Abstract
Introduction
Pathological examination of histopathological slides is a routine clinical procedure for lung cancer diagnosis and prognosis. Although the classification of lung cancer has been updated to become more specific, only a small subset of the total morphological features are taken into consideration. The vast majority of the detailed morphological features of tumor tissues, particularly tumor cells’ surrounding microenvironment, are not fully analyzed. The heterogeneity of tumor cells and close interactions between tumor cells and their microenvironments are closely related to tumor development and progression. The goal of this study is to develop morphological feature–based prediction models for the prognosis of patients with lung cancer.
Method
We developed objective and quantitative computational approaches to analyze the morphological features of pathological images for patients with NSCLC. Tissue pathological images were analyzed for 523 patients with adenocarcinoma (ADC) and 511 patients with squamous cell carcinoma (SCC) from The Cancer Genome Atlas lung cancer cohorts. The features extracted from the pathological images were used to develop statistical models that predict patients’ survival outcomes in ADC and SCC, respectively.
Results
We extracted 943 morphological features from pathological images of hematoxylin and eosin–stained tissue and identified morphological features that are significantly associated with prognosis in ADC and SCC, respectively. Statistical models based on these extracted features stratified NSCLC patients into high-risk and low-risk groups. The models were developed from training sets and validated in independent testing sets: a predicted high-risk group versus a predicted low-risk group (for patients with ADC: hazard ratio = 2.34, 95% confidence interval: 1.12–4.91, p = 0.024; for patients with SCC: hazard ratio = 2.22, 95% confidence interval: 1.15–4.27, p = 0.017) after adjustment for age, sex, smoking status, and pathologic tumor stage.
Conclusions
The results suggest that the quantitative morphological features of tumor pathological images predict prognosis in patients with lung cancer.
Keywords: Pathological image, Prognosis, Statistical modeling, Morphological features, Lung adenocarcinoma, Lung squamous cell carcinoma
Introduction
NSCLC is the most common cause of lung cancer mortality, accounting for approximately 85% of such deaths.1 Within NSCLC, adenocarcinoma (ADC) and squamous cell carcinoma (SCC) are the two major subtypes, with distinct prognoses and therapeutic remedies.2,3
Recent studies have shown that the growth patterns of lung cancer are associated with patients’ survival outcomes.4–7 A new classification system based on the predominant tumor histological patterns was recommended for lung ADC,8 and its prognostic impacts have been verified in several studies.9–11 Furthermore, these newly defined ADC subtypes have different responses to chemotherapies.12 Despite its predictive and prognostic value, the new ADC subtype classification system requires extensive information processing by a pathologist to interpret highly complex pathological images, which is time-consuming and subjective, and generates considerable interobserver and intraobserver variations,13,14 Furthermore, there are no clear distinctions among ADC subtypes and most ADC cases are mixtures of several subtypes, so determining the ADC subtypes is challenging and prone to error.
Image features derived from computer-aided pathological analysis have been used to predict the survival of patients with breast cancer and to complement cancer molecular testing.15–17 These studies demonstrated the feasibility of using digital pathological image analysis for objective and unbiased clinical prognosis. However, there is a lack of such comprehensive pathological image analysis for lung cancer owing to the complexity and heterogeneity of the disease.
In this study, we developed a pathological image analysis pipeline to automatically extract morphological features and a statistical model based on these extracted features to predict survival outcomes of patients with lung cancer. The pathological images for 523 patients with ADC and 511 patients with SCC were downloaded from The Cancer Genome Atlas (TCGA) data set and analyzed using this pipeline. Prediction models were developed from training sets and validated in independent testing sets for ADC and SCC separately. The results showed that the pathological imaging–based algorithms predicted the prognoses of patients with lung cancer.
Materials and Methods
Collection and Preprocessing of Patient Histological Samples
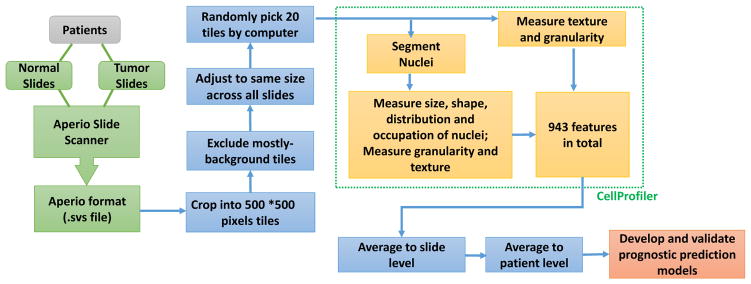
We acquired hematoxylin and eosin (HE)-stained histological images for patients with ADC and patients with SCC from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). All available HE pathological images from the TCGA data portal were included in this study. These images were captured at ×20 or ×40 magnification and include both frozen and formalin-fixed, paraffin-embedded slides. The corresponding patients’ clinical information was also obtained from the TCGA database. Detailed information on the slides and patients is summarized in Table 1. Image data in .svs file format were read and processed in MATLAB, version 2015a (The MathWorks, Natick, MA). To make the downstream computation feasible and prepare the data in the correct format for morphological features extraction, SVS images were cropped into millions of 500 × 500-pixel tiff images. The tiff image patches that contained mostly background white space were filtered out by a computer algorithm. From the remaining imaging patches, 20 representative tiff images were randomly selected by a computer algorithm to avoid potential subjective bias and reduce computational time for each SVS image.
Table 1.
Patient Data Summary
| Characteristic | Histologic Classes | |
|---|---|---|
|
| ||
| ADC | SCC | |
| No. patients | 523 | 511 |
|
| ||
| No. slides (tumor/nontumor) | 1581 (1337/244) | 1625 (1272/353) |
|
| ||
| Median age at diagnosis (LQ–HQ), y | 66 (59–73) (NA = 22) | 68 (62–73) (NA = 24) |
|
| ||
| Median follow-up (LQ–HQ), d | 230 [46–730] (NA = 22) | 272 [52–937] (NA = 26) |
|
| ||
| Vital status, n (%) | ||
| Alive | 394 (75.3) | 335 (65.6) |
| Deceased | 126 (24.1) | 160 (31.3) |
| NA | 3 (0.6) | 16 (3.1) |
|
| ||
| Sex, n (%) | ||
| M | 242 (46.3) | 367 (71.8) |
| F | 278 (53.2) | 128 (25) |
| NA | 3 (0.6) | 16 (3.1) |
|
| ||
| Cancer stage, n (%) | ||
| I | 281 (53.7) | 242 (47.4) |
| II | 126 (24.1) | 156 (30.5) |
| III | 85 (16.3) | 87 (17) |
| IV | 27 (5.2) | 7 (1.4) |
| NA | 4 (0.8) | 19 (3.7) |
|
| ||
| Smoking status, n (%) | ||
| Current smoker | 121 (23.1) | 134 (26.2) |
| Former smoker | 310 (59.3) | 336 (65.8) |
| Nonsmoker | 75 (14.3) | 18 (3.5) |
| NA | 17 (3.3) | 23 (4.5) |
Note: Summary of the number of slides in ADC and SCC, respectively, and the patient demographic information.
ADC, adenocarcinoma; SCC, squamous cell carcinoma; LQ, the lower quartile, 25th percentile; HQ, the higher quartile, 75th percentile; NA, not available; M, male, F, female.
Extraction of Morphological Features
The morphological features of the image patches were extracted and measured using CellProfiler, an open source cell image analysis software program developed by the Broad Institute (Cambridge, MA).18,19 Features were extracted by choosing and setting the parameters for different analysis modules (see Supplementary Materials for module parameter settings), and finally batch files were created to be able to process all images in batches by using a computational cluster. All 943 extracted morphological features of the 20 representative tiff images for each SVS slide were summarized by using the sample mean to slide level. When a patient had multiple tissue slides, the summarized value of the morphological features from the multiple slides were averaged to represent the value in the patient for further statistical analyses.
Development of Prediction Models
The TCGA data set was randomly partitioned with the R package caret (version 6.0-47) into two data sets: two-thirds of the patients were used in the training set and one-third were used in the testing set. To ensure the robustness and validity of our results, the random partition was repeated 100 times. Cox proportional hazards analysis by the R package survival (version 2.38-1) was used to select the morphological features that were significantly associated (p < 0.05) with patient overall survival in the training set. A random survival forest method20,21 (R package randomForest SRC 1.6.1) was used to develop the prediction models for patient survival outcomes in ADC and SCC, respectively. For each tumor subtype (ADC or SCC), a prediction model was developed from the training set and validated in the testing set. The model assigned a risk score for each patient in the testing set on the basis of the morphological features for that specific patient. The patients in the testing set were then separated into high- and low-risk groups on the basis of their predicted risk scores (with the median risk score used as a cutoff). All the analyses were performed with R version 3.1.1., using the aforementioned packages.
Survival Analysis
Overall survival time was calculated from the date of surgery until death or the date of last follow-up contact. Survival curves were estimated by the Kaplan-Meier product limit method.22 Differences in the survival outcomes between the predicted high- and low-risk groups were compared by a log-rank test to evaluate the performance of the prediction model in the testing sets. A multivariate Cox proportional hazards model23 was used to determine the association between predicted risk groups and the overall survival adjusted for other clinical variables, including age, sex, smoking status, and stage.
Results
Comprehensive Extraction of Morphological Features of Lung Pathological Images
Image data obtained from the TCGA cohort contained 1581 slides from 523 patients with ADC and 1625 slides from 511 patients with SCC. Of the 1581 ADC slides, 1337 were tumor samples and 224 were nonmalignant. Of the 1625 SCC slides, 1272 were tumor samples and 353 were nonmalignant. Age, sex, vital status, cancer stage, and smoking status were summarized in Table 1. After primary processing and filtering, morphological phenotypes of images were analyzed to extract regional features, such as cell and tissue texture and granularity. Next, cell nuclei were detected and segmented so that the cell-level features, such as cell size, shape, distribution, and texture of nuclei, could be specifically measured and analyzed. In total, we extracted 943 morphological features of tissue texture, cells, nuclei, and neighboring architecture. These features contained objective and quantitative morphological information provided by the histological images. The data processing procedures are summarized in Figure 1.
Figure 1.
Pathological image data processing and analyses pipeline. Image data obtained from The Cancer Genome Atlas lung cancer cohort contain slides of both nonmalignant lung and tumor. After initial filtering and normalization, morphological phenotypes of images were analyzed by using CellProfiler, and 943 morphological features of tissue texture, cells, nuclei, and neighboring architecture were extracted. These features were used to develop prediction models for patient survival, and cross-validations were implemented for model evaluation.
Morphological Features Predict Patient Survival in Both the ADC and SCC Subtypes
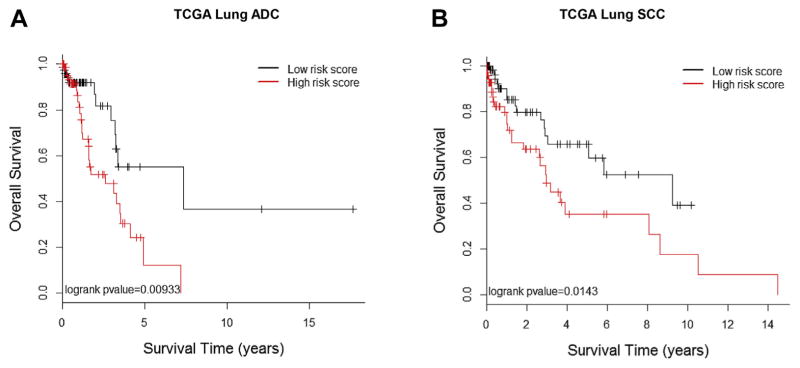
Among the extracted morphological features, 18 features were significantly associated (p < 0.05) with the overall survival of the patients with ADC in the training set (Table 2). These features were highly enriched (p = 0.00035) by tissue texture–related features (Supplementary Table 1). A prediction model based on these 18 morphological features was developed from the training set and validated in the testing set. The predicted high- and low-risk groups showed significant difference in survival (p = 0.009, log-rank test). The median survival time for the predicted low-risk group was 7.35 years versus 2.60 years for the predicted high-risk group (Fig. 2A). Multivariate analysis indicated that the predicted risk group was significantly associated with patients’ overall survival (HR = 2.34, 95% confidence interval: 1.12–4.91, p = 0.024) after adjustment for age, sex, smoking status, and tumor stage (Table 3).
Table 2.
Top Morphological Features Associated with Survival Outcomes of Patients with Lung ADC
| Feature Name from CellProfiler | p Value |
|---|---|
| Granularity_9_MaskedEosin | 0.0041 |
| Mean_Tissue_Texture_Correlation_MaskedEosin_80_45 | 0.0042 |
| Mean_Tissue_Texture_Correlation_MaskedEosin_40_135 | 0.0079 |
| Mean_Nuclei_AreaShape_Zernike_5_3 | 0.011 |
| Granularity_15_MaskedHema | 0.013 |
| Mean_Nuclei_AreaShape_Zernike_5_1 | 0.017 |
| Texture_InfoMeas2_Inverted_80_45 | 0.017 |
| Texture_Correlation_MaskedEosin_80_45 | 0.024 |
| Granularity_8_MaskedEosin | 0.025 |
| Mean_Tissue_Texture_Correlation_MaskedEosin_80_0 | 0.026 |
| Mean_Tissue_Texture_Correlation_MaskedEosin_40_0 | 0.031 |
| Texture_Contrast_NucleiNeighborCount_100_90 | 0.032 |
| Texture_Correlation_PercentNucleiTouching_100_0 | 0.035 |
| Mean_Nuclei_AreaShape_Zernike_3_1 | 0.043 |
| Mean_Tissue_Texture_Correlation_MaskedEosin_40_45 | 0.044 |
| Mean_Tissue_Texture_Gabor_MaskedEosin_40 | 0.045 |
| Mean_Nuclei_AreaShape_EulerNumber | 0.049 |
| Mean_Tissue_Texture_InfoMeas1_Inverted_40_45 | 0.049 |
Note: The 18 morphological features associated with survival outcomes of the Cancer Genome Atlas patients with ADC ranked by lowest p value from univariate analysis on the training set of data.
ADC, adenocarcinoma.
Figure 2.
Associations between survival outcomes and morphological feature–defined risk groups in The Cancer Genome Atlas (TCGA) cohorts of patients with adenocarcinoma (ADC) and squamous cell carcinoma (SCC). Kaplan-Meier survival curves for patients in the high-risk group (red) and low-risk group classified by morphological feature–based prediction models for both ADC (p = 0.009) (A) and SCC (p = 0.0143) (B). The features and prediction model were derived from the training data, and the survival analysis was performed in the testing data set.
Table 3.
Multivariate Analyses of Predicted Risk Groups Adjusted for Clinical Characteristics
| Histological Classes | ADC | SCC | ||
|---|---|---|---|---|
|
|
|
|
||
| Predictor | HR (95% CI) | p Value | HR (95% CI) | p Value |
| High- vs. low-risk group | 2.34 (1.12–4.91) | 0.024 | 2.22 (1.15–4.27) | 0.017 |
|
| ||||
| Age | 1.02 (0.99–1.06) | 0.21 | 1.03 (1.00–1.07) | 0.086 |
|
| ||||
| Sex | 1.28 (0.63–2.64) | 0.50 | 0.99 (0.49–2.00) | 0.97 |
|
| ||||
| Smoker vs. nonsmoker | 0.62 (0.22–1.79) | 0.38 | 9.92e+05 (0–inf) | 1.00 |
|
| ||||
| Pathological stage (II vs. I) | 1.39 (0.61–3.17) | 0.44 | 1.22 (0.59–2.54) | 0.59 |
|
| ||||
| Pathological stage (III and IV vs. I) | 2.02 (0.94–4.36) | 0.073 | 1.12 (0.51–2.45) | 0.78 |
Note: The prediction performances of the morphological feature–based models adjusted for clinical variables in the testing data sets for ADC and SCC, respectively. Risk groups were defined on the basis of the risk scores of individual patients predicted by morphological feature–based models.
ADC, adenocarcinoma; SCC, squamous cell carcinoma; HR, hazard ratio; CI, confidence interval.
For the SCC cohort, 12 morphological features were significantly associated with survival (p < 0.05) in the training set (Table 4). These features were significantly enriched (p = 0.0036) by granularity-related features (Supplementary Table 2). SCC is morphologically and clinically heterogeneous; further subclassification on the basis of morphology and molecular profiles may stratify the heterogeneity and lead to more accurate clinical prognosis.24,25 Furthermore, the differentiation level of SCC may be relevant for tumor prognosis. The morphological features are likely to reflect the different differentiation levels and subtypes of SCCs and thus be associated with patient prognosis. Again, a prediction model based on these 12 morphological features was developed from the training set and validated in the testing set. The predicted high- and low-risk groups showed significant difference in survival (p = 0.014, log-rank test). The median survival time for the predicted low-risk group was 9.25 years versus 2.95 years for the predicted high-risk group (Fig. 2B). Multivariate analysis indicated that predicted risk group was significantly associated with patients’ overall survival (HR = 2.22, 95% confidence interval: 1.15–4.27, p value = 0.017) after adjustment for age, sex, smoking status, and tumor stage (see Table 3).
Table 4.
Top Morphological Features Associated with Survival Outcomes of Patients with Lung SCC
| Feature Name from CellProfiler | p Value |
|---|---|
| Granularity_7_MaskedEosin | 0.005282 |
| Texture_Contrast_NucleiNeighborCount_100_0 | 0.006297 |
| Granularity_14_MaskedHema | 0.009601 |
| Granularity_6_MaskedEosin | 0.010949 |
| Mean_Tissue_Texture_Gabor_MaskedEosin_80 | 0.017315 |
| Granularity_8_MaskedHema | 0.023477 |
| Texture_Gabor_MaskedEosin_80 | 0.029167 |
| Texture_Correlation_PercentNucleiTouching_100_135 | 0.029596 |
| Texture_Correlation_NucleiNeighborCount_100_0 | 0.031148 |
| Mean_Tissue_Texture_AngularSecondMoment_MaskedEosin_40_135 | 0.031895 |
| Granularity_2_Inverted | 0.03556 |
| Granularity_8_MaskedEosin | 0.039156 |
Note: The top 12 morphological features associated with survival outcomes of the Cancer Genome Atlas patients with SCC ranked by lowest p value from univariate analysis on the training set of data.
SCC, squamous cell carcinoma.
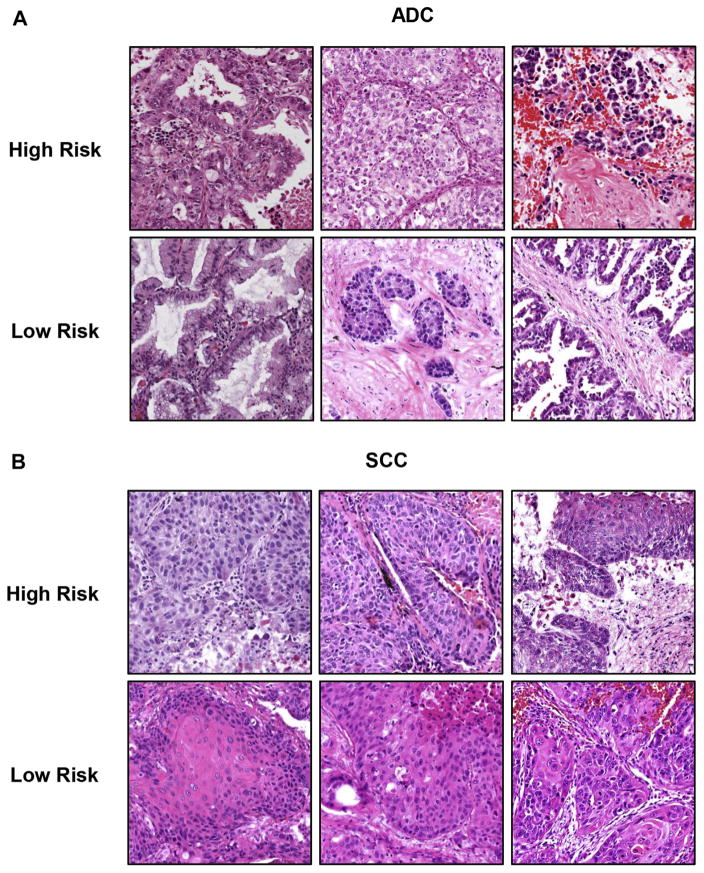
Representative images for high- and low-risk patients were shown both for patients with ADC (Fig. 3A) and for patients with SCC (Fig. 3B). Some images from low-risk patients showed a higher level of differentiation compared with images from high-risk patients, such as mucus secretion in ADC and keratinization in SCC. Moreover, some low-risk images show large areas of stroma, which could potentially restrict the tumors from further expansion. Interestingly, many images that are challenging for pathologists to distinguish solely by eye were separated into high- and low-risk groups by the computational image analysis, which provides meaningful additional diagnostic value.
Figure 3.
Representative images for high- and low-risk patients with adenocarcinoma (ADC) and squamous cell carcinoma (SCC). The patients with ADC and SCC in the testing set were classified into high- and low-risk groups on the basis of prediction from extracted morphological features. Three representative images are shown for each risk group for patients with ADC (A) and SCC (B).
To test the robustness of the prognosis features in both the ADC and SCC data sets, we repeated the random partitioning of training and testing sets 100 times. We performed the univariate survival analysis for each feature in each randomly selected training set and calculated the frequency of each feature showing a significant association (p < 0.05) with survival outcomes out of these 100 random partitions. The analysis showed that the result is robust. For example, a top feature Texture_Correlation_MaskedEosin_80_45 associated with ADC prognosis in the original training data set showed a significant association with survival in 87 of 100 bootstrapping data sets. In addition, all of the highly robust features for SCC prognosis (significant in >50% bootstrapping data sets) were included in our reported 12 SCC prognostic features from the training data set, and 70% of such features were included in our ADC prognostic features. All these results showed the robustness of the feature sets reported in the study.
Discussion
Comprehensive Exploration of the Morphological Information in Pathological Images
Current lung cancer classification focuses on several distinct tumor cell morphological features. However, even within the same type of cancer cells, the tumor cell morphology can be quite heterogeneous.26 The tumor microenvironment, which plays an essential role in tumor prognosis and response to therapy, has been largely overlooked.27 For example, macrophage and lymphocyte filtration have dual roles in immune defense and tumor progression.28 Increased abnormal vasculature may be associated with tumor expansion and metastasis.29,30 Keratinization of squamous tumor cells indicates good differentiation and is associated with better prognosis.31,32 Pathological images harbor essential information on the histological organization and morphological characteristics of tumor cells and their surrounding tumor microenvironment, both of which affect tumor growth patterns and contribute to tumor prognosis.33 However, this information is hard to distinguish and extract using only the human eye and brain. Therefore, systematic analyses of pathological images using computer algorithms could reveal hidden information in decoding tumor development and progression in lung cancer. The 943 features we extracted from the pathological images provide objective and quantitative information harbored in the images, which makes them well suited for downstream statistical analysis and modeling.
Computation-Based Image Analyses Predict Patient Survival in Both ADC and SCC
Clinicopathologic staging is a standard clinical procedure for tumor diagnosis and prognosis for lung cancer. However, it does not fully capture the complexity of the disease, so heterogeneous clinical outcomes within the same stage are common. Although the classification of lung cancer has been updated to become increasingly specific,34,35 the disease progression and response to treatment vary widely even among patients with the same histological subtype.26,36 Therefore, it is of substantial clinical importance to be able to predict patients’ clinical outcomes and thereby “tailor” the treatment for each individual patient. In this study, we showed that prognostic models developed for lung cancer morphological features predicted the prognosis of patients with lung cancer. In independent validation sets, the patients in the predicted low-risk group had significantly better survival compared with those in the predicted high-risk group.
Previously, Wang et al.37 identified pathological image markers for ADC versus SCC classification and survival prediction. However, that study was based on only 122 patients with lung cancer. More importantly, the prediction model developed from that study was not subtype specific. Because the growth patterns, prognoses, and therapeutic remedies are distinct between ADC and SCC,2,38 subtype-specific prediction models are more clinically meaningful.
A recent independent research by Yu et. al.39 also used CellProfiler software to extract features from pathological images of the TCGA lung cancer cohorts and reached a main conclusion similar to that of our study: that computational pathological image analysis is effective and powerful in predicting the prognosis of patients with ADC and patients with SCC. Despite the similarities of our studies, there are some differences and unique perspectives in our approaches and focus. First, our multivariate analysis (shown in Table 3) indicated that the prognostic performance of the identified morphological features is independent of other clinical variables, including age, sex, smoking status, and pathological stages, whereas Yu et. al.39 focused only on stage I patients and did not consider other clinical variables in the analysis. Second, we identified 18 features belonging to four morphological categories associated with prognosis in patients with ADC (see Table 2) and found that tissue texture–related morphological features are enriched in ADC prognostic features (Supplementary Table 1). Similarly, we identified 12 features belonging to three morphological categories associated with the prognosis of patients with SCC (see Table 4), and tissue granularity–related morphological features are enriched in SCC prognostic features (Supplementary Table 2). No specific prognostic morphological features or categories were reported in the study by Yu et. al.39 Third, we repeated the random partitioning of training and testing sets 100 times to demonstrate the robustness of the selected features. In addition, we analyzed the associations between ADC subtypes (solid, papillary, micropapillary, lepidic, and acinar) and the morphological features. We found that although some of the ADC prognostic morphological features are associated with ADC subtypes, the predictive power of the extracted morphological features could not be solely explained by ADC subtypes. Thus, our study demonstrated the feasibility and robustness of using computerized pathological imaging analysis in lung cancer prognosis to take NSCLC histological subtypes into consideration.
Expanding Our Understanding of Tumor Development and Progression
Systematic analyses of the morphological information in tumor pathological images provide information on tumor purity and tumor heterogeneity, which could facilitate normalization and standardization in molecular analyses of tumor genetic alterations.17 Furthermore, studies on the morphology features of certain molecular subtypes could improve our understanding of the disease mechanisms and how different molecular compositions lead to different morphological characteristics.40 Moreover, tumor morphology and molecular profiles (including gene mutation, copy number variation, mRNA, and protein expression) provide complementary views of cell activities at different levels. In combination with molecular assays and other clinical tests, the morphological analysis of tumor cells and their surrounding environment could predict the clinical outcomes of individual patients more precisely. With the advance of technology and analytical methods, it could be possible to combine cancer imaging analysis with molecular analyses and all other clinical tests to depict a comprehensive picture of the disease mechanisms, tumor grade, and patient prognosis, and to eventually guide in the therapeutic decision for “precision medicine.”
Limitations and Future Research
Our research serves as a pioneer study to explore the feasibility of using digital pathological image analysis for objective and unbiased clinical prognosis of patients with lung cancer. The image feature–based prediction models predict the prognosis for patients with ADC and patients with SCC. However, our current approach still faces some short-term limitations and suggests interesting directions for future research and applications. First of all, CellProfiler was developed to analyze cell images, but not specifically to analyze HE-stained pathological images. This makes it rather difficult to translate the features defined in CellProfiler into clinical terms used among pathologists. To overcome this limitation, computational algorithms specifically designed for pathological image analyses with quantitative features directly associated with clinical pathology are needed. Second, the implementation and interpretation of a digital pathology analysis pipeline currently requires relatively large data storage space and computational specialists to run the analysis. These may prevent the implementation of such a method in smaller facilities and hospitals. Therefore, cloud-based storage, user-friendly software, and centralized image-based diagnostic services will facilitate the application of digital pathology. Furthermore, the goal of this study was to show the feasibility of pathological image analysis for the prognosis of lung cancer, and only existing TCGA data were used in this study. Therefore, we could not systematically evaluate how different types of samples and image acquisition procedures affect the quality of data. A prospective study designed to evaluate the effects of different resolutions, specimen sizes, and types of samples, such as resection, biopsy, or cytological samples, will be important for future applications of computational pathological imaging analysis in diagnosis and medical decision making.
In summary, our computational approach performed objective and quantitative analysis of HE-stained pathological images and successfully predicted the prognosis of patients with lung ADC and SCC, respectively. Our predictive model can improve the current clinical practice and assist pathologists and clinicians in diagnosis and decision making for patients with lung cancer. It can be extended to analysis of other types of images and other cancer types. In the future, such pathological image–based analysis could be integrated with molecular analyses and clinical tests to guide in the therapeutic decision for precision medicine.
Supplementary Material
Footnotes
Disclosure: The authors declare no conflict of interest.
Note: To access the supplementary material accompanying this article, visit the online version of the Journal of Thoracic Oncology at www.jto.org and at http://dx.doi.org/10.1016/j.jtho.2016.10.017.
References
- 1.Tsuboi M, Ohira T, Saji H, et al. The present status of postoperative adjuvant chemotherapy for completely resected non-small cell lung cancer. Ann Thorac Cardiovasc Surg. 2007;13:73–77. [PubMed] [Google Scholar]
- 2.Cross MD. Advances in NSCLC: histologic distinction between adenocarcinoma and squamous cell carcinoma. MLO Med Lab Obs. 2012;44:40–42. 44. [PubMed] [Google Scholar]
- 3.Maeda H, Matsumura A, Kawabata T, et al. Adenosquamous carcinoma of the lung: surgical results as compared with squamous cell and adenocarcinoma cases. Eur J Cardiothorac Surg. 2012;41:357–361. doi: 10.1016/j.ejcts.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 4.Gleason DF, Mellinger GT. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol. 1974;111:58–64. doi: 10.1016/s0022-5347(17)59889-4. [DOI] [PubMed] [Google Scholar]
- 5.Amin MB, Tamboli P, Merchant SH, et al. Micropapillary component in lung adenocarcinoma: a distinctive histologic feature with possible prognostic significance. Am J Surg Pathol. 2002;26:358–364. doi: 10.1097/00000478-200203000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Barletta JA, Yeap BY, Chirieac LR. Prognostic significance of grading in lung adenocarcinoma. Cancer. 2010;116:659–669. doi: 10.1002/cncr.24831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borczuk AC, Qian F, Kazeros A, et al. Invasive size is an independent predictor of survival in pulmonary adenocarcinoma. Am J Surg Pathol. 2009;33:462–469. doi: 10.1097/PAS.0b013e318190157c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyoshi T, Satoh Y, Okumura S, et al. Early-stage lung adenocarcinomas with a micropapillary pattern, a distinct pathologic marker for a significantly poor prognosis. Am J Surg Pathol. 2003;27:101–109. doi: 10.1097/00000478-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 9.Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sica G, Yoshizawa A, Sima CS, et al. A grading system of lung adenocarcinomas based on histologic pattern is predictive of disease recurrence in stage I tumors. Am J Surg Pathol. 2010;34:1155–1162. doi: 10.1097/PAS.0b013e3181e4ee32. [DOI] [PubMed] [Google Scholar]
- 11.Russell PA, Wainer Z, Wright GM, Daniels M, Conron M, Williams RA. Does lung adenocarcinoma subtype predict patient survival?: A clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J Thorac Oncol. 2011;6:1496–1504. doi: 10.1097/JTO.0b013e318221f701. [DOI] [PubMed] [Google Scholar]
- 12.Tsao MS, Marguet S, Le Teuff G, et al. Subtype classification of lung adenocarcinoma predicts benefit from adjuvant chemotherapy in patients undergoing complete resection. J Clin Oncol. 2015;33:3439–3446. doi: 10.1200/JCO.2014.58.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective. Acta Neuropathol. 2010;120:297–304. doi: 10.1007/s00401-010-0725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper LA, Kong J, Gutman DA, Dunn WD, Nalisnik M, Brat DJ. Novel genotype-phenotype associations in human cancers enabled by advanced molecular platforms and computational analysis of whole slide images. Lab Invest. 2015;95:366–376. doi: 10.1038/labinvest.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabesh A, Teverovskiy M, Pang H-Y, et al. Multifeature prostate cancer diagnosis and Gleason grading of histological images. IEEE Trans Med Imaging. 2007;26:1366–1378. doi: 10.1109/TMI.2007.898536. [DOI] [PubMed] [Google Scholar]
- 16.Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 17.Yuan Y, Failmezger H, Rueda OM, et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med. 2012;4:157ra143. doi: 10.1126/scitranslmed.3004330. [DOI] [PubMed] [Google Scholar]
- 18.Carpenter AE, Jones TR, Lamprecht MR, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 20.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann App Statist. 2008;2:841–860. [Google Scholar]
- 21.Taylor JM. Random survival forests. J Thorac Oncol. 2011;6:1974–1975. doi: 10.1097/JTO.0b013e318233d835. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 23.Collett D. Modelling Survival Data in Medical Research. Boca Raton, FL: Chapman and Hall/CRC; 2003. [Google Scholar]
- 24.Wilkerson MD, Yin X, Hoadley KA, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16:4864–4875. doi: 10.1158/1078-0432.CCR-10-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Moreno P, Brambilla E, Thomas R, Soria JC. Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin Cancer Res. 2012;18:2443–2451. doi: 10.1158/1078-0432.CCR-11-2370. [DOI] [PubMed] [Google Scholar]
- 26.Errico A. Lung cancer: heterogeneity in space and time. Nat Rev Clin Oncol. 2014;11:684. doi: 10.1038/nrclinonc.2014.186. [DOI] [PubMed] [Google Scholar]
- 27.Dudas J. Supportive and rejective functions of tumor stroma on tumor cell growth, survival, and invasivity: the cancer evolution. Front Oncol. 2015;5:44. doi: 10.3389/fonc.2015.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kataki A, Scheid P, Piet M, et al. Tumor infiltrating lymphocytes and macrophages have a potential dual role in lung cancer by supporting both host-defense and tumor progression. J Lab Clin Med. 2002;140:320–328. doi: 10.1067/mlc.2002.128317. [DOI] [PubMed] [Google Scholar]
- 29.Kaessmeyer S, Bhoola K, Baltic S, Thompson P, Plendl J. Lung cancer neovascularisation: cellular and molecular interaction between endothelial and lung cancer cells. Immunobiology. 2014;219:308–314. doi: 10.1016/j.imbio.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Ushijima C, Tsukamoto S, Yamazaki K, Yoshino I, Sugio K, Sugimachi K. High vascularity in the peripheral region of non-small cell lung cancer tissue is associated with tumor progression. Lung Cancer. 2001;34:233–241. doi: 10.1016/s0169-5002(01)00246-x. [DOI] [PubMed] [Google Scholar]
- 31.Nhung NV, Mirejovsky P, Mirejovsky T, Melinova L. Cytokeratins and lung carcinomas. Cesk Patol. 1999;35:80–84. [PubMed] [Google Scholar]
- 32.Cheung WK, Nguyen DX. Lineage factors and differentiation states in lung cancer progression. Oncogene. 2015;34:5771–5780. doi: 10.1038/onc.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vannucci L. Stroma as an active player in the development of the tumor microenvironment. Cancer Microenviron. 2015;8:159–166. doi: 10.1007/s12307-014-0150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spiro SG, Tanner NT, Silvestri GA, et al. Lung cancer: progress in diagnosis, staging and therapy. Respirology. 2010;15:44–50. doi: 10.1111/j.1440-1843.2009.01674.x. [DOI] [PubMed] [Google Scholar]
- 35.Petersen I, Warth A. Lung cancer: developments, concepts, and specific aspects of the new WHO classification. J Cancer Res Clin Oncol. 2016;142:895–904. doi: 10.1007/s00432-015-2004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yaqub F. Intratumour heterogeneity in lung cancer. Lancet Oncol. 2014;15:e536. doi: 10.1016/s1470-2045(14)71030-7. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Xing F, Su H, Stromberg A, Yang L. Novel image markers for non-small cell lung cancer classification and survival prediction. BMC Bioinformatics. 2014;15:310. doi: 10.1186/1471-2105-15-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas A, Liu SV, Subramaniam DS, Giaccone G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol. 2015;12:511–526. doi: 10.1038/nrclinonc.2015.90. [DOI] [PubMed] [Google Scholar]
- 39.Yu KH, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimm J, Kirsch DG, Windsor SD, et al. Use of gene expression profiling to direct in vivo molecular imaging of lung cancer. Proc Natl Acad Sci USA. 2005;102:14404–14409. doi: 10.1073/pnas.0503920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.