Abstract
Treatment of urinary tract infections is today a challenge due to the increasing prevalence of multidrug-resistant ESBL-producing uropathogenic Escherichia coli (UPEC). There is an urgent need for new treatment strategies for multidrug-resistant UPEC and preferably with targets that have low potential for development of resistance. Carbon monoxide-releasing molecules (CORMs) are novel and potent antibacterial agents. The present study examines the transcriptomic targets of CORM-2 in a multidrug-resistant ESBL-producing UPEC isolate in response to a single exposure to CORM-2 and after repeated exposure to CORM-2. The bacterial viability and minimal inhibitory concentration (MIC) were also examined after repeated exposure to CORM-2. Microarray analysis revealed that a wide range of processes were affected by CORM-2, including a general trend of down-regulation in energy metabolism and biosynthesis pathways and up-regulation of the SOS response and DNA repair. Several genes involved in virulence (ibpB), antibiotic resistance (marAB, mdtABC) and biofilm formation (bhsA, yfgF) were up-regulated, while some genes involved in virulence (kpsC, fepCEG, entABE), antibiotic resistance (evgA) and biofilm formation (artIP) were down-regulated. Repeated exposure to CORM-2 did not alter the gene expression patterns, the growth inhibitory response to CORM-2 or the MIC values for CORM-2, cefotaxime, ciprofloxacin and trimethoprim. This study identifies several enriched gene ontologies, modified pathways and single genes that are targeted by CORM-2 in a multidrug-resistant UPEC isolate. Repeated exposure to CORM-2 did not change the gene expression patterns or fold changes and the susceptibility to CORM-2 remained after repeated exposure.
Introduction
Nearly one-fifth of all uropathogenic strains of E. coli (UPEC) are resistant to the most commonly used antibiotics [1]. Therapeutic options are limited for extended spectrum beta-lactamase (ESBL)-producing E. coli, where the bacteria have acquired a plasmid with genes that code for the enzyme ESBL. ESBL-producing Enterobacteriaceae spp. contain genes that code for the ESBL enzyme, and several different ESBL enzyme variants (TEM, SHV, CTX-M) have been identified. ESBL-producing E. coli can inactivate most of the beta-lactam antibiotics and cephalosporins and frequently demonstrate co-resistance to other antibiotics, such as aminoglycosides and quinolones [2]. The most significant factor for the development of antimicrobial resistance has been found to be selection pressure caused by antibiotics [3]. In Europe, an association between use of antimicrobial drugs and occurrence of resistance has been described at a country level [4]. Development of resistance may arise after mutations through stable genetic alterations or be an adaptive phenomenon characterised by induced tolerance when the drug is present [5]. Mechanisms of antibiotic resistance include enzymatic modification of the antibiotic, reprogramming or camouflaging the target by mutation and efflux pumps which pump the antibiotic out of the cell [6].
Carbon monoxide (CO) has been ascribed a novel role as a host defence molecule with bactericidal effects [7]. CO is produced endogenously as a result of heme metabolism through the enzyme heme oxygenase (HO) and acts as a potent regulatory and protective molecule with e.g., anti-apoptotic, anti-inflammatory and anti-proliferative effects [8]. Metal carbonyl compounds or CO-releasing molecules, CORMs, for temporal and spatial CO-delivery have been developed for therapeutic applications [9]. CO easily diffuses through membranes, while CO derived from metal carbonyl compounds may be internalized into bacteria through a Trojan horse mechanism [10], [11]. The effect of CORMs on non-pathogenic E. coli seems extensive, including actions on heme-containing proteins, and a wide range of transcriptional modifications in key metabolic pathways have been observed by CORMs [11], [12], [13], [14]. A synergistic effect of CO and the metal ion co-ligand in CORMs seems to be required for full bactericidal effect [14], [15]. Our previous results show that CORM-2 has bactericidal effects against multidrug-resistant ESBL-producing UPEC [16].
There is an urgent need for new treatment strategies suitable for targeting bacteria that are resistant to traditional antibiotics. One strategy for overcoming resistance may be to develop inhibitors of novel targets, assuming that new chemical entities are not susceptible to existing resistance mechanisms [17]. Interestingly, CORMs may be less likely to cause development of resistance mechanisms, due to multiple and different targets than existing antibiotics [9]. One of the few known carbon monoxide resistance genes is cor, which counteracts CO toxicity in Mycobacterium tuberculosis [18]. In addition, deletion of genes implicated in the process of biofilm formation (tqsA and bhsA) results in higher resistance to CORM-2 in non-pathogenic E. coli, while strains mutated in methionine related genes are hypersensitive to CORM-2 [12]. Gene profiling studies on CORMs have up to now only been carried out in non-pathogenic E. coli K12 strains [11], [12], [13], [14]. The effects of CORMs on gene expression in pathogenic bacteria, such as UPEC strains, are therefore unknown. Moreover, studies addressing the potential for bacteria to develop resistance to CORMs have not yet been performed.
The aim of the present study was to use global gene profiling to assess the transcriptomic impact of CORM-2 in a multidrug-resistant ESBL-producing UPEC isolate. In addition, possible changes in gene expression, antibiotic susceptibility and virulence properties were evaluated after repeated exposure to CORM-2.
Materials and methods
Reagents
CORM-2 (tricarbonyldichlororuthenium (II) dimer ([Ru(CO)3Cl2]2)) (Sigma-Aldrich, St. Louis, MO, USA) and trimethoprim (Sigma-Aldrich) were prepared by dissolution in dimethyl sulfoxide (DMSO). Cefotaxime and ciprofloxacin (Sigma-Aldrich) were prepared in sterile water. All reagents were freshly prepared or used from stock solutions.
Bacterial strains
Two clinical UPEC strains, the ESBL-producing E. coli isolate 7 (ESBL7) and the non-ESBL-producing isolate UPEC2, were subjected to primary susceptibility testing through the disk diffusion method at the Department of Laboratory Medicine, Microbiology, Örebro University Hospital. ESBL7 was confirmed as ESBL-producing by detecting clavulanic acid reversible resistance for oxyiminocephalosporins and found to belong to the CTX-M-15 enzyme type and sequence type 131 [19]. ESBL7 showed resistance to cefotaxime (CTX), ceftazidime (CAZ), trimethoprim (TMP), ciprofloxacin (CIP) and mecillinam (MEL). UPEC2 was susceptible to CTX, CIP, MEL, TMP and nitrofurantoin (NIT). The commensal E. coli K12 strain MG1655 was used from laboratory stocks. The study did not involve analysis of human data, specimens or tissue samples.
Bacterial media and growth conditions
Cultures were maintained on tryptic soy agar (TSA) (Becton Dickinson, Le Pont Claix, France). Overnight cultures were grown in Difco Luria-Bertani (LB) broth (Lennox; Franklin Lakes, NJ, USA) at 37°C aerobically on a shaker at 200 rpm.
Repeated exposures to CORM-2 or vehicle
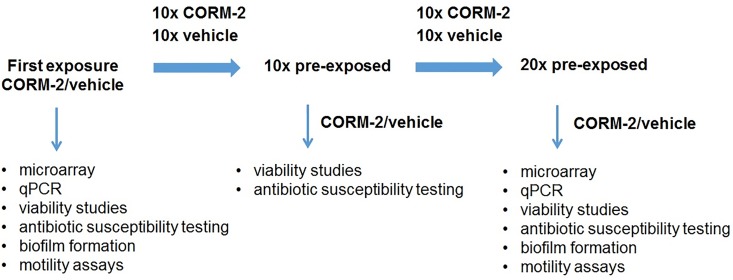
Bacteria (ESBL7, UPEC2, MG1655; picked from 5–10 colonies) were suspended in 1 ml of PBS, yielding a suspension corresponding to the turbidity of McFarland 0.5, and diluted 1:100 in minimal salt (MS)-medium (~106 CFU/ml). MS-medium was prepared as previously described [20] (1.3% [wt/vol] Na2HPO4, 0.3% KH2PO4, 0.05% NaCl, and 0.1% NH4Cl supplemented with 20 mM glucose, 2 mM MgSO4, 100 μM CaCl2, and 0.25% Casamino Acids). The suspension was exposed to CORM-2 (250 μM) or vehicle (2.5% DMSO) for 4 hours at 37°C. A volume (10 μl) was spread onto TSA-agar plates and incubated at 37°C overnight. This procedure was repeated 10 times (10x, ~45 generations) or 20 times (20x, ~90 generations). For experimental design, see Fig 1.
Fig 1. Summary of the experimental design.
RNA isolation
Overnight cultures of ESBL7 from the original isolate, or isolates pre-exposed 20 times to CORM-2 or vehicle, were used to inoculate MS-medium to an optical density (OD620) of 0.1, followed by exposure to CORM-2 (250 μM) or vehicle (2.5% DMSO) for 30 min at 37°C. RNA isolation was performed using an RNeasy mini kit (Qiagen Technologies, Hilden, Germany), according to the manufacturer’s protocol. DNA decontamination treatment was performed using Turbo DNase (Qiagen) and the quantity and purity of the purified RNA samples were determined using a spectrophotometer Nanodrop-1000 (Nanodrop Technologies Inc., Wilmington, DE, USA) by measuring the absorbance (A260, 230, 280) and calculating absorbance ratios (A260/A230 and A260/A280). All samples had A260/A230 and A260/A280 ratios above 1.9. The RNA integrity was analysed using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) in conjunction with RNA 6000 Nano LabChip kit (Agilent Technologies) according to the manufacturer’s protocol. RNA integrity number (RIN) values were > 8.5 for all samples.
Microarray analysis
High-quality total RNA was used to prepare labelled cRNA with One-color Low Input Quick Amp WT Labelling Kit (Agilent) according to the manufacturer’s instructions. The cDNA synthesis was performed by using WT Primer Mix and cDNA Master Mix (Agilent). Labelled samples were hybridised onto G4813A E. coli gene expression Microarray 8×15K glass slides (Agilent) containing 15 208 E. coli probes. Microarrays were scanned with a G2565 CA array laser scanner (Agilent) followed by image analysis and data extraction with Feature Extraction Software version 10.7.3.1 (Agilent). Four experimental groups with 4 biological replicates in each group were analysed (total of 16 RNA samples).
Quantitative real-time PCR (qPCR)
cDNA synthesis (0.1 μg of total RNA) was performed by using High Capacity cDNA Reverse Transcription Kit for single-stranded cDNA synthesis (Applied Biosystems, CA, USA) according to manufacturer`s protocol. qPCR was performed with Maxima SYBR Green qPCR Master Mix (ThermoFisher Scientific, MA, USA) according to manufacturer’s instructions. 200–300 nM of primer and 5 ng template cDNA was added to each supermix. Primers were ordered from Eurofins MWG Synthesis GmbH (Ebersberg, Munich, Germany) (S1 Table). The RT-PCR amplification was performed in a CFX96 Touch™ Real-Time PCR Detection System (Biorad, CA, USA) using the following protocol: initial denaturation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s followed by annealing at 60°C for 30 s and extension at 72°C for 30 s. Each PCR was followed by a dissociation curve analysis between 60–95°C. The Ct values were analysed by the comparative Ct (ΔΔCt) method and normalized to the endogenous control gapA (encoding glyceraldehyde 3-phosphate dehydrogenase A). Fold difference was calculated as 2-ΔΔCt.
Determination of bacterial viability after exposure to CORM-2
Overnight culture grown in LB broth was diluted 1/1000 in MS-medium (to ~106 CFU/ml) and further incubated at 37°C on a shaker at 200 rpm to early log phase (OD620 = 0.1). The bacterial concentrations of the initial inocula used in these experiments were in the range of 107−108 CFU/ml. Thereafter, the bacteria were exposed to CORM-2 (250–500 μM) and grown for up to 24 h in darkness at 37°C. Time-zero samples (starting inocula) were taken and the number of viable colonies determined as described below. Samples were taken at different times after addition of CORM-2 (1, 2, 4, 8 and 24 h) depending on the experimental protocol. All samples were diluted in PBS and at least three serial dilutions were plated on TSA-plates. Following overnight culture at 37°C, bacterial CFU/ml was determined as the mean of two dilutions. Viability was calculated as the CFU/ml in CORM-2 exposed cultures divided by the number of CFU/ml formed upon plating of the initial starting inocula and expressed as log CFU/ml.
Determination of minimum inhibitory concentration (MIC)
MIC (minimum inhibitory concentration) for CORM-2, cefotaxime, ciprofloxacin and trimetophrim was determined using the broth dilution test. The test substances were inoculated with a bacterial suspension (~106 CFU/ml) in LB-broth or MS-medium (CORM-2) on 96-well plates for 18–20 h at 37°C. All MIC tests were performed in duplicate and at least twice. The MIC was read as the lowest concentration yielding no visible growth.
Analysis of biofilm formation
Overnight cultures in LB-broth were used to inoculate (at 0.1%) fresh MS-medium to an OD620 of approximately 0.05. The bacteria were seeded into 96-well plastic plates (Nunc C96 Microwell plate, Nunc A/S, Roskilde, Denmark) and exposed to CORM-2 (250 μM) or vehicle (2.5% DMSO). After 24 h of incubation under static conditions at 37°C, biofilm formation was quantified by the crystal violet method as previously described [12]. The absorbance at 540 nm was measured by spectrophotometer (Multiscan Ascent, Thermo Labsystems, Helsingfors, Finland). The experiments were repeated three times in quadruplicate.
Motility assays
Overnight cultures in LB-broth were used to inoculate (at 0.1%) fresh MS-medium to an OD620 of approximately 0.1, followed by exposure to 250 μM CORM-2 or vehicle (2.5% DMSO). Swimming motility plates (0.3% agar) and swarming motility plates (0.5% agar) were prepared as previously described [21] and bacterial suspensions were inoculated on the plates. One μl of bacterial suspension was stabbed into the swimming agar plates and 5 μl bacterial suspension spotted on swarming agar plates. The distance of migration (the diameter of the growth around the inoculation site) was measured after incubation for 14 h (swimming plates) or 20 h (swarming plates) at 37°C. The experiments were repeated three times in duplicate.
Host renal cell activation
The human renal epithelial cell line A498 (HTB-44) was obtained from American Type Culture Collection (Manassas, USA) and cultured in Dulbecco's modified eagle medium (DMEM, Sigma-Aldrich) containing 10% fetal bovine serum (FBS), 2 mM L-glutamine, 1 mM non-essential amino acids (all from Invitrogen Ltd, Paisley, UK) at 37°C in a 5% CO2 atmosphere. During experiments, the FBS concentration was reduced to 2%. The A498 epithelial cells were stimulated with overnight cultures of ESBL7 representing the original isolate, or isolates pre-exposed 20 times to CORM-2 or vehicle. A multiplicity of infection (MOI) of 10 was used. Cell supernatants were collected after stimulation for 6 h and centrifuged for 5 min at 5000 x g and stored at– 80°C. IL-6 and IL-8 cytokine production were measured using human IL-8 and IL-6 kits (ELISA MAX™ Deluxe Sets, BioLegend, San Diego, CA, USA) according to manufacturer's protocol and measured on a spectrophotometer (Multiscan Ascent). Cell cytotoxicity was determined using the Pierce™ LDH Cytotoxicity Assay Kit (TermoFisher Scientific, MA, USA) and absorbance measured on a spectrophotometer (Multiscan Ascent). Samples were normalized to unstimulated and lysed control cells.
Statistical analysis and microarray data processing
Data are expressed as mean ± SEM. Student’s t-test was used to compare two groups and a one-way analysis of variance (ANOVA) parametric test was used for comparison of multiple groups, followed by Bonferroni multiple testing correction using the software GraphPad Prism (GraphPad Software Inc., La Jolla, CA, USA). Results were considered statistically significant at p-values < 0.05. Microarray data analysis was performed using GeneSpring GX version 12.1 (Agilent) after per chip and 75th percentile shift gene normalization of samples. Statistical significant entities were obtained using the one-way ANOVA parametric test, followed by Tukey HSD post hoc test and Bonferroni FWER multiple testing correction, with a statistical significance set at a corrected p-value < 0.05 and a biological significance set at a fold change ≥ 2. Significant GO term enrichment and single experiment pathway analysis (SEA), was set at a p-value < 0.05 and < 0.1, respectively. n = number of independent experiments.
Genes that exhibited a two-fold or higher increase or decrease (p < 0.05) were further classified by use of gene annotations in NCBI http://www.ncbi.nlm.nih.gov, EcoCyc http://ecocyc.org and literature mining. In addition, a virulence factor list for E. coli was generated through the PATRIC database (www.patricbrc.org), MESH virulence term association and literature mining. Gene expression data is available in the GEO database with the accession number GSE87627.
Results
Analysis of transcriptional alterations in response to CORM-2
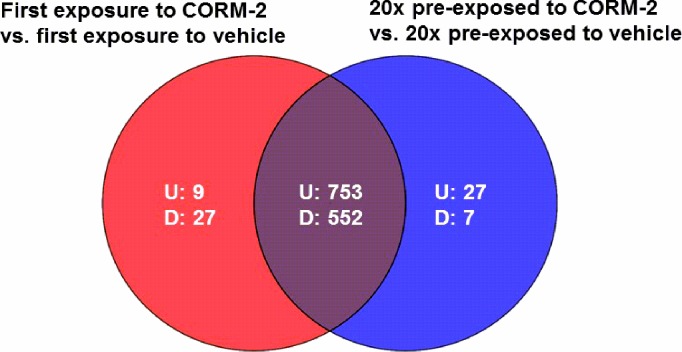
Microarray analysis was performed to analyse the gene expression alterations of ESBL7 in response to first-time exposure to CORM-2 and after pre-exposure 20 times to CORM-2. A total of 1 305 entities, common for both experimental settings, were differentially expressed with at least a two-fold change compared with vehicle (Fig 2). Of the 1 305 entities with altered transcription, 753 entities were up-regulated and 552 entities were down-regulated. Some differentially expressed gene entities were not shared between the experimental settings and were only found in response to first-time exposure to CORM-2 or after pre-exposure 20 times to CORM-2. Specific alterations in gene expression in response to first-time exposure and after exposure 20 times to CORM-2 showed 9 and 27 up-regulated and 27 and 7 down-regulated entities, respectively (Fig 2).
Fig 2. Venn diagram of differently expressed entities in ESBL isolate 7.
Shown in red, first-time exposure to CORM-2 (250 μM) versus first-time exposure to vehicle (2.5% DMSO); in blue, pre-exposed 20 times to CORM-2 versus pre-exposed 20 times to vehicle. Up- and down-regulated entities are designated U and D respectively (n = 4 in each group). Overlapping regions represent entities present in both experimental conditions.
Gene ontology analysis
Gene ontology (GO) analysis were performed on gene entities for each of the experimental settings. In total, 9 gene ontologies were enriched by the differentially expressed entities (Table 1). The enriched gene ontologies were common and found both in response to first-time exposure to CORM-2 and after pre-exposure 20 times to CORM-2. The enriched gene ontology classes were cell communication, SOS response, cellular response to external stimulus, cellular response to extracellular stimulus, response to extracellular stimulus, fermentation, cellular response to DNA damage stimulus, DNA repair and cellular response to stress (Table 1). The differentially expressed genes enriching the different gene ontology classes are summarized in Table 2and S2 Table.
Table 1. Enriched gene ontologies, common for both first-time exposed and 20 times repeated exposure to CORM-2 (250 μM) versus vehicle (2.5% DMSO) in ESBL-producing E. coli.
| GO ID | GO term | Adjusted p-value | Count in selection | Count in total | No of genes up-/down-regulated |
|---|---|---|---|---|---|
| 7154 | cell communication | 0.000 | 16 | 24 | +16 |
| 9432 | SOS response | 0.000 | 16 | 24 | +16 |
| 71496 | cellular response to external stimulus | 0.000 | 16 | 24 | +16 |
| 31668 | cellular response to extracellular stimulus | 0.000 | 16 | 24 | +16 |
| 9991 | response to extracellular stimulus | 0.000 | 22 | 41 | +18/-4 |
| 6113 | fermentation | 0.005 | 17 | 33 | +6/-11 |
| 6974 | cellular response to DNA damage stimulus | 0.01 | 20 | 45 | +17/-3 |
| 6281 | DNA repair | 0.01 | 20 | 45 | +17/-3 |
| 33554 | cellular response to stress | 0.01 | 22 | 52 | +19/-3 |
Table 2. Differentially expressed genes of ESBL-producing E. coli following exposure to CORM-2 (250 μM) versus vehicle (2.5% DMSO).
| Gene | Fold change | Fold change | Gene product |
|---|---|---|---|
| symbol | First exposure | 20x pre-exposed | |
| CORM-2 vs | CORM-2 vs | ||
| first exposure | 20x pre-exposed | ||
| vehicle | vehicle | ||
| Represented in all enriched ontologies | |||
| polB | 22.5 | 25.6 | DNA polymerase II |
| sulA | 13.9 | 15.6 | suppressor of lon; inhibits cell division and ftsZ ring formation |
| recAa | 13.6 | 14.2 | DNA strand exchange and renaturation, DNA-dependent ATPase |
| yebG | 12.5 | 15.3 | DNA damage-inducible protein regulated by LexA |
| dinI | 11.8 | 17.6 | damage-inducible protein I |
| recN | 11.4 | 13.0 | protein used in recombination and DNA repair |
| umuD | 10.2 | 14.5 | SOS mutagenesis; error-prone repair |
| umuC | 9.1 | 10.3 | SOS mutagenesis and repair |
| lexA | 8.2 | 10.8 | regulator for SOS |
| ruvB | 3.8 | 3.8 | Holliday junction helicase subunit A |
| uvrD | 3.8 | 2.9 | DNA-dependent ATPase I and helicase II |
| ruvA | 2.5 | 3.3 | Holliday junction helicase subunit B |
| uvrA | 2.6 | 2.5 | excision nuclease subunit A |
| uvrB | 2.1 | 2.4 | DNA repair; excision nuclease subunit B |
| Represented only in cell communication, SOS response, cellular response to external stimulus, cellular response to extracellular stimulus, response to extracellular stimulus, DNA repair or cellular response to stress | |||
| ydjM | 21.3 | 18.2 | inner membrane protein regulated by LexA |
| dinB | 5.2 | 6.1 | damage-inducible protein P; putative tRNA synthetase |
| Represented only in cellular response to DNA damage stimulus | |||
| mutM | 25.2 | 25.8 | formamidopyrimidine/5-formyluracil/ 5-hydroxymethyluracil DNA glycosylase |
| recF | 7.5 | 5.7 | ssDNA and dsDNA binding, ATP binding |
| mug | 3.2 | 3.5 | G/U mismatch-specific DNA glycosylase |
| phr | -3.0 | -2.3 | deoxyribodipyrimidine photolyase |
| alkB | -2.9 | -2.6 | DNA repair system specific for alkylated DNA |
| Represented only in cellular response to DNA damage stimulus | |||
| ung | -2.6 | -2.0 | uracil-DNA-glycosylase |
| Represented only in response to extracellular stimulus | |||
| sspB | 2.9 | 2.4 | stringent starvation protein B |
| sspA | 2.3 | 2.0 | regulator of transcription; stringent starvation protein A |
| yjiY | -22.9 | -19.2 | putative carbon starvation protein |
| slp | -8.2 | -7.1 | outer membrane protein induced after carbon starvation |
| psiF | -3.3 | -2.3 | induced by phosphate starvation |
| rspB | -2.9 | -2.7 | starvation sensing protein |
Presented genes are derived from significant enrichment in the gene ontologies cell communication, SOS response, cellular response to external stimulus, cellular response to extracellular stimulus, response to extracellular stimulus, cellular response to DNA damage stimulus, DNA repair or cellular response to stress. n = 4
a also represented in virulence
Pathway analysis
Single experiment pathway analysis (SEA) was performed in order to discover affected pathways and to further categorize the altered gene entities according to biological function. A total of 15 pathways were enriched and all were related to metabolism. Fourteen of these affected pathways were common and found both in response to first-time exposure to CORM-2 and after pre-exposure 20 times to CORM-2 (Table 3). The pathway carnitine degradation I was enriched only in response to first-time exposure to CORM-2.
Table 3. Single experiment pathway analysis of ESBL-producing E. coli gene entities.
| Fold change | ||||
|---|---|---|---|---|
| Common in CORM-2 vs vehicle | ||||
| Pathway | p-value | Matched | Pathway | No of genes |
| entities | entities | up-/down-regulated | ||
| glycolysis I (from glucose-6P) | 0.045 | 2 | 16 | -2 |
| glycolysis II (from fructose-6P) | 0.045 | 2 | 3 | -2 |
| gluconeogenesis I | 0.083 | 2 | 14 | -2 |
| glucose and xylose degradation | 0.003 | 4 | 6 | -3/+1 |
| mixed acid fermentation | 0.002 | 3 | 3 | -1/+1a |
| superpathway of N-acetylneuraminate degradation | 0.003 | 4 | 6 | -4 |
| superpathway of 5-aminoimidazole ribonucleotide biosynthesis | 0.083 | 2 | 4 | -2 |
| superpathway of chorismate metabolism | 0.079 | 6 | 24 | -5/+1 |
| superpathway of histidine, purine and pyrimidine biosynthesis | 0.043 | 4 | 12 | -4 |
| superpathway of lysine degradation | 0.017 | 2 | 11 | -2 |
| superpathway of phenylalanine, tyrosine and tryptophan biosynthesis | 0.050 | 3 | 7 | -3 |
| superpathway of pyrimidine deoxyribonucleotides de novo biosynthesis | 0.045 | 2 | 15 | -1/+1 |
| superpathway of tryptophan biosynthesis | 0.050 | 3 | 7 | -3 |
| tryptophan biosynthesis | 0.002 | 3 | 9 | -3 |
Presented pathways are affected following exposure to CORM-2 (250 μM) versus vehicle (2.5% DMSO).
a part of protein complex
Alterations in gene expression common for first-time exposure to CORM-2 and pre-exposure 20 times to CORM-2
A more detailed study of the alterations in expression of virulence, antibiotic resistance and biofilm genes was performed. Some genes involved in virulence were induced following exposure to CORM-2 (such as ibpB, recA, ycfQ), but many genes were repressed (such as kpsC, ompW, ompT, fepEG) (Table 4). Several antibiotic resistance-associated genes, such as genes coding for different multidrug efflux systems, were induced following exposure to CORM-2 (such as mdtABC, marAB, acrD) and some were repressed (such as evgA, mdtE) (Table 5). Some genes involved in biofilm formation, such as bhsA and yfgF encoding the anaerobic cyclic-di-GMP phosphodiesterase, were induced and some biofilm genes were repressed (Table 6). Genes involved in defence, stress response or repair, such as the gene encoding the heat shock chaperone ibpAB were markedly induced following exposure to CORM-2, while hdeA and evgA were repressed (Table 7). Three genes, hdeA, cusF (Table 7) and cusX (-58.2 first exposure; -26.3 pre-exposure 20 times) showed a significantly lower repression after pre-exposure 20 times to CORM-2 compared to first-time exposure. Differentially expressed genes associated with fimbriae and flagella are shown in S3 Table. CORM-2 is known to affect respiration and the majority of the differentially expressed genes involved in regulation of respiration were down-regulated (S4 Table).
Table 4. ESBL-producing E. coli genes associated with the functional category virulence that are differentially expressed following exposure to CORM-2 (250 μM) versus vehicle (2.5% DMSO).
| Gene | Fold change | Fold change | Gene product |
|---|---|---|---|
| symbol | First exposure | 20x pre-exposed | |
| CORM-2 vs | CORM-2 vs | ||
| first exposure | 20x pre-exposed | ||
| vehicle | vehicle | ||
| ibpBa | 2920.4 | 2409.3 | heat shock protein |
| recAb | 13.6 | 14.2 | DNA strand exchange and renaturation, DNA-dependent ATPase |
| ycfQ | 9.9 | 10.6 | repressor for bhsA |
| degP | 8.2 | 6.2 | periplasmic serine protease Do; heat shock protein HtrA |
| oxyR | 7.5 | 8.8 | activator, hydrogen peroxide-inducible genes |
| flu | 7.2 | 7.0 | outer membrane fluffing protein |
| rdoA | 6.6 | 6.6 | Thr/Ser kinase involved in Cpx stress response |
| flhE | 4.8 | 3.3 | flagellar protein flhE precursor |
| sat | 4.7 | 3.3 | secreted auto transporter toxin |
| hfq | 4.6 | 5.0 | host factor I for bacteriophage Q beta replication |
| flhB | 2.9 | 3.0 | putative part of export apparatus for flagellar proteins |
| sbmA | 2.6 | 2.5 | sensitivity to microcin B17, possibly envelop protein |
| dsbA | 2.3 | 3.1 | protein disulfide isomerase I |
| fepE | -18.5 | -14.6 | ferric enterobactin transport protein fepE |
| kpsC | -16.9 | -21.7 | KpsC protein |
| ompW | -16.4 | -13.5 | outer membrane protein W precursor |
| carA | -11.7 | -22.3 | carbamoyl-phosphate synthetase, glutamine |
| carB | -10.3 | -10.6 | carbamoyl-phosphate synthase large subunit |
| ompT | -10.1 | -9.2 | outer membrane protein 3b |
| chuT | -9.7 | -14.1 | putative periplasmic binding protein |
| iucA | -8.1 | -7.3 | IucA protein |
| serA | -7.7 | -3.9 | D-3-phosphoglycerate dehydrogenase |
| iucB | -7.1 | -6.3 | IucB protein |
| trpB | -6.9 | -13.4 | tryptophan synthase, beta protein |
| pyrD | -6.8 | -10.4 | dihydro-orotate dehydrogenase |
| fepG | -6.5 | -11.1 | ferric enterobactin transport protein |
| rfaL | -6.5 | -5.6 | O-antigen ligase |
| iucC | -6.3 | -6.1 | IucC protein |
| papX | -6.0 | -8.5 | PapX protein |
| flhD | -5.3 | -7.5 | regulator of flagellar biosynthesis |
| entA | -5.2 | -6.0 | 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase |
| fepC | -5.1 | -5.6 | ATP-binding component of ferric enterobactin transport |
| entB | -5.0 | -6.2 | 2,3-dihydro-2,3-dihydroxybenzoate synthetase |
| evgS | -4.6 | -6.4 | putative sensor for regulator EvgA |
| entE | -4.6 | -6.2 | 2,3-dihydroxybenzoate-AMP ligase |
| chuU | -4.0 | -4.8 | putative permease of iron compound ABC transport |
| chuA | -3.1 | -3.7 | outer membrane heme/hemoglobin receptor |
| csgE | -3.0 | -3.3 | curli production assembly/transport component |
| rfaP | -2.4 | -2.1 | lipopolysaccharide core biosynthesis |
Table 5. ESBL-producing E. coli genes associated with the functional category antibiotic resistance that are differentially expressed following exposure to CORM-2 (250 μM) versus vehicle (2.5% DMSO).
| Gene | Fold change | Fold change | Gene product |
|---|---|---|---|
| symbol | First exposure | 20x pre-exposed | |
| CORM-2 vs | CORM-2 vs | ||
| first exposure | 20x pre-exposed | ||
| vehicle | vehicle | ||
| marA | 43.9 | 31.4 | multiple antibiotic resistance transcriptional regulator |
| mdtA | 42.4 | 47.4 | multidrug efflux system, subunit A |
| marR | 37.8 | 33.1 | multiple antibiotic resistance protein; repressor of mar operon |
| marB | 29.5 | 21.0 | multiple antibiotic resistance protein |
| mdtB | 16.0 | 17.4 | multidrug efflux system, subunit B |
| acrD | 9.7 | 7.1 | aminoglycoside/multidrug efflux system |
| ECs1864 | 6.7 | 5.1 | multidrug-efflux transport protein |
| mdtC | 5.6 | 8.8 | multidrug efflux system, subunit C |
| hslJ | 3.8 | 4.1 | heat-inducible lipoprotein involved in novobiocin resistance |
| nfsA | 3.1 | 3.5 | nitroreductase A, modulator of drug activity A |
| gyrB | 2.4 | 2.2 | DNA gyrase subunit B, type II topoisomerase |
| rcsB | 2.0 | 2.0 | response regulator in two-component regulatory system with RcsC and YojN |
| evgAa | -38.9 | -25.0 | response regulator in two-component regulatory system with EvgS |
| mdtE | -5.3 | -4.2 | anaerobic multidrug efflux transporter |
| tehB | -2.5 | -2.8 | tellurite, selenium resistance protein |
n = 4
a also represented in defence, stress response or repair, Table 7
Table 6. ESBL-producing E. coli genes associated with the functional category biofilm that are differentially expressed following exposure to CORM-2 (250 μM) versus vehicle (2.5% DMSO).
| Gene | Fold change | Fold change | Gene product |
|---|---|---|---|
| symbol | First exposure | 20x pre-exposed | |
| CORM-2 vs | CORM-2 vs | ||
| first exposure | 20x pre-exposed | ||
| vehicle | vehicle | ||
| bhsA | 191.8 | 242.8 | biofilm, cell surface and signalling protein |
| ydeH | 93.7 | 100.2 | diguanylate cyclase, zinc-sensing |
| bssS | 19.7 | 31.3 | biofilm regulator |
| tqsA | 13.3 | 16.5 | pheromone autoinducer 2 |
| ybiJ | 5.8 | 5.8 | DUF1471 family putative periplasmic protein |
| yfgF | 5.3 | 4.6 | cyclic-di-GMP phosphodiesterase |
| yfaL | 2.4 | 2.4 | adhesin |
| artP | -5.1 | -7.1 | ATP-binding component of arginine transport system |
| bscB | -3.7 | -3.8 | regulator of cellulose synthase, cyclic di-GMP binding |
| artI | -2.9 | -3.1 | arginine transport system, periplasmic binding protein |
| csgF | -2.2 | -2.9 | curli production assembly/transport component |
n = 4
Table 7. ESBL-producing E. coli genes associated with defence, stress response or repair that are differentially expressed following exposure to CORM-2 (250 μM) versus vehicle (2.5% DMSO).
| Gene | Fold change | Fold change | Gene product |
|---|---|---|---|
| symbol | First exposure | 20x pre-exposed | |
| CORM-2 vs | CORM-2 vs | ||
| first exposure | 20x pre-exposed | ||
| vehicle | vehicle | ||
| ibpBa | 2920.4 | 2409.3 | heat shock protein |
| ibpA | 1424.7 | 1292.7 | heat shock chaperone |
| spy | 138.9 | 197.5 | periplasmic ATP-independent protein refolding chaperone |
| frmB | 86.5 | 48.5 | S-formylglutathione hydrolase |
| zraP | 71.5 | 158.7 | zinc resistance protein |
| soxS | 49.5 | 49.4 | superoxide response regulon transcriptional activator; autoregulator |
| pspB | 48.5 | 55.2 | psp operon transcription co-activator |
| pspC | 48.5 | 52.8 | psp operon transcription co-activator |
| pspG | 39.9 | 64.8 | phage shock protein G |
| htpG | 36.1 | 36.4 | protein refolding molecular co-chaperone Hsp90, heat-shock protein |
| yhcN | 29.9 | 27.2 | cadmium and peroxide resistance protein |
| clpB | 29.4 | 34.9 | protein disaggregation chaperone |
| dnaK | 29.2 | 27.2 | chaperone Hsp70; DNA biosynthesis |
| dnaJ | 28.5 | 21.0 | chaperone with DnaK; heat shock protein |
| htpX | 27.8 | 25.7 | heat shock protein, integral membrane protein |
| pspA | 18.0 | 21.6 | regulatory protein for phage-shock-protein operon |
| hslU | 14.8 | 13.9 | heat shock protein hslVU, ATPase subunit |
| norR | 13.7 | 12.5 | anaerobic nitric oxide reductase DNA-binding transcriptional activator |
| iscR | 10.0 | 10.7 | isc operon transcriptional repressor; suf operon transcriptional activator |
| grpE | 8.6 | 11.6 | heat shock protein |
| hslO | 8.4 | 6.4 | heat shock protein Hsp33 |
| rpoH | 8.3 | 10.4 | RNA polymerase, sigma |
| loiP | 7.9 | 9.0 | Phe-Phe periplasmic metalloprotease, OM lipoprotein |
| iscS | 7.2 | 4.9 | putative aminotransferase |
| groL | 6.9 | 7.6 | GroEL, chaperone Hsp60, peptide-dependent ATPase |
| groS | 4.6 | 4.4 | GroES, chaperone binds to Hsp60 |
| norW | 3.6 | 4.3 | NADH:flavorubredoxin oxidoreductase |
| hdeAb | -108.5 | -42.6 | stress response acid-resistance protein |
| evgAc | -38.9 | -25.0 | response regulator in two-component regulatory system with EvgS |
| gadB | -30.9 | -24.5 | glutamate decarboxylase B, PLP-dependent |
| gadX | -17.6 | -16.4 | acid resistance regulon transcriptional activator |
| cusFb | -16.6 | -8.6 | periplasmic copper- and silver-binding protein |
| gadA | -10.9 | -8.0 | glutamate decarboxylase A, PLP-dependent |
| aidB | -5.3 | -4.0 | DNA alkylation damage repair protein |
| katE | -4.2 | -4.3 | catalase HPII |
| katG | -2.2 | -2.6 | catalase HPI |
In order to confirm the microarray results, qPCR was carried out on five genes belonging to the functional category “antibiotic resistance” (marABR and mdtAB) and the two genes (hdeA and cusF) that showed significant differences between first-time and 20 times repeated exposure to CORM-2. In agreement with the microarray data, a marked up-regulation of marABR and mdtAB was found by qPCR (Table 8). Fold changes in mdtA expression was significantly higher after repeated exposure than after first-time exposure based on qPCR, which was not found in the microarray analysis. The microarray data showed that cusF and hdeA were significantly less repressed in cells after repeated exposure to CORM-2. qPCR data confirmed a repression of these genes but could not confirm a statistical difference between first-time and repeated exposure to CORM-2 (Table 8).
Table 8. Quantitative real-time PCR data for ESBL-producing E. coli genes following exposure to CORM-2 (250 μM) versus vehicle (2.5% DMSO).
| Gene | First exposure | 20x pre- exposed | Gene product |
|---|---|---|---|
| symbol | CORM-2 vs | CORM-2 vs | |
| first exposure | 20x pre-exposed | ||
| vehicle | vehicle | ||
| Fold change ± | Fold change ± | ||
| SEM | SEM | ||
| marA | 26 ± 4.5 | 30 ± 5.9 | multiple antibiotic resistance transcriptional regulator |
| marB | 24 ± 4.1 | 24 ± 4.9 | multiple antibiotic resistance protein |
| marR | 40 ± 3.8 | 41 ± 18 | multiple antibiotic resistance protein; repressor of mar operon |
| mdtAa | 240 ± 21 | 370 ± 59 | multidrug efflux system, subunit A |
| mdtB | 105 ± 32 | 170 ± 18 | multidrug efflux system, subunit B |
| cusF | -1.2 ± 1.4 | -0.37 ± 0.84 | periplasmic copper- and silver-binding protein |
| hdeA | -0.72 ± 1.0 | 0.24 ± 0.91 | stress response acid-resistance protein |
n = 3
a significant difference between first exposure and 20x pre-exposure
Alterations in gene expression specific for first-time exposure to CORM-2 or pre-exposure 20 times to CORM-2
Although the vast majority of the differentially expressed genes were common and found both in response to first-time exposure to CORM-2 and after pre-exposure 20 times to CORM-2, some specific changes were noted. Overall, the specific changes were modest with a fold change close to 2 (S5 Table).
Bacterial viability in response to repeated exposure to CORM-2
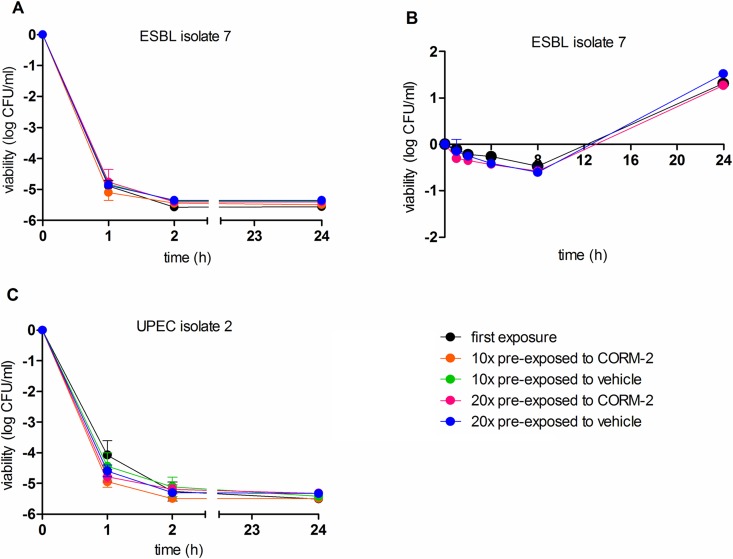
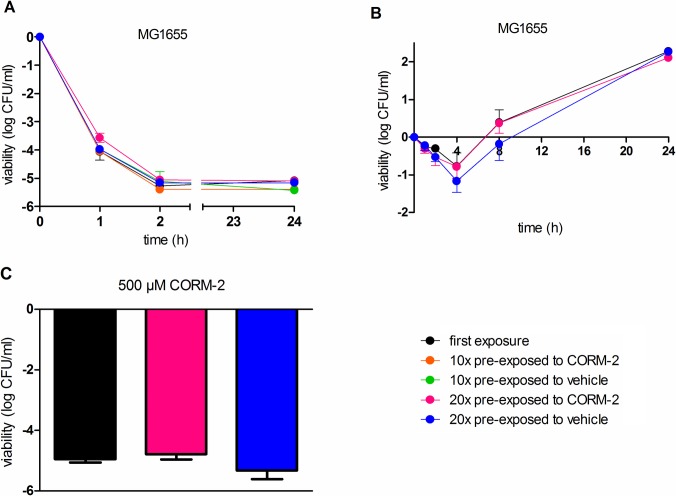
Bacterial viability studies were performed to compare the growth inhibitory effect of CORM-2 (500 μM) after first-time exposure with the inhibitory effect after pre-exposure 10 or 20 times to CORM-2 or vehicle. In the viability studies, three different bacterial isolates were used: ESBL7, a non ESBL-producing UPEC isolate (UPEC2) and a commensal E. coli K12 strain (MG1655). CORM-2 (500 μM) showed a fast bactericidal effect with a reduction of bacterial counts by 4–5 log units after 1 hour of exposure in all three isolates (Figs 3A, 3C and 4A). Growth inhibition peaked after 2 hours with no resumed growth during the 24-hour study period. Untreated controls showed an increased growth response of ~2 log units during the 24-hour study period (data not shown). The inhibitory effect of CORM-2 (500 μM) did not differ significantly between samples pre-exposed to CORM-2 or samples pre-exposed to vehicle. Neither were there any significant differences in response to CORM-2 (500 μM) between bacteria exposed once, 10 or 20 times to CORM-2 (Figs 3A, 3C and 4A). A sub-MIC concentration of CORM-2 (250 μM) was examined in ESBL7 and MG1655, showing a bacteriostatic response for 4–8 hours with a recovered growth after 24 hours (Figs 3B and 4B). No significant difference in viability was found between the first-time exposure and after pre-exposure 20 times to CORM-2. To study if the recovered growth observed 24 h after exposure to 250 μM CORM-2 was caused by survival of a resistant phenotype, the bacteria were immediately re-exposed to a higher concentration of CORM-2 (500 μM). However, the sensitivity to CORM-2 (500 μM) was not reduced or dependent on the previous exposures to CORM-2 (Fig 4C).
Fig 3. Effects of CORM-2 exposure on viability in ESBL isolate 7 and UPEC isolate 2.
A) ESBL7 and C) UPEC2 were grown to early log phase in MS-medium and then exposed to CORM-2 (500 μM) for 1, 2 or 24 h. B) ESBL7 were grown to early log phase in MS-broth and then exposed to CORM-2 (250 μM) for 1, 2, 4, 8 or 24 h. Data show viability after first-time exposure and after pre-exposure 10 or 20 times to CORM-2 (250 μM) or vehicle (2.5% DMSO). Viability is presented as log CFU/ml of CORM-2 exposed bacteria compared with the initial starting inoculum. The data are shown as mean ± SEM from three independent experiments.
Fig 4. Effect of CORM-2 exposure on viability in E. coli K12 strain MG1655.
MG1655 was grown to early log phase in MS-medium and then exposed to A) CORM-2 (500 μM) for 1, 2 or 24 h or to B) CORM-2 (250 μM) for 1, 2, 4, 8 or 24 h. C) Bacteria with a recovered growth after 24 h of exposure to 250 μM CORM-2 (see panel B) were re-exposed to a higher concentration of CORM-2 (500 μM) and the viability evaluated. Data show viability after first-time exposure and after pre-exposure 10 or 20 times to CORM-2 (250 μM) or vehicle (2.5% DMSO). Viability is presented as log CFU/ml of CORM-2 exposed bacteria compared with the initial starting inoculum. The data are shown as mean ± SEM from three independent experiments.
Effect of repeated exposure to CORM-2 on cefotaxime, ciprofloxacin and trimethoprim susceptibility
Determination of MIC values was performed using the broth dilution test. The MIC value for CORM-2 was determined to be 500 μM for all strains (ESBL7, UPEC2, MG1655) and MIC did not differ between first-time or repeated exposures (Table 9). Evaluation of MIC values was also performed to address whether repeated CORM-2 exposure affected the bacterial susceptibility to cefotaxime, ciprofloxacin and trimethoprim, antibiotics that are used to treat UTI. ESBL7 was resistant to cefotaxime and trimethoprim as expected, but the response to ciprofloxacin was indeterminate (Table 9). The MIC values for cefotaxime, ciprofloxacin and trimethoprim in ESBL7 did not differ between the first-time exposure and after repeated exposure to CORM-2 or vehicle. The isolates UPEC2 and MG1655 were sensitive to cefotaxime, ciprofloxacin and trimethoprim. The MIC values for cefotaxime and ciprofloxacin did not change after repeated exposure to CORM-2 in UPEC2 or MG1655 (Table 9). The MIC value for trimethoprim in isolate UPEC2 remained unchanged, but strain MG1655 showed a higher MIC value (1 μg/ml vs 0.5 μg/ml) for trimethoprim after pre-exposure 20 times to CORM-2 or vehicle (Table 9).
Table 9. MIC values for CORM-2, ciprofloxacin, cefotaxime and trimethoprim for ESBL-producing E. coli isolate 7, uropathogenic UPEC isolate 2 or non-pathogenic MG1655 in response to first-time exposure to CORM-2 and after 10 or 20 times pre-exposure to CORM-2 (250 μM) or vehicle (2.5% DMSO).
| Antibiotic susceptibility testing, MIC values | |||||
|---|---|---|---|---|---|
| First exposure CORM-2 | 10x CORM-2 | 10x vehicle | 20x CORM-2 | 20x vehicle | |
| CORM-2 (μM) | |||||
| ESBL7 | 500 | 500 | 500 | 500 | 500 |
| UPEC2 | 500 | 500 | 500 | 500 | 500 |
| MG1655 | 500 | 500 | 500 | 500 | 500 |
| Breakpointa - | |||||
| CIP (μg/ml) | |||||
| ESBL7 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| UPEC2 | 0.031 | 0.031 | 0.031 | 0.031 | 0.031 |
| MG1655 | 0.031 | 0.031 | 0.031 | 0.031 | 0.031 |
| Breakpointa 0.5/1 | |||||
| CTX (μg/ml) | |||||
| ESBL7 | >32 | >32 | >32 | >32 | >32 |
| UPEC2 | 0.062 | 0.062 | 0.062 | 0.062 | 0.062 |
| MG1655 | 0.062 | 0.062 | 0.062 | 0.062 | 0.062 |
| Breakpointa 1/2 | |||||
| TMP (μg/ml) | |||||
| ESBL7 | >32 | >32 | >32 | >32 | >32 |
| UPEC2 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| MG1655 | 0.5 | 0.5 | 0.5 | 1 | 1 |
| Breakpointa 2/4 | |||||
Abbreviations: ciprofloxacin (CIP), cefotaxime (CTX), trimethoprim (TMP)
a Clinical MIC breakpoint for Enterobacteriaceae set by the SRGA and the European Committee on Antimicrobial Susceptibility Testing (EUCAST). S, susceptibility/ R, resistant. -, No clinical breakpoint.
Effect of repeated exposure to CORM-2 on biofilm formation and motility
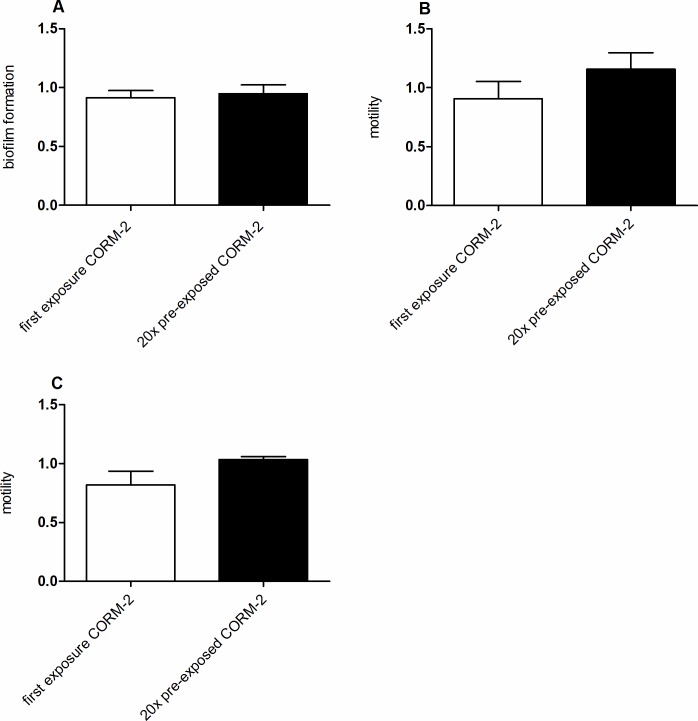
Many genes encoding biofilm were altered in response to CORM-2 and quantification of biofilm formation was performed in ESBL7 by the crystal violet method. The basal biofilm formation in ESBL7 was low (A540 ~ 0.1) and the effect of CORM-2 (250 μM) on biofilm formation was minor and not significantly different from the effect evoked by the vehicle (Fig 5A). Neither were there any significant differences in biofilm formation between bacteria exposed once or 20 times to CORM-2 (Fig 5A). Several genes encoding flagella were affected by CORM-2 and to determine whether changes in expression resulted in changed motility two motility assays were performed. The swimming motility assay measures flagella driven individual cell movement and the swarming motility assay the flagella driven multicellular surface movement. ESBL7 developed the typical colonial patterns associated with swimming and swarming migration (data not shown). The effect of CORM-2 (250 μM) per se on motility was not significantly different from the effect evoked by the vehicle. There were no significant differences in swimming (Fig 5B) or swarming (Fig 5C) motility between bacteria exposed once or 20 times to CORM-2.
Fig 5. The phenotypic effect of CORM-2 on biofilm formation and motility in ESBL isolate 7.
A) Biofilm formation measured after first-time exposure to CORM-2 and after pre-exposure 20 times to CORM-2 (250 μM). The biofilm formation is presented as relative changes compared to the formation evoked by the vehicle (2.5% DMSO). Motility measured on B) swimming plates and C) swarming plates after first-time exposure and after pre-exposure 20 times to CORM-2 (250 μM) or vehicle (2.5% DMSO). The motility data are presented as relative changes compared to the motility evoked by the vehicle (2.5% DMSO). Data are shown as mean ± SEM from three independent experiments.
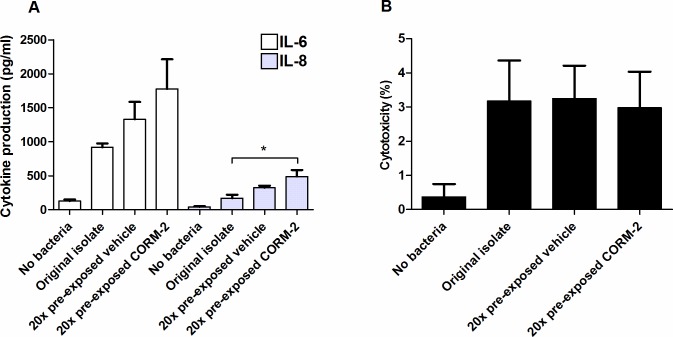
Effect of repeated exposure to CORM-2 on host renal cell production of cytokines
Uroepithelial cells contribute to the initiation of host defense against UPEC through the production of various cytokines and chemokines [22]. We next addressed whether repeated CORM-2 exposure affected the ability of EBL7 to evoke IL-6 and IL-8 production from host renal epithelial cells. The original ESBL7 isolate stimulated production of IL-6 and IL-8 compared to un-stimulated A498 cells (Fig 6A). The cytokine production was further increased in ESBL7 pre-exposed 20 times to CORM-2 or vehicle, but only significantly higher than the original isolate for IL-8 and CORM-2 pre-exposed bacteria (Fig 6A). The host renal cell cytotoxicity was low (~ 3%) for all experimental conditions (Fig 6B).
Fig 6. Host renal cell production of cytokines and cytotoxicity in response to ESBL isolate 7.
A) IL-6 and IL-8 production from A498 renal epithelial cells after stimulation for 6 h with the original ESBL7 isolate or with ESBL7 that have been pre-exposed 20 times to CORM-2 (250 μM) or vehicle (2.5% DMSO). B) Host renal cell cytotoxicity measured as LDH-release during the same conditions as in panel A and normalized to unstimulated and lysed control cells. Data are presented as mean ± SEM from three independent experiments. Asterisk denotes statistical significance (*p<0.05).
Discussion
Gene profiling of a multidrug-resistant ESBL-producing UPEC isolate demonstrated a significant alteration of a large number of genes after exposure to the CO-donor CORM-2. In all, close to 9% of the entities on the array were altered. However, this does not correspond to a fixed number of altered genes in the genome, since multiple entities sometimes represent the same gene. Our results are in agreement with a previous transcriptome analysis of non-pathogenic E. coli where ~9% of the total genome for anaerobically grown cells and ~4% for aerobically grown cells were altered in response to CORM-2 [12]. Thus, it appears to be an extensive flux in the transcriptome of the bacteria in order to cope with the altered environment induced by CORM-2 exposure.
The vast majority of the identified gene changes were common for bacteria exposed one time or repeatedly (20 times) to CORM-2. The enriched gene ontologies and pathway analysis stratified at the level common for both first-time exposed and repeatedly exposed samples showed that cellular responses and adaptions in metabolism genes are substantially affected. CORM-2 caused a general trend of down-regulation in energy metabolism, biosynthesis pathways, catabolism and up-regulation of the SOS response and DNA damage and repair mechanisms. A reduced fermentation is indicated by down-regulation of several genes, e.g., frdB and fumB [23], [24]. In E. coli, the reduced cofactor NADH plays a key role and without NADH reoxidation, cellular metabolism and growth are halted [25], [26]. Many genes coding for the NADH:ubiquinone oxidoreductase subunits (nuoABCEFGHIJMN) and trimethylamine N-oxide reductase (torACD) were repressed by CORM-2. A down-regulation of the nuo-operon (11 genes) by CORM-3 was also found in E. coli MG1655 [11], indicating that the NADH dehydrogenase complex is a target for CORMs. It is presumed that CO gas and CORMs will interact with quinol oxidase protein complexes of E. coli [27], but no significant changes in gene expression of cydAB (encoding cytochrome oxidase bd-I) or cyoABCDE (encoding cytochrome bo oxidase) were observed in our study. Previous transcriptomic analysis with E. coli and CORM-3, performed in defined medium under aerobic conditions, demonstrated down-regulation of cyoABCDE genes and a modest increase followed by a decrease in the cydAB genes [11], [13]. It is possible that the less defined growth conditions used in our study may explain the lack of altered expression of cytochrome genes.
There was a general trend of up-regulated genes in the enriched gene ontologies SOS response, cellular response to DNA damage stimulus, DNA repair and cellular response to stress. The SOS response is an extensive and effective response to DNA damage and the SOS response is regulated by LexA/RecA [28]. The induction of genes for recA and repressor lexA indicates an increased need for repair mechanisms in bacteria exposed to CORM-2. Many LexA regulated genes were induced (recN, recA, sulA, uvrD, umuC, umuD, polB), as were the damage-inducible genes dinI and dinB. DNA polymerase II, encoded by polB, is proposed to have a role in repair of oxidative damage and also to increase the rate of mutations during the SOS response [29]. Thus, another outcome of inducing the SOS response is increased genetic variability and the acquisition of bacterial mutations that may lead to resistance to some antibiotic drugs [30]. Moreover, bactericidal antibiotics may induce mutagenesis by stimulating the production of reactive oxygen species (ROS) [31]. Although controversial, the bactericidal cell death caused by CORM-2 may involve generation of intracellular ROS and a subsequent induction of DNA damage [32], [33].
CORM-2 has been shown to increase biofilm formation in an E. coli K12 strain [12]. BhsA is a small outer membrane protein involved in biofilm formation and stress response [34], and bhsA was induced ~200-fold by CORM-2 in our study. However, the overall biofilm formation, at least on plastic abiotic surface, was low in ESBL7 and the effect of CORM-2 on biofilm formation was minor. The reduced gene expression of artP and artI is in agreement with previous studies showing repression of these transport-encoding genes during biofilm formation [12], [35]. In agreement with previous results in an E. coli K12 strain [12], exposure to CORM-2 increased expression of several genes known to encode cytoplasmic adaptions and stress responses, like ibBA, ibpA and spy (approximately 3000-, 1500- and 140-fold increases, respectively). Notably, the fold changes of these genes were found to be considerably higher in our study. The spy gene is implicated in both zinc homeostasis and envelope stress responses [36], and spy has been identified as one of the main non-heme targets for CORM-3 [14]. Chaperone ibpA/B activities seem to promote disaggregation of protein aggregates [37], and ibpB has been associated with hyper-colonization of UPEC in the mouse urinary tract [38].
In the present study, a pathogenic E. coli strain was used which increases the clinical relevance of the acquired transcriptional data, including information of potential virulence genes, compared to previous studies using non-pathogenic E. coli K12 strains. Some virulence genes were up-regulated but the majority were down-regulated, like genes encoding products involved in iron transport and acquisition (fepEG, entABE). These genes have previously been reported to be up-regulated by CORM-3 exposure in an E. coli K12 strain [14]. However, there are known discrepancies when comparing transcriptomic data after CORM-2 and CORM-3 exposure and, in addition, different growth conditions, growth rates and exposure times between studies may influence the results [9]. We found a reduced expression of kpsC that is involved in group II capsule biosynthesis in UPEC. This gene is absent from the genome of E. coli K12 strains [39]. Interestingly, a previous study has shown that synthesis of extracellular polysaccharides, including group II capsular polysaccharide, is necessary for optimal urovirulence in the murine urinary tract [39]. Taken together, CORM-2 exposure appears to reduce the expression of many UPEC virulence factors. UPEC attachment to and/or invasion of epithelial cells are the initial steps in the pathogenesis of UTI [22]. UPEC employ multiple strategies to attenuate the initiation of the host response in order to evade the recruitment and activity of phagocytic neutrophils. Compared with commensal strains of E. coli, UPEC are able to suppress epithelial cytokine and chemokine production and many UPEC isolates elicit lower levels of IL-6 and IL-8 secretion from uroepithelial cells [40]. Our functional host renal cell experiments demonstrated an enhanced IL-8 production in response to bacteria repeatedly exposed to CORM-2, which may support the transcriptional data showing reduced expression of many virulence genes.
Overall, the alterations and fold-changes in gene expression were markedly consistent between bacteria exposed one time to CORM-2 or 20 times to CORM-2. Three genes hdeA, cusF and cusX were significantly less repressed after repeated exposure to CORM-2 compared with a single exposure. The hdeA gene encodes an HDEA protein that confers acid resistance [41] and it is possible that the intracellular pH homeostasis is altered in response to CORM-2 exposure due to impairment of the respiratory chain [20]. However, measurement of pH during bacterial growth before and during exposure to 250 μM CORM-2 for up to 4 h did not reveal any pH changes, at least not in the extracellular space (data not shown). CusCFBA is an efflux system protecting the periplasm from transition metal-mediated damage using a proton gradient [42], while the cusX gene encodes a hypothetical protein. qPCR analysis confirmed that hdeA and cusF were repressed by CORM-2, but no difference between single and repeated exposures was found by qPCR.
Several genes encoding efflux pump systems were altered by CORM-2. An induced expression of the genes mdtABC was found in agreement with a previous study performed in an E. coli K12 strain exposed to CORM-3 [14]. Further, an increased expression of marABR was found; marA is known to control expression of resistance to antibiotics like tetracycline, chloramphenicol and cephalosporins [43] and oxidative stress agents [44], by altering the expression of multiple genes on the bacterial chromosome. Consistent with microarray data, marABR and mdtAB were markedly upregulated by CORM-2 based on qPCR data. Many multidrug-resistant intestinal bacteria show an increased expression of genes for efflux pumps of the Resistance-Nodulation-cell Division (RND) family (as acrAB-TolC) involved in the reduction of antibiotic susceptibility [45]. In our study, the gene acrA was only induced in response to first-time exposure to CORM-2, while the gene acrD was induced both in first-time exposed and repeatedly exposed bacteria. The gene acrD encodes the efflux pump acrD which participates in aminoglycoside efflux [46]. However, it should be noted that the alterations in gene expression specific for first-time exposure or pre-exposure 20 times to CORM-2 were all small (approximately 2-fold), which indicates that these differences are rather uncertain and may be biological insignificant. Multidrug efflux pumps are known to confer low-level intrinsic resistance to drugs, and when mutations in regulatory genes appear, high expression levels of multidrug efflux pumps may interfere with therapeutic treatments [47]. Any new antibiotic seems to be favoured by not inducing overexpression of efflux pumps [48]. Thus, the CORM-2 evoked expression of genes for different efflux pumps may suggest a possibility for development of an antibiotic resistant phenotype. The finding that CORM-2 up-regulated genes for multidrug efflux pumps may support the notion that CORMs enter the bacteria through a specific, although yet unknown, transport mechanism [9]. However, CORMs may not per se activate the AcrAB-TolC multidrug efflux system since gene expression of efflux pumps is also enhanced by the SOS response [49].
The results regarding genes that encode flagella and fimbriae were not conclusive and both induced and reduced genes were noted. One of the genes exclusively induced by repeated exposure to CORM-2 was csgC, a gene encoding a potential curli assembly protein [50]. In previous E. coli K12 studies, an increased expression of the flagellar repressor lrhA was reported in response to CORM-2 [12], and expression of motility genes and functional motility was diminished in response CORM-3 [13]. RecA, one of the most up-regulated genes by CORM-2, promotes swarming motility in E. coli K12 by a yet unclear mechanism [21]. However, in our study, the changes in expression of genes responsible for flagella function did not correspond to any changes in functional cell motility, as evaluated by swimming and swarming motility assays.
Functional viability studies showed a bactericidal effect by 500 μM CORM-2 on ESBL7, the non-ESBL-producing UPEC isolate 2 and the non-pathogenic E. coli K12 strain MG1655, in agreement with previous studies [20], [16]. CORM-2, at 250 μM, caused a short-lasting growth inhibitory effect that was fully recovered after 24 h. The CORM-2-evoked growth inhibition achieved after first-time exposure was similar to the inhibition noted in samples pre-exposed 10 times or 20 times to CORM-2. Taken together, these data show that the growth inhibitory response was not attenuated after repeated exposure to CORM-2, neither in a multidrug-resistant E. coli strain nor in two antibiotic susceptible strains. In a previous study [16], we addressed whether multidrug-resistant ESBL-producing UPEC isolates were less sensitive to CORM-2 than non-pathogenic E. coli MG1655. However, no correlation between sensitivity towards CORM-2 and the pathogenic potential or antibiotic resistance of the strains was observed.
A comparison of MIC values for CORM-2 after first-time and repeated exposure to CORM-2 revealed no changes in MIC for any of the tested strains. Thus, the results obtained for CORM-2 by the antimicrobial susceptibility test supported the results from the viability studies. Increased gene expression of efflux pumps or increased mutation rates from repeated CORM-2 exposure could presumably lead to increased resistance towards other antibiotic classes. However, there were no indications of altered susceptibility (MIC values) for three classes of traditional UTI antibiotics (cefotaxime, trimethoprim or ciprofloxacin) after repeated exposure for 10 or 20 times to CORM-2. The non-pathogenic strain MG1655 showed slightly increased MIC values for trimethoprim after exposure for 20 times; however, this was also found for the DMSO vehicle. The use of DMSO in medium (0.1–10%) has been shown to cause reversion of sensitivity in E. coli strains [51] and it can therefore not be excluded that DMSO has affected the susceptibility. Moreover, an occasional two-fold difference in MIC values is expected in two-fold dilution assay. A limitation of the present study is that the applied protocol for repeated exposure to CORM-2 may not have been optimal to detect possible changes in susceptibility to CORM-2. A protocol with a gradual increase in the antibiotic concentration, starting from a very low sub-inhibitory concentration may allow sufficient time for mutations and selection [52]. In addition, extension of the experiments to include more generations may display resistance development [52]. Further studies with more extensive and different exposure protocols are certainly needed to fully evaluate development of resistance to CORMs.
Conclusions
This is the first study addressing the potential for bacteria to develop resistance to CORMs. Repeated exposure to CORM-2 did not change the gene expression patterns or fold changes and the viability studies showed a sustained phenotypic susceptibility to CORM-2. CORM-2 caused a pronounced activation of the potentially mutagenic SOS response and an increased expression of efflux pumps that may suggest that CORM-2 has a potential for resistance development. However, CORMs seem to be favoured by interactions with multiple and novel target sites that are different from those of traditional antibiotics and pre-existing resistance mechanisms. More comprehensive studies, including sequencing of the genome and mutation analysis, are needed to evaluate the likelihood for CORMs to develop resistance. Multidrug-resistance among uropathogenic bacteria is today prominent and the risk of treatment failure is an emerging threat and a public health concern. CORMs are interesting candidate molecules for development of new antibiotics to treat UTI. In addition, adjuvant treatment strategies where CORMs are combined with established antibiotics [15] are one interesting approach that should be further evaluated.
Supporting information
(DOCX)
Presented genes are derived from significant enrichment in the gene ontology fermentation. n = 4
(DOCX)
n = 4
(DOCX)
n = 4
(DOCX)
n = 4
(DOCX)
Acknowledgments
The authors would like to thank R.K. Poole for critically reading the manuscript.
Data Availability
All microarray data files are available from the Gene Expression Omnibus database (accession number GSE87627).
Funding Statement
We gratefully acknowledge the support from the Faculty of Medicine and Health at Örebro University and Nyckelfonden at Örebro University Hospital. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stamm WE. Scientific and clinical challenges in the management of urinary tract infections. The American journal of medicine. 2002;113 Suppl 1A:1S–4S. [DOI] [PubMed] [Google Scholar]
- 2.Song S, Lee EY, Koh EM, Ha HS, Jeong HJ, Bae IK, et al. Antibiotic resistance mechanisms of Escherichia coli Isolates from urinary specimens. The Korean journal of laboratory medicine. 2009;29(1):17–24. doi: 10.3343/kjlm.2009.29.1.17 [DOI] [PubMed] [Google Scholar]
- 3.Aryee A, Price N. Antimicrobial stewardship—can we afford to do without it? Br J Clin Pharmacol. 2015;79(2):173–81. doi: 10.1111/bcp.12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van de Sande-Bruinsma N, Grundmann H, Verloo D, Tiemersma E, Monen J, Goossens H, et al. Antimicrobial drug use and resistance in Europe. Emerg Infect Dis. 2008;14(11):1722–30. doi: 10.3201/eid1411.070467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez L, Hancock RE. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev. 2012;25(4):661–81. doi: 10.1128/CMR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406(6797):775–81. doi: 10.1038/35021219 [DOI] [PubMed] [Google Scholar]
- 7.Chin BY, Otterbein LE. Carbon monoxide is a poison… to microbes! CO as a bactericidal molecule. Curr Opin Pharmacol. 2009;9(4):490–500. doi: 10.1016/j.coph.2009.06.025 [DOI] [PubMed] [Google Scholar]
- 8.Ryter SW, Alam J, Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev. 2006;86(2):583–650. doi: 10.1152/physrev.00011.2005 [DOI] [PubMed] [Google Scholar]
- 9.Wareham LK, Poole RK, Tinajero-Trejo M. CO-releasing Metal Carbonyl Compounds as Antimicrobial Agents in the Post-antibiotic Era. J Biol Chem. 2015;290(31):18999–9007. doi: 10.1074/jbc.R115.642926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLean S, Mann BE, Poole RK. Sulfite species enhance carbon monoxide release from CO-releasing molecules: implications for the deoxymyoglobin assay of activity. Anal Biochem. 2012;427(1):36–40. doi: 10.1016/j.ab.2012.04.026 [DOI] [PubMed] [Google Scholar]
- 11.Davidge KS, Sanguinetti G, Yee CH, Cox AG, McLeod CW, Monk CE, et al. Carbon monoxide-releasing antibacterial molecules target respiration and global transcriptional regulators. J Biol Chem. 2009;284(7):4516–24. doi: 10.1074/jbc.M808210200 [DOI] [PubMed] [Google Scholar]
- 12.Nobre LS, Al-Shahrour F, Dopazo J, Saraiva LM. Exploring the antimicrobial action of a carbon monoxide-releasing compound through whole-genome transcription profiling of Escherichia coli. Microbiology. 2009;155(Pt 3):813–24. doi: 10.1099/mic.0.023911-0 [DOI] [PubMed] [Google Scholar]
- 13.McLean S, Begg R, Jesse HE, Mann BE, Sanguinetti G, Poole RK. Analysis of the bacterial response to Ru(CO)3Cl(Glycinate) (CORM-3) and the inactivated compound identifies the role played by the ruthenium compound and reveals sulfur-containing species as a major target of CORM-3 action. Antioxid Redox Signal. 2013;19(17):1999–2012. doi: 10.1089/ars.2012.5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson JL, Wareham LK, McLean S, Begg R, Greaves S, Mann BE, et al. CO-Releasing Molecules Have Nonheme Targets in Bacteria: Transcriptomic, Mathematical Modeling and Biochemical Analyses of CORM-3 [Ru(CO)3Cl(glycinate)] Actions on a Heme-Deficient Mutant of Escherichia coli. Antioxid Redox Signal. 2015;23(2):148–62. doi: 10.1089/ars.2014.6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wareham LK, Begg R, Jesse HE, Van Beilen JW, Ali S, Svistunenko D, et al. Carbon Monoxide Gas Is Not Inert, but Global, in Its Consequences for Bacterial Gene Expression, Iron Acquisition, and Antibiotic Resistance. Antioxid Redox Signal. 2016;24(17):1013–28. doi: 10.1089/ars.2015.6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bang CS, Kruse R, Demirel I, Onnberg A, Soderquist B, Persson K. Multiresistant uropathogenic extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli are susceptible to the carbon monoxide releasing molecule-2 (CORM-2). Microb Pathog. 2014;66:29–35. doi: 10.1016/j.micpath.2013.12.003 [DOI] [PubMed] [Google Scholar]
- 17.Silver LL, Bostian KA. Discovery and development of new antibiotics: the problem of antibiotic resistance. Antimicrob Agents Chemother. 1993;37(3):377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zacharia VM, Manzanillo PS, Nair VR, Marciano DK, Kinch LN, Grishin NV, et al. cor, a novel carbon monoxide resistance gene, is essential for Mycobacterium tuberculosis pathogenesis. MBio. 2013;4(6):e00721–13. doi: 10.1128/mBio.00721-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahlberg Bang C, Kruse R, Johansson K, Persson K. Carbon monoxide releasing molecule-2 (CORM-2) inhibits growth of multidrug-resistant uropathogenic Escherichia coli in biofilm and following host cell colonization. BMC microbiology. 2016;16:64 doi: 10.1186/s12866-016-0678-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nobre LS, Seixas JD, Romao CC, Saraiva LM. Antimicrobial action of carbon monoxide-releasing compounds. Antimicrob Agents Chemother. 2007;51(12):4303–7. doi: 10.1128/AAC.00802-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez-Gomez JM, Manfredi C, Alonso JC, Blazquez J. A novel role for RecA under non-stress: promotion of swarming motility in Escherichia coli K-12. BMC biology. 2007;5:14 doi: 10.1186/1741-7007-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ragnarsdottir B, Lutay N, Gronberg-Hernandez J, Koves B, Svanborg C. Genetics of innate immunity and UTI susceptibility. Nature reviews Urology. 2011;8(8):449–68. doi: 10.1038/nrurol.2011.100 [DOI] [PubMed] [Google Scholar]
- 23.Hirsch CA, Rasminsky M, Davis BD, Lin EC. A fumarate reductase in Escherichia coli distinct from succinate dehydrogenase J Biol Chem. 1963;238:3770–4. [PubMed] [Google Scholar]
- 24.van Vugt-Lussenburg BM, van der Weel L, Hagen WR, Hagedoorn PL. Biochemical similarities and differences between the catalytic [4Fe-4S] cluster containing fumarases FumA and FumB from Escherichia coli. PloS one. 2013;8(2):e55549 doi: 10.1371/journal.pone.0055549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonardo MR, Dailly Y, Clark DP. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol. 1996;178(20):6013–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holm AK, Blank LM, Oldiges M, Schmid A, Solem C, Jensen PR, et al. Metabolic and transcriptional response to cofactor perturbations in Escherichia coli. J Biol Chem. 2010;285(23):17498–506. doi: 10.1074/jbc.M109.095570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jesse HE, Nye TL, McLean S, Green J, Mann BE, Poole RK. Cytochrome bd-I in Escherichia coli is less sensitive than cytochromes bd-II or bo'' to inhibition by the carbon monoxide-releasing molecule, CORM-3: N-acetylcysteine reduces CO-RM uptake and inhibition of respiration. Biochimica et biophysica acta. 2013;1834(9):1693–703. doi: 10.1016/j.bbapap.2013.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley WL. Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol Microbiol. 2006;62(5):1228–38. doi: 10.1111/j.1365-2958.2006.05444.x [DOI] [PubMed] [Google Scholar]
- 29.Escarceller M, Hicks J, Gudmundsson G, Trump G, Touati D, Lovett S, et al. Involvement of Escherichia coli DNA polymerase II in response to oxidative damage and adaptive mutation. J Bacteriol. 1994;176(20):6221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foster PL. Stress-induced mutagenesis in bacteria. Critical reviews in biochemistry and molecular biology. 2007;42(5):373–97. doi: 10.1080/10409230701648494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Molecular cell. 2010;37(3):311–20. doi: 10.1016/j.molcel.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tavares AF, Teixeira M, Romao CC, Seixas JD, Nobre LS, Saraiva LM. Reactive oxygen species mediate bactericidal killing elicited by carbon monoxide-releasing molecules. J Biol Chem. 2011;286(30):26708–17. doi: 10.1074/jbc.M111.255752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nobre LS, Jeremias H, Romao CC, Saraiva LM. Examining the antimicrobial activity and toxicity to animal cells of different types of CO-releasing molecules. Dalton transactions (Cambridge, England: 2003). 2016;45(4):1455–66. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XS, Garcia-Contreras R, Wood TK. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J Bacteriol. 2007;189(8):3051–62. doi: 10.1128/JB.01832-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Gene expression in Escherichia coli biofilms. Applied microbiology and biotechnology. 2004;64(4):515–24. doi: 10.1007/s00253-003-1517-y [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto K, Ogasawara H, Ishihama A. Involvement of multiple transcription factors for metal-induced spy gene expression in Escherichia coli. Journal of biotechnology. 2008;133(2):196–200. doi: 10.1016/j.jbiotec.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 37.Ratajczak E, Zietkiewicz S, Liberek K. Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. Journal of molecular biology. 2009;386(1):178–89. doi: 10.1016/j.jmb.2008.12.009 [DOI] [PubMed] [Google Scholar]
- 38.Haugen BJ, Pellett S, Redford P, Hamilton HL, Roesch PL, Welch RA. In vivo gene expression analysis identifies genes required for enhanced colonization of the mouse urinary tract by uropathogenic Escherichia coli strain CFT073 dsdA. Infect Immun. 2007;75(1):278–89. doi: 10.1128/IAI.01319-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bahrani-Mougeot FK, Buckles EL, Lockatell CV, Hebel JR, Johnson DE, Tang CM, et al. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol Microbiol. 2002;45(4):1079–93. [DOI] [PubMed] [Google Scholar]
- 40.Olson PD, Hunstad DA. Subversion of Host Innate Immunity by Uropathogenic Escherichia coli. Pathogens (Basel, Switzerland). 2016;5(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gajiwala KS, Burley SK. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. Journal of molecular biology. 2000;295(3):605–12. doi: 10.1006/jmbi.1999.3347 [DOI] [PubMed] [Google Scholar]
- 42.Bagai I, Rensing C, Blackburn NJ, McEvoy MM. Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry. 2008;47(44):11408–14. doi: 10.1021/bi801638m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hachler H, Cohen SP, Levy SB. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1991;173(17):5532–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alekshun MN, Levy SB. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends in microbiology. 1999;7(10):410–3. [DOI] [PubMed] [Google Scholar]
- 45.Nikaido H, Pages JM. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev. 2012;36(2):340–63. doi: 10.1111/j.1574-6976.2011.00290.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberg EY, Ma D, Nikaido H. AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol. 2000;182(6):1754–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Misra R, Bavro VN. Assembly and transport mechanism of tripartite drug efflux systems. Biochimica et biophysica acta. 2009;1794(5):817–25. doi: 10.1016/j.bbapap.2009.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Bambeke F, Glupczynski Y, Plesiat P, Pechere JC, Tulkens PM. Antibiotic efflux pumps in prokaryotic cells: occurrence, impact on resistance and strategies for the future of antimicrobial therapy. J Antimicrob Chemother. 2003;51(5):1055–65. doi: 10.1093/jac/dkg224 [DOI] [PubMed] [Google Scholar]
- 49.Demple B. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene. 1996;179(1):53–7. [DOI] [PubMed] [Google Scholar]
- 50.Taylor JD, Zhou Y, Salgado PS, Patwardhan A, McGuffie M, Pape T, et al. Atomic resolution insights into curli fiber biogenesis. Structure (London, England: 1993). 2011;19(9):1307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szydlowska T. In vitro and in vivo studies on the role of dimethylsulfoxide (DMSO) in resensibilization of bacterial strains resistant to antibiotics and chemotherapeutic agents. Zentralbl Bakteriol Orig A. 1977;239(2):270–4. [PubMed] [Google Scholar]
- 52.Hein-Kristensen L, Franzyk H, Holch A, Gram L. Adaptive evolution of Escherichia coli to an alpha-peptide/beta-peptoid peptidomimetic induces stable resistance. PloS one. 2013;8(9):e73620 doi: 10.1371/journal.pone.0073620 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Presented genes are derived from significant enrichment in the gene ontology fermentation. n = 4
(DOCX)
n = 4
(DOCX)
n = 4
(DOCX)
n = 4
(DOCX)
Data Availability Statement
All microarray data files are available from the Gene Expression Omnibus database (accession number GSE87627).