require the production of a meristemoid through an asymmetric division. These early mechanisms may have been initiated in the expanding sporophyte via the actions of an ancestral heterodimeric bHLH complex consisting of SMF and SCRM orthologs, regulating transcriptional activity in specific protodermal cells and promoting GMC and stomatal fate. To enforce stomatal patterning by cell-cell signaling prior to (and perhaps during) GMC formation, an ancestral EPF, TMM, and ERECTA module arose or was co-opted. Once formed, GMCs could then undergo differentiation and finally a symmetric division to form a pair of guard cells. The same ancestral SMF/SCRM bHLH heterodimers responsible for lineage initiation may have also orchestrated the lineage conclusion. We propose that the richness and complexity that now governs plant epidermal development arose from this relatively simple program.
CONCLUSION
Occam’s razor is a powerful tool to guide research into the origins of stomatal form and function. A single origin of a core genetic module for stomatal development in the common ancestor to hornworts, mosses, and vascular plants is arguably the most parsimonious explanation for the wealth of evidence from the fossil record and from the taxonomic, genomic, transcriptomic, and morphological data amassing from across extant land plants.
The Arabidopsis model has provided insight into dicot stomatal development and patterning. By applying this knowledge to outstanding evolutionary questions, we are reaping the rewards of decades of molecular and genetic Arabidopsis research. These insights, from the base of the land plant tree to the most recently divergent taxa, are testament to the power of this approach. We will improve our understanding of the origins and evolutionary development of stomata as we obtain better resolution of the early land plant phylogeny and expand the range of genetic models available (see Outstanding Questions). The development of molecular genetic techniques for the liverwort Marchantia (Ishizaki et al., 2008) and the hornwort Anthoceros (Szövényi et al., 2015) will permit a greater understanding of the relationships between ancestral clades and the acquisition of those traits that permitted the colonization of the land. With the identification of new genes that potentially act on stomatal development, we now have an updated roadmap with which to interrogate some of the unanswered questions relating to the evolution of stomata.
Based on the current land plant phylogeny, developmental studies and phylogenies of the key genes involved in stomatal development and patterning, it would seem that the core regulatory network overseeing these processes first evolved prior to the divergence of the hornworts from the ancestral lineage. This appraisal, based on the current phylogeny, points to a single origin of stomata in land plants with subsequent losses in the liverworts and early diverging mosses. Exciting times lie ahead in truly understanding from where stomata arose nearly half a million years ago.
Accession Numbers
These data are under accessions number SRX538621 and are accessible at: https://www.ncbi.nlm.nih.gov/sra/SRX538621.
Supplemental Data
Supplemental Table S1. Protein accession IDs and sequences used for A. punctatos and S. moellendorffii.
Supplementary Material
Acknowledgments
We thank Professor J. Langdale, Dr. Eftychios Frangedakis, and Dr. Steve Kelly (University of Oxford) for kindly providing access to A. punctatus sequence data prior to publication (these data are now available at https://www.ncbi.nlm.nih.gov/sra/SRX538621. We also thank Joanna Landymore and Jessica Dunn (University of Sheffield) for providing some of the stomatal images used in Figure 1.
Glossary
- MMC
meristemoid mother cell
- SLGC
stomatal lineage ground cell
- GMC
guard mother cell
Footnotes
This work was funded by the Biotechnology and Biological Sciences Research Council and the Newton Fund-Mexican Academy of Sciences-Consejo Nacional de Ciencia y Tecnología.
These authors contributed equally to the article.
Articles can be viewed without a subscription.
References
- Assmann SM, Jegla T (2016) Guard cell sensory systems: Recent insights on stomatal responses to light, abscisic acid, and CO2. Curr Opin Plant Biol 33: 157–167 [DOI] [PubMed] [Google Scholar]
- Baars C, Edwards D (2008) Effects of elevated atmospheric CO2 on spore capsules of the moss Leptobryum pyriforme. J Bryol 30: 36–40 [Google Scholar]
- Berry JA, Beerling DJ, Franks PJ (2010) Stomata: Key players in the earth system, past and present. Curr Opin Plant Biol 13: 233–240 [DOI] [PubMed] [Google Scholar]
- Bowman JL. (2011) Stomata: Active portals for flourishing on land. Curr Biol 21: R540–R541 [DOI] [PubMed] [Google Scholar]
- Caine RS, Chater CC, Kamisugi Y, Cuming AC, Beerling DJ, Gray JE, Fleming AJ (2016) An ancestral stomatal patterning module revealed in the non-vascular land plant Physcomitrella patens. Development 143: 3306–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson SA, Franklin KA, Gray JE, Grierson CS, Whitelam GC, Hetherington AM (2009) phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr Biol 19: 229–234 [DOI] [PubMed] [Google Scholar]
- Chater C.C., Caine R.S., Tomek M., Wallace S., Kamisugi Y., Cuming A.C., Lang D., MacAlister C.A., Casson S., Bergmann D.C., et al. (2016) Origin and function of stomata in the moss Physcomitrella patens. Nat Plants 2: 16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater C, Gray JE, Beerling DJ (2013) Early evolutionary acquisition of stomatal control and development gene signalling networks. Curr Opin Plant Biol 16: 638–646 [DOI] [PubMed] [Google Scholar]
- Chater C, Kamisugi Y, Movahedi M, Fleming A, Cuming AC, Gray JE, Beerling DJ (2011) Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr Biol 21: 1025–1029 [DOI] [PubMed] [Google Scholar]
- Chater CCC, Oliver J, Casson S, Gray JE (2014) Putting the brakes on: Abscisic acid as a central environmental regulator of stomatal development. New Phytol 202: 376–391 [DOI] [PubMed] [Google Scholar]
- Chater C, Peng K, Movahedi M, Dunn JA, Walker HJ, Liang YK, McLachlan DH, Casson S, Isner JC, Wilson I, et al. (2015) Elevated CO2-induced responses in stomata require ABA and ABA signaling. Curr Biol 25: 2709–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z-H, Chen G, Dai F, Wang Y, Hills A, Ruan Y-L, Zhang G, Franks PJ, Nevo E, Blatt MR (2017) Molecular evolution of grass stomata. Trends Plant Sci 22: 124–139 [DOI] [PubMed] [Google Scholar]
- Cullen E, Rudall PJ (2016) The remarkable stomata of horsetails (Equisetum): Patterning, ultrastructure and development. Ann Bot (Lond) 118: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KA, Bergmann DC (2014) Functional specialization of stomatal bHLHs through modification of DNA-binding and phosphoregulation potential. Proc Natl Acad Sci USA 111: 15585–15590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow GJ, Bergmann DC (2014) Patterning and processes: How stomatal development defines physiological potential. Curr Opin Plant Biol 21: 67–74 [DOI] [PubMed] [Google Scholar]
- Dow GJ, Bergmann DC, Berry JA (2014) An integrated model of stomatal development and leaf physiology. New Phytol 201: 1218–1226 [DOI] [PubMed] [Google Scholar]
- Duckett JG, Pressel S, P’ng KMY, Renzaglia KS (2009) Exploding a myth: the capsule dehiscence mechanism and the function of pseudostomata in Sphagnum. New Phytol 183: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D, Davies KL, Axe L (1992) A vascular conducting strand in the early land plant Cooksonia. Nature 357: 683–685 [Google Scholar]
- Edwards D, Morris JL, Richardson JB, Kenrick P (2014) Cryptospores and cryptophytes reveal hidden diversity in early land floras. New Phytol 202: 50–78 [DOI] [PubMed] [Google Scholar]
- Egunyomi A. (1982) On the stomata of some tropical african mosses. Lindbergia 8: 121–124 [Google Scholar]
- Engineer CB, Ghassemian M, Anderson JC, Peck SC, Hu H, Schroeder JI (2014) Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513: 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field KJ, Duckett JG, Cameron DD, Pressel S (2015) Stomatal density and aperture in non-vascular land plants are non-responsive to above-ambient atmospheric CO2 concentrations. Ann Bot (Lond) 115: 915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks PJ, Beerling DJ (2009) Maximum leaf conductance driven by CO2 effects on stomatal size and density over geologic time. Proc Natl Acad Sci USA 106: 10343–10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks PJ, Britton-Harper ZJ (2016) No evidence of general CO2 insensitivity in ferns: one stomatal control mechanism for all land plants? New Phytol 211: 819–827 [DOI] [PubMed] [Google Scholar]
- Franks PJ, Casson S (2014) Connecting stomatal development and physiology. New Phytol 201: 1079–1082 [DOI] [PubMed] [Google Scholar]
- Franks PJ, W Doheny-Adams T, Britton-Harper ZJ, Gray JE (2015) Increasing water-use efficiency directly through genetic manipulation of stomatal density. New Phytol 207: 188–195 [DOI] [PubMed] [Google Scholar]
- Garner DLB, Paolillo DJ (1973) On the functioning of stomates in Funaria. Bryologist 76: 423–427 [Google Scholar]
- Geisler M, Nadeau J, Sack FD (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat GE, Torres PS, Vojnov AA (2009) Stomata and pathogens: Warfare at the gates. Plant Signal Behav 4: 1114–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. (2013) Filial mistletoes: The functional morphology of moss sporophytes. Ann Bot (Lond) 111: 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S-K, Torii KU (2016) Lineage-specific stem cells, signals and asymmetries during stomatal development. Development 143: 1259–1270 [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T (2007) The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T (2009) Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol 50: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Hunt L, Bailey KJ, Gray JE (2010) The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol 186: 609–614 [DOI] [PubMed] [Google Scholar]
- Hunt L, Gray JE (2009) The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Chiyoda S, Yamato KT, Kohchi T (2008) Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol 49: 1084–1091 [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Mizutani M, Shimamura M, Masuda A, Nishihama R, Kohchi T (2013) Essential role of the E3 ubiquitin ligase nopperabo1 in schizogenous intercellular space formation in the liverwort Marchantia polymorpha. Plant Cell 25: 4075–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewaria PK, Hara T, Tanaka H, Kondo T, Betsuyaku S, Sawa S, Sakagami Y, Aimoto S, Kakimoto T (2013) Differential effects of the peptides Stomagen, EPF1 and EPF2 on activation of MAP kinase MPK6 and the SPCH protein level. Plant Cell Physiol 54: 1253–1262 [DOI] [PubMed] [Google Scholar]
- Jones VAS, Dolan L (2012) The evolution of root hairs and rhizoids. Ann Bot (Lond) 110: 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones VA, Dolan L (February 7, 2017) MpWIP regulates air pore complex development in the liverwort Marchantia polymorpha. Development 10.1242/dev.144287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Kajita R, Miyazaki A, Hokoyama M, Nakamura-Miura T, Mizuno S, Masuda Y, Irie K, Tanaka Y, Takada S, et al. (2010) Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol 51: 1–8 [DOI] [PubMed] [Google Scholar]
- Lampard GR, Lukowitz W, Ellis BE, Bergmann DC (2009) Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21: 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, Macalister CA, Bergmann DC (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Lau OS, Davies KA, Chang J, Adrian J, Rowe MH, Ballenger CE, Bergmann DC (2014) Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345: 1605–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, Lin YCL, Putarjunan A, Han SK, Avila J, Torii KU (2015) Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU (2012) Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev 26: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann P, Or D (2015) Effects of stomata clustering on leaf gas exchange. New Phytol 207: 1015–1025 [DOI] [PubMed] [Google Scholar]
- Liu T, Ohashi-Ito K, Bergmann DC (2009) Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136: 2265–2276 [DOI] [PubMed] [Google Scholar]
- Luo L, Zhou WQ, Liu P, Li CX, Hou SW (2012) The development of stomata and other epidermal cells on the rice leaves. Biol Plant 56: 521–527 [Google Scholar]
- MacAlister CA, Bergmann DC (2011) Sequence and function of basic helix-loop-helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol Dev 13: 182–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister CA, Ohashi-Ito K, Bergmann DC (2007) Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540 [DOI] [PubMed] [Google Scholar]
- McAdam SAM, Brodribb TJ (2016) Linking turgor with ABA biosynthesis: Implications for stomatal responses to vapor pressure deficit across land plants. Plant Physiol 171: 2008–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwain JC, Chaloner WG (1995) Stomatal density and index of fossil plants track atmospheric carbon dioxide in the Palaeozoic. Ann Bot 76: 389–395 [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L (2007) An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480 [DOI] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii KU, He P, Shan L (2015) Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr Biol 25: 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merced A, Renzaglia KS (2013) Moss stomata in highly elaborated Oedipodium (Oedipodiaceae) and highly reduced Ephemerum (Pottiaceae) sporophytes are remarkably similar. Am J Bot 100: 2318–2327 [DOI] [PubMed] [Google Scholar]
- Merced A, Renzaglia K (2014) Developmental changes in guard cell wall structure and pectin composition in the moss Funaria: Implications for function and evolution of stomata. Ann Bot (Lond) 114: 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merced A, Renzaglia KS (2016) Patterning of stomata in the moss Funaria: A simple way to space guard cells. Ann Bot (Lond) 117: 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JL, Rouzé P, Verhelst B, Lin YC, Bayer T, Collen J, Dattolo E, De Paoli E, Dittami S, Maumus F, et al. (2016) The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530: 331–335 [DOI] [PubMed] [Google Scholar]
- Papanatsiou M, Amtmann A, Blatt MR (2016) Stomatal spacing safeguards stomatal dynamics by facilitating guard cell ion transport independent of the epidermal solute reservoir. Plant Physiol 172: 254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton JA, Pearce JV (1957) The occurrence, structure and functions of the stomata in British bryophytes. Trans Brit Bryol Soc 3: 228–259 [Google Scholar]
- Payne WW. (1979) Stomatal patterns in embyrophytes—Their evolution, ontogeny and interpretation. Taxon 28: 117–132 [Google Scholar]
- Peterson KM, Rychel AL, Torii KU (2010) Out of the mouths of plants: The molecular basis of the evolution and diversity of stomatal development. Plant Cell 22: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Bogenschutz NL, Torii KU (2008) The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant Cell Physiol 49: 934–943 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU (2007) Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505 [DOI] [PubMed] [Google Scholar]
- Pressel S, Goral T, Duckett JG (2014) Stomatal differentiation and abnormal stomata in hornworts. J Bryol 36: 87–103 [Google Scholar]
- Qi X, Han S-KI, Dang JH, Garrick JM, Ito M, Hofstetter AK, Torii KU (2017) Autocrine regulation of stomatal differentiation potential by EPF1 and ERECTA-LIKE1 ligand-receptor signaling. eLife 6: e24102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu YL, Li L, Wang B, Chen Z, Knoop V, Groth-Malonek M, Dombrovska O, Lee J, Kent L, Rest J, et al. (2006) The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA 103: 15511–15516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Abrash E, Bettadapur A, Vogel JP, Bergmann DC (2016) Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc Natl Acad Sci USA 113: 8326–8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Matos JL, Gil MXA, Kornfeld A, Bettadapur A, Abrash E, Allison HR, Badgley G, Vogel JP, Berry JA, et al. (2017) Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355: 1215–1218 [DOI] [PubMed] [Google Scholar]
- Ran JH, Shen TT, Liu WJ, Wang XQ (2013) Evolution of the bHLH genes involved in stomatal development: implications for the expansion of developmental complexity of stomata in land plants. PLoS One 8: e78997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA. (2002) Selection pressures on stomatal evolution. New Phytol 153: 371–386 [DOI] [PubMed] [Google Scholar]
- Remy W, Gensel PG, Hass H (1993) The gametophyte generation of some early Devonian land plants. Int J Plant Sci 154: 35–58 [Google Scholar]
- Rudall PJ, Hilton J, Bateman RM (2013) Several developmental and morphogenetic factors govern the evolution of stomatal patterning in land plants. New Phytol 200: 598–614 [DOI] [PubMed] [Google Scholar]
- Rudall PJ, Knowles EVW (2013) Ultrastructure of stomatal development in early-divergent angiosperms reveals contrasting patterning and pre-patterning. Ann Bot (Lond) 112: 1031–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG (2014) From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol 14: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszala EM, Beerling DJ, Franks PJ, Chater C, Casson SA, Gray JE, Hetherington AM (2011) Land plants acquired active stomatal control early in their evolutionary history. Curr Biol 21: 1030–1035 [DOI] [PubMed] [Google Scholar]
- Rychel AL, Peterson KM, Torii KU (2010) Plant twitter: Ligands under 140 amino acids enforcing stomatal patterning. J Plant Res 123: 275–280 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Shaw AJ, Schmutz J, Devos N, Shu S, Carrell AA, Weston DJ. 2016. Chapter five: The Sphagnum Genome Project: A new model for ecological and evolutionary genomics. In Stefan A.R., ed, Advances in Botanical Research. Academic Press, Cambridge, MA, pp 167–187 [Google Scholar]
- Shimamura M. (2016) Marchantia polymorpha: Taxonomy, phylogeny and morphology of a model system. Plant Cell Physiol 57: 230–256 [DOI] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Simmons AR, Bergmann DC (2016) Transcriptional control of cell fate in the stomatal lineage. Curr Opin Plant Biol 29: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V, Kosarev P, Seledsov I, Vorobyev D (2006) Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol 7 Suppl 1: S10.11–S10.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I (2010) Stomagen positively regulates stomatal density in Arabidopsis. Nature 463: 241–244 [DOI] [PubMed] [Google Scholar]
- Szövényi P, Frangedakis E, Ricca M, Quandt D, Wicke S, Langdale JA (2015) Establishment of Anthoceros agrestis as a model species for studying the biology of hornworts. BMC Plant Biol 15: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata N, Yokota K, Ohki S, Mori M, Taniguchi T, Kurita M (2013) Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS One 8: e65183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam THY, Catarino B, Dolan L (2015) Conserved regulatory mechanism controls the development of cells with rooting functions in land plants. Proc Natl Acad Sci USA 112: E3959–E3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU. (2015) Stomatal differentiation: The beginning and the end. Curr Opin Plant Biol 28: 16–22 [DOI] [PubMed] [Google Scholar]
- Trembath-Reichert E, Wilson JP, McGlynn SE, Fischer WW (2015) Four hundred million years of silica biomineralization in land plants. Proc Natl Acad Sci USA 112: 5449–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villagarcia H, Morin A-C, Shpak ED, Khodakovskaya MV (2012) Modification of tomato growth by expression of truncated ERECTA protein from Arabidopsis thaliana. J Exp Bot 63: 6493–6504 [DOI] [PubMed] [Google Scholar]
- Villarreal A JC, Renzaglia KS (2006) Structure and development of Nostoc strands in Leiosporoceros dussii (Anthocerotophyta): A novel symbiosis in land plants. Am J Bot 93: 693–705 [DOI] [PubMed] [Google Scholar]
- Villarreal JC, Renzaglia KS (2015) The hornworts: Important advancements in early land plant evolution. J Bryol 37: 157–170 [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. (2014) Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA 111: E4859–E4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Jiang Y, Jia B, Zhou G (2016) Elevated-CO2 response of stomata and its dependence on environmental factors. Front Plant Sci 7: 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K-Z, Jiang M, Wang M, Xue S, Zhu L-L, Wang H-Z, Zou J-J, Lee E-K, Sack F, Le J (2015) Phosphorylation of serine 186 of bHLH transcription factor SPEECHLESS promotes stomatal development in Arabidopsis. Mol Plant 8: 783–795 [DOI] [PubMed] [Google Scholar]
- Yang T, Liu X (2015) Comparing photosynthetic characteristics of Isoetes sinensis Palmer under submerged and terrestrial conditions. Sci Rep 5: 17783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Sack FD (1995) The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7: 2227–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger E, Farquhar GD, Cowan IR. 1987. Stomatal Function, Stanford University Press, Stanford, CA [Google Scholar]
- Zhao L, Sack FD (1999) Ultrastructure of stomatal development in Arabidopsis (Brassicaceae) leaves. Am J Bot 86: 929–939 [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L (1965) Molecules as documents of evolutionary history. J Theor Biol 8: 357–366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.