Abstract
The CONSTANS/FLOWERING LOCUS T (CO/FT) regulon plays a central role in the control of flowering time in photoperiod-sensitive plants. Flowering time in wild cotton (Gossypium spp.) has strict photoperiod sensitivity, but domesticated cotton is day-neutral. Information on the molecular characterization of the CO and CO-like (COL) genes in cotton is very limited. In this study, we identified 42 COL homologs (GhCOLs) in the G. hirsutum genome, and many of them were previously unreported. We studied their chromosome distribution, phylogenetic relationships, and structures of genes and proteins. Our results showed that GhCOLs were classified into three groups, and 14 COLs in group I showed conserved structure when compared with other plants. Two homoeologous pairs, GhCOL1-A and GhCOL1-D in Group I, showed the highest sequence similarity to Arabidopsis thaliana CO and rice CO homologous gene Heading date1 (Hd1). Tissue-specific expression showed that 42 GhCOL genes may function as tissue-specific regulators in different cells or organs. We cloned and sequenced the 14 GhCOL genes in Group I related to flowering induction to study their diurnal expression pattern, and found that their expression showed distinct circadian regulation. Most of them peaked at dawn and decreased rapidly to their minima at dusk, then started to accumulate until following dawn under long- or short-day conditions. Transgenic study in the Arabidopsis co-2 mutant demonstrated that GhCOL1-A and GhCOL1-D fully rescued the late-flowering phenotype, whereas GhCOL3-A, GhCOL3-D, GhCOL7-A, and GhCOL7-D partially rescued the late-flowering phenotype, and the other five homoeologous pairs in Group I did not promote flowering. These results indicate that GhCOL1-A and GhCOL1-D were potential flowering inducers, and are candidate genes for research in flowering regulation in cotton.
Introduction
Seasonal and diurnal variations of day length in nature are consistent from year to year. Many plants perceive photoperiodic information to predict upcoming environmental changes and precisely regulate flowering time in favorable conditions [1]. In plants, the circadian clock regulates a wide range of biological processes and represents the plant’s endogenous timekeeper. Two proteins, CONSTANS (CO) and FLOWERING LOCUS T (FT), are the central integrator of the photoperiod pathway in Arabidopsis thaliana [2]. AtCO induces the expression of FT in the leaf under long-day (LD) inductive conditions [1,3,4]. In rice, heading date 1 (Hd1, the CO ortholog) promotes heading date 3a (Hd3a, the FT ortholog) expression under short-day (SD) conditions, but inhibits Hd3a expression under non-inductive LD conditions [5]. Many studies have shown that flowering time is governed by the CO/FT module which is highly conserved among photoperiod-sensitive plants although its action models are inconsistent in different species [6–8]. CO encodes a putative B-box zinc finger transcription factor unique to plants and mediates between the circadian clock and the flowering time control [9–11]. High CO levels activate the expression of FT, which encodes a member of the phosphatidylethanolamine-binding protein that is a major component of florigen [3,4,12].
It has been documented that the accumulation of CO mRNA and CO protein is regulated at the transcriptional and posttranslational level through a number of proteins. Cycling of CO mRNA is regulated transcriptionally through circadian clock-regulated components, such as GIGANTIA (GI), CYCLING DOF FACTORS (CDFs), and the F-box protein FLAVIN BINDING, KELCHREPEAT (FKF1) [13–17]. The GI-FKF1 complex modulates CO protein stability, which degrades a family of CO repressors, the CDFs, resulting in maximum CO transcription at the end of the day [11,18]. Plants can perceive specific light quality by multiple photoreceptors to trigger posttranslational regulation of CO protein. In the early morning under LD conditions, the red-light receptor phytochrome B (PHYB) promotes degradation of CO protein and plays a major role in the regulation early in the day [19,20]. The E3 ubiquitin ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1) that physically interacts with CO is involved in the red light-mediated degradation of CO that occurs early in the daylight period [21,22]. In the evening, blue light prevents CO proteolysis by CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) [19,23,24]. The far-red receptor phytochrome (PHYA) and the blue-light receptors Cryptochrome 1 (CRY1) and CRY2 stabilize CO protein toward the end of the day through inhibition of proteasome-dependent CO degradation [25,26,27].
CONSTANS-like (COL) proteins in this family are characterized by the presence of one or two zinc finger B-box domains at the N-terminus or a C-terminal CCT (CO, CO-like, and TOC1) domain [10]. The COL gene family in both monocots and dicots has many members, for example 17 in Arabidopsis [28], 16 in rice [29], 9 in barley [30], 10 in sugar beet [31], 11 in Medicago [32], 26 in soybean [33], 25 in Chinese cabbage [34], 11 in Chrysanthemum lavandulifolium [35], 6 in ramie [36], and 25 in banana [37]. Phylogenetic analysis divided COL proteins in plants into three major groups [30]. Group I COLs contain two B-box domains, one CCT domain, and an additional VP motif (valine-proline motif involved in the interaction with COP1). Group II COLs contain only one B-box and a CCT domain. Group III have one full B-box, a second diverged zinc finger, and a CCT domain [30,38,39].
The cotton genus (Gossypium) contains approximately 50 species and five allopolyploid species [40]. Wild cotton species are perennial plants and mostly SD-photoperiodic, with a diversity of architecture and flowering time. However, domesticated cotton species underwent extensive artificial selection and gradually lost their photoperiodic sensitivity. Upland cotton (G. hirsutum L.) is the most extensively cultivated Gossypium species and numerous elite types have been bred successfully, which have been widely grown in more than 80 countries and account for more than 95% of commercial cotton production worldwide [41,42]. However, the molecular mechanisms regulating the transition from vegetative to reproductive growth in cotton are less well characterized than in other plant species, mostly due to the complexity of the cotton genome and scarcity of cotton flowering time mutants. Zhang et al. [43] reported identification of 23 putative COL genes in G. raimondii based on its genome sequence data. They studied their structures, phylogenetic relationships, and molecular evolution, and found that COL1, COL2, and COL8 experienced greater selective pressures during the domestication process [43]. To date, information on the numbers and characterizations of COL genes in G. hirsutum is not clear. However, successful sequencing of the G. hirsutum genome provides a valuable resource for genome evolution, fiber improvement, and gene identification [44,45]. Because of the lack of good information on the numbers and characterizations of COL genes in G. hirsutum, we aim to characterize COL family members in G. hirsutum using its genome sequence data. We identified and characterized 42 GhCOL genes and their chromosomal distribution, phylogenetic relationship, gene structure, conserved motif, and tissue specificity expression profiles. Additionally, we focused on the 14 GhCOLs in Group I–which has been characterized in many plant species–and this cluster with AtCO and rice Hd1. We respectively examined the diurnal expression of the 14 GhCOLs under LD or SD conditions. We further performed complement experiments to analyze their putative functions in the flowering signal pathway. Our results support the conclusion that GhCOL1-A and GhCOL1-D homoeologs may be the key inducers of flowering in cotton. Our data also provide a broader understanding of the COL gene family in upland cotton.
Materials and methods
Plant material and growth conditions
Cotton seeds (G. hirsutum L. cv. XLZ 42) were field-grown under natural conditions during the summer of 2015 in Shihezi (Xinjiang, China). The seeds of Arabidopsis ecotype Ler and mutant co-2 (in the Ler background) obtained from the Arabidopsis Biology Resources Center (ABRC, Columbus, OH, USA) were surface sterilized for 20 min with 2.8% sodium hypochlorite solution containing 0.1% surfactant (Triton X-100; Sigma-Aldrich, Munich, Germany) and rinsed several times with sterile water. The sterilized seeds were stratified for 3 d at 4°C in darkness and then plated on Petri dishes with half-strength Murashige-Skoog (MS) salt mixture (pH 5.7; Duchefa, Haarlem, the Netherlands), 1% (w/v) sucrose, and 0.8% (w/v) agar. Petri dishes were then placed in a phytotron at 22°C for 10 d under LD conditions (16 h light/8 h dark), and the seedlings were transplanted into pots containing peat soil and vermiculite (1:1) and kept in a growth chamber with a 16-h photoperiod. The light intensity for Arabidopsis growth was 200 μmol m–2 s–1.
For tissue expression analysis, roots, stems, leaves, and shoot apical meristems (SAM) were collected at the third true-leaf expanding stage (approximately 20 d after planting). During the cotton flowering period, tissues of sepals and petals were collected at 0 d of anthesis (DOA), and fibers were sampled at 15 d post-anthesis (DPA). For diurnal rhythmic expression analyses, the plants were grown in a 25°C chamber in LD and SD conditions (8 h light/16 h dark photoperiod) with 150 μmol m–2 s–1 of light intensity, respectively. The third true leaves were sampled every 4 h at 13 different time points from zeitgeber time (ZT) 0 h for 2 d. For gene expression analyses, the fresh leaves of 20 d Ler, co-2 and all the transgenic lines were sampled under LD conditions. All samples were frozen immediately in liquid nitrogen and stored at –80°C.
Identification of COL family genes from G. hirsutum
In an effort to identify all COL family genes in the upland cotton genome, a batch Basic Local Alignment Search Tool (BLAST) search was performed against the G. hirsutum genome (v1.1) [45] downloaded from CottonGen (https://www.cottongen.org/) using the full-length amino acid sequences of Arabidopsis CO and G. raimondii COLs [43] as queries with an E-value cut of 1 × 10−15. All retrieved proteins were then submitted to PFAM (http://pfam.xfam.org/) databases for annotating of the domain structure. Only candidates encoding both one or two zinc-binding B-box domains at the N-terminus and a CCT domain at the C terminus were regarded as “true” G. hirsutum COs (GhCOLs). The Blast search continued until no more new COL homologs were matched. As a result, 42 genomic sequences of GhCOLs were obtained. We found that GhCOL1-A and GhCOL8-A genes were not annotated in the G. hirsutum genome (v1.1) [45], whereas their homoeologous GhCOL1-D and GhCOL8-D genes were. Therefore, we amplified the coding sequences of GhCOL1-A and GhCOL8-A by PCR using gene-specific primers based on the GhCOL1-D and GhCOL8-D sequences. The detailed information of the upland cotton COL genes was supplied in S1 Table. We found that no sequences of GhCOL2-A, GhCOL2-D, GhCOL18-D, and GhCOL23-D were annotated in the G. hirsutum genome database, and so these four GhCOL genes were not identified.
Chromosomal mapping and phylogenetic analysis
Chromosomal position and gene structure information of GhCOLs were obtained from G. hirsutum gene annotation (v1.1) [45], and these putative COL genes were mapped on the corresponding At (‘t’ indicates tetraploid) or Dt chromosomes using the MapInspect software (http://mapinspect.software.informer.com/). In total, 122 COL homologs (S2 Table), including 16 Arabidopsis COLs, 14 rice COLs, 26 soybean COLs, 23 G. raimondii COLs, and 42 GhCOLs were used to construct a phylogenetic tree. Multiple sequence alignments were performed by ClustalW [46] under default parameters with a gap opening penalty of 10 and gap extension penalty of 0.2. MEGA5.1 [47] was used to make a phylogeny reconstruction analysis using the Neighbor-Joining (NJ) method and Poisson correction distance model. The bootstrap analysis was performed to estimate nodal support on the basis of 1000 re-samplings.
Gene structure and protein profile analysis
Gene exon–intron structure information for GhCOLs was retrieved from G. hirsutum gene annotation (v1.1) [45], and a gene structure schematic diagram was drawn using the Gene Structure Display Server [48]. Protein length, molecular weight, and isoelectric point of GhCOLs were analyzed using Lasergene v7.1 software (http://www.dnastar.com/) with default parameters. Protein subcellular localization was predicted by WoLF PSORT (www.genscript.com/wolf-psort.html).
RNA preparation, cDNA synthesis, and qRT-PCR analyses
Total RNA was isolated using the RNAprep pure Plant Kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. The quality and quantity of each RNA sample were determined using gel electrophoresis and a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The cDNA synthesis reactions were performed using the Superscript First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions with 1 μg of total RNA per reaction used as a template.
Quantitative real-time PCR (qRT-PCR) was carried out on an Applied Biosystems 7500 Fast Real-Time PCR System (Life Technologies, Foster City, CA, USA) in a 25-μl volume containing 10 ng of cDNA, 5 pM of each primer, and 25 μl of Fast SYBR Green Master Mixture (CWBIO, Beijing, China) according to the manufacturer’s protocol. The PCR conditions were as follows: primary denaturation at 95°C for 20 s followed by 40 amplification cycles of 3 s at 95°C, and 30 s at 60°C. Melting curve analysis was performed to ensure there was no primer-dimer formation. Amplicons were also loaded on a 2% agarose gel for visual inspection. Primer information of qRT-PCR for gene expression analysis and gene cloning used in this study is listed in S3 Table. The nucleotide sequences of GhCOLs in Group I marked with primer location for qRT-PCR were shown in S1 Fig. Three replicate assays were performed with independently isolated RNAs, and each RT reaction was loaded in triplicate. Relative expression levels of each GhCOL gene are presented using the 2–ΔCt method [49].
The heatmap of tissue expression for all GhCOLs was performed as described by Deng et al. [50]. All the 2–ΔCt values calculated from Ct data by qRT-PCR in different tissues, including root, stem, true leaf, flower, sepal, SAM, and fiber were saved in a Microsoft Excel spreadsheet (.xls). This file can be loaded into a Heat map Illustrator tool named HemI 1.0 (http://hemi.biocuckoo.org/) for visualizing a heatmap of gene expression. Given a selected color scale, the total color space will be automatically processed into a numerical matrix. HemI project contains all information needed to draw a heatmap and will generate a heatmap after loaded. Last, a publication-quality heatmap of gene expression can be exported directly.
Cloning of GhCOL genes in Group I and transformation of Arabidopsis
The complete open reading frame cDNAs for seven pairs of homoeologs in Group I were obtained from G. hirsutum cv. XLZ42 by PCR amplification using gene-specific primers designed according to the putative A- or D-homoeologous sequences in G. hirsutum genome database (S3 Table), and then subcloned into the pMD-19 vector (TaKaRa, Dalian, China) following the manufacturer’s instructions. Several independent clones for each COL gene were sequenced for validation of A- or D-homoeologous COLs by comparing sequences with TM-1 genome. Finally, 14 coding sequences of COLs were separately transferred into the overexpression binary vector pCAMBIA 2300-35S-OCS [51] to construct 35S:GhCOLs. The later flowering Arabidopsis co-2 mutant plants were separately infected with Agrobacterium tumefaciens strain GV3101 transformed with the obtained 35S:GhCOLs clones using the floral dip method [52]. Transgenic plants were selected on half-strength MS culture medium containing 50 μg/ml kanamycin. Homozygotes were replanted and subsequently monitored for flowering using non-transgenic wild type seedlings as controls. Flowering time was recorded as the number of rosette leaves per plant at the time the first flower bloomed from at least 20 individuals for each T3 lines and control [53]. Statistical analysis of the number of rosette leaves was performed using Student’s t-test.
Results
Identification and chromosomal distribution of COL family genes in upland cotton
To identity the COL family genes in the upland cotton genome, we carried out a genome-wide analysis of the putative GhCOL genes in the TM-1 genome database. We obtained 42 putative genomic sequences of upland COL homologs, and each GhCOL was then assigned a name based on its similarity level to Arabidopsis CO and COLs (S1 Table), with a designation of A or D for A- or D-subgenome chromosome. The methods of classification and nomenclature for G. hirsutum GhCOLs in our study were consistent with G. raimondii COLs [43].
Subcellular localization prediction showed that most GhCOL proteins mainly located in nucleus or both nucleus and cytoplasm, which are correlated to their functions as transcription factors (S1 Table). However, GhCOL3, GhCOL5 and GhCOL6 homoeologous protein located only in cytoplasm, and GhCOL17 homoeologs located only in chloroplast.
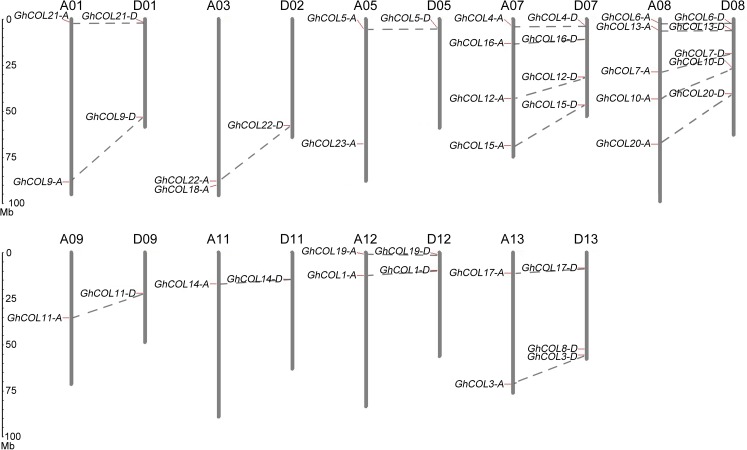
Chromosome mapping reveals that the 42 GhCOLs were not evenly distributed on the 18 chromosomes (Fig 1). There were 1–5 genes on each chromosome: one gene on chromosomes D02, D05, A09, D09, A11, and D11; two genes on chromosomes A01, D01, A03, A05, A12, D12, and A13; three genes on chromosome D13; four genes on chromosomes A 07 and D 07; and five genes on chromosomes A 08 and D 08. The distribution ratio for each chromosome was in the range of 2.38–11.91%.
Fig 1. Chromosomal distributions of the identified CONSTANS-like (COL) genes in upland cotton (G. hirsutum acc. TM-1).
Chromosomal locations were shown from top to bottom on corresponding chromosomes according to G. hirsutum genome (v1.1) annotation [45]. Duplicated gene pairs were linked by dotted lines.
Phylogenetic tree, gene structure, and conserved motif analyses of GhCOLs
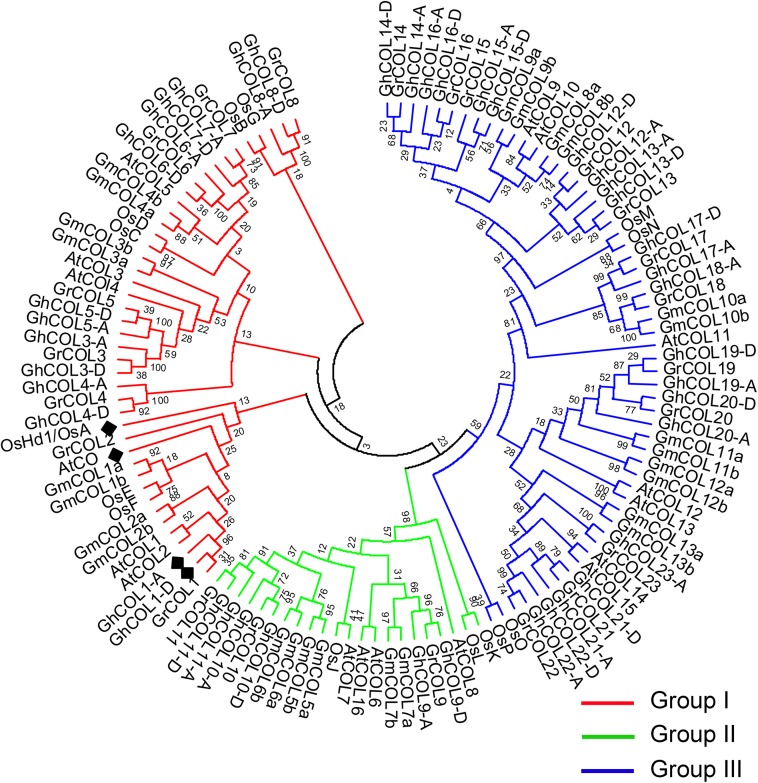
To investigate the phylogenetic relationships among COL family genes, we constructed a NJ phylogenetic tree with 122 COL protein amino-acid sequences retrieved from Arabidopsis, rice, soybean, and cotton databases based on multiple alignment analyses. These COL homologs were classified into three major clades, and cotton COLs were divided into Group I–III (Fig 2), consistent with results for diploid cotton (G. raimondii) [43]. Of the 42 GhCOLs in G. hirsutum, 14 genes were in Group I, six were in Group II, and the remaining 22 were in Group III. Among the 14 GhCOLs in Group I, the homoeologs of GhCOL1-A and GhCOL1-D had 98.7% amino acid sequence similarity and were clustered with the known functional flowering inducers, Arabidopsis CO [10] and rice Hd1 [29].
Fig 2.
A Neighbor Joining phylogenetic analysis of the CO and COLs family from Arabidopsis (At), rice (Os), soybean (Gm), G. raimondii (Gr) and G. hirsutum (Gh). Multiple alignments were generated using ClustalW [46]. The phylogenetic tree was constructed using MEGA5.1 [47]. Bootstrap values for 1,000 re-samplings were shown on each branch. The 122 CO homologs from five plant species were identified by homology searches in GenBank database using Arabidopsis CO and COL proteins as entry. The clades were divided into three groups, branches of which were marked in differently colors. AtCO, OsHd1, GhCOL1-A and GhCOL1-D were indicated using a black prism.
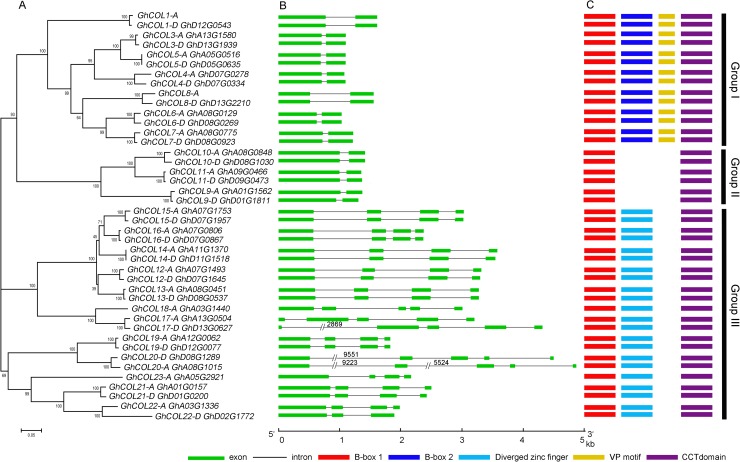
Phylogenetic analysis of 42 GhCOL proteins showed that cotton GhCOLs were categorized into three groups more obviously (Fig 3A). We analyzed the genome structure of the 42 GhCOL genes by aligning the genomic and cDNA sequences (Fig 3B). The 14 genes in Group I and six in Group II were highly conserved, containing two exons and one intron, and their full-length genomic DNA sequences ranged from 1,031 bp (GhCOL6-D) to 1,611 bp (GhCOL1-D). Of the 14 COLs in Group I, the intron lengths in GhCOL1-D and GhCOL8-D were obviously longer than in other family members, whereas exon I in COL8-D was shorter than others, leading to variation in gene length. However, 17 genes in Group III had different gene structure. GhCOL17-A, GhCOL17-D, GhCOL18-A, GhCOL20-A, and GhCOL20-D contained five exons, while the other 12 genes contained four exons. The genome lengths of these 17 genes ranged from 1,824 bp (GhCOL19-A and GhCOL19-A) to 5,613 bp (GhCOL17-D) except for GhCOL20-A and GhCOL20-D which had an abnormally-sized first intron.
Fig 3. Phylogenetic relationships and structures of GhCOL genes and GhCOL proteins.
(A) A Neighbor-Joining (NJ) phylogenetic tree of the 42 COL homologs from G. hirsutum was constructed using MEGA5.1 [47]. The bootstrap consensus tree was inferred from 1,000 replicates. (B) The gene structures were drawn using the Gene Structure Display Server [48]. Green boxes and black lines were exonic and intronic regions, respectively. (C) The domain structure of GhCOL proteins. Colorful boxes indicated B-box 1, B-box 2, diverged zinc finger, VP motif and CCT domain, respectively.
The annotation of the domain structure and multiple alignments of amino acid sequences showed that the COLs of Group I contained one B-box 1, one B-box 2, one VP motif, and one CCT domain. However, B-box 1 in GhCOL6-A and GhCOL6-D, and B-box 1 and the VP motif in GhCOL8-A and GhCOL8-D, were incomplete. Six COLs in Group II had one B-box 1 and one CCT domain, whereas the remaining 22 COLs in Group III contained one B-box 1, one diverged zinc finger, and one CCT domain (Fig 3C and S2 Fig).
Tissue-specific expression patterns of GhCOLs in upland cotton
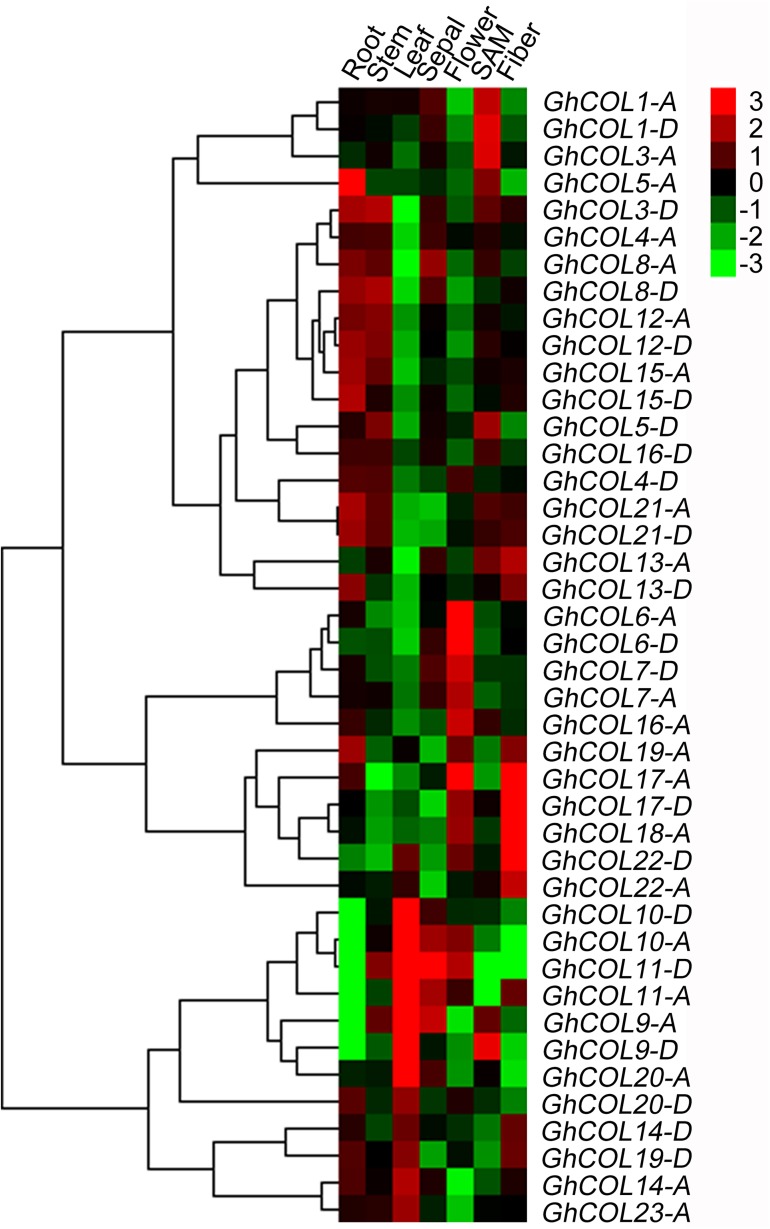
To understand the temporal and spatial transcriptional patterns of GhCOLs, we first analyzed their transcriptional levels in different tissues, including root, stem, true leaf, flower, sepal, SAM, and fiber using qRT-PCR. There were 42 GhCOLs expressed in various tissues with different expression levels (Fig 4). The expression patterns of GhCOLs were not consistent with their phylogenetic relationship of clustering into Group I–III (Fig 2). GhCOL5-A, GhCOL12-D, GhCOL15-A/D, and GhCOL21-A/D were mainly expressed in roots. GhCOL3-D, GhCOL5-D, GhCOL8-D, and GhCOL12-A/D were mainly expressed in stems. GhCOL9-A/D, GhCOL10-A/D, GhCOL11-A/D, and GhCOL20-A were predominantly expressed in leaves; and GhCOL14-A/D, GhCOL19-D, GhCOL20-D, and GhCOL23-A were also expressed in leaves with low expression. GhCOL9-A and GhCOL11-D were expressed significantly in sepals, and GhCOL9-D was also highly expressed in SAM. The GhCOL6 and GhCOL7 homoeologs, GhCOL16-A, and GhCOL17-A, were highly expressed in flowers, and GhCOL17-A was also highly expressed in fibers. The highest expressions of GhCOL1 homoeologs and GhCOL3-A were only in SAM. GhCOL17 homoeologs, GhCOL18-A and GhCOL22-D, were highly expressed in fibers. GhCOL4 homoeologs and GhCOL16-D showed very low expression in various tissues. Our results showed that GhCOLs had specific transcript accumulation in seven different tissues, suggesting that they may function as tissue-specific regulators in different cells or organs of cotton.
Fig 4. Heat map of GhCOL genes expression profiles in cotton different tissues.
Quantitative real time-PCR (qRT-PCR) was used to analyze the relative expression levels of 42 GhCOL genes in various tissues, and cotton UBQ7 (GenBank accession no. DQ116441) was used as an internal control. Roots, stems, leaves and shoot apical meristems (SAM) were sampled at the third true-leaf stage, and sepals were collected at the flowering stage, respectively. Fibers were sampled on 15 d post anthesis (DPA). The patterns were clustered and visualized using heatmap program HemI 1.0 [50]. The color scale at the right-above of the heat map is given in log2 -transformed 2–ΔCt value.
Diurnal expression pattern of Group I GhCOLs in LD and SD conditions
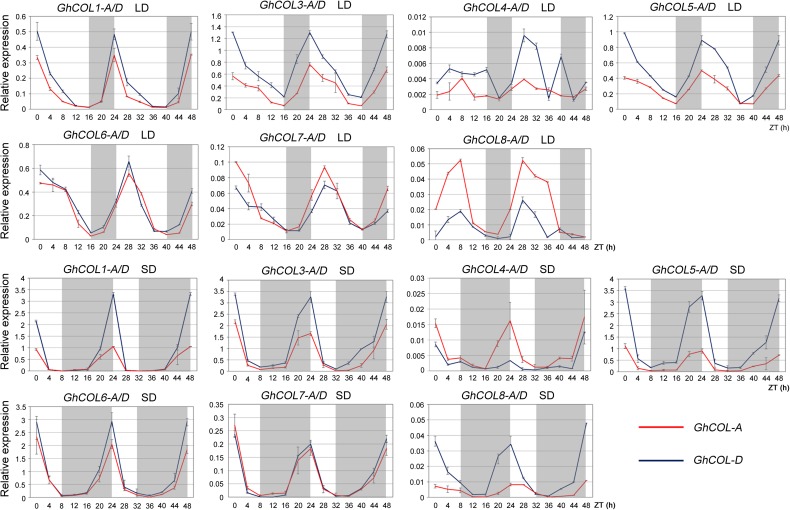
COL genes in Group I have been documented to play key roles in regulating flowering time and show obvious circadian rhythm characteristics in all plants studied [37]. We next focused on investigating the seven pairs of homoeologous COL genes in Group I for respective diurnal expression patterns over a 48-h period at 4-h intervals in LD or SD conditions. In both light conditions, six homoeologous gene pairs, except for GhCOL4-A/D in LD, showed a clear diurnal expression pattern with biased A- or D-homoeologs expression (Fig 5). GhCOL1, GhCOL3, and GhCOL5 homoeologs exhibited similar diurnal rhythms, and their expression peaked at dawn and then decreased rapidly to a minimum at dusk, then began to increase until the following dawn.
Fig 5. Diurnal expression pattern of the seven homoeologous COL gene pairs form Group I under LD or SD conditions.
Sample collection started at the beginning of the light period at zeitgeber time (ZT) 0 and continued every 4 h for 48 h in LD and SD conditions. The x-axis shows the time points and y-axis represents relative gene expression against cotton UBQ7 (DQ116441) as the control. Gray boxes over each chart indicate night. Data represent the mean ± SE obtained from three independent biological repeats.
Under LD, GhCOL6 and GhCOL7 homoeologs showed cyclic expression patterns with light/dark induction treatment, but the expression peak occurred at dawn or 4 h later. Interestingly, under SD both showed similar expression patterns with peaks at dawn and a rapid decline to their minima, which was similar to Arabidopsis CO under SD conditions [11]. The COL4 and COL8 homoeologs showed obviously different expression patterns in both light conditions. In LD, COL4-D expression peaked more than twice, and COL4-A peaked once. However, in SD, COL4 homoeologs had clear diurnal expression similar to COL1, COL3 and COL5 homoeologs. Expression of COL8 homoeologs peaked at dawn under SD but 4 h later for LD.
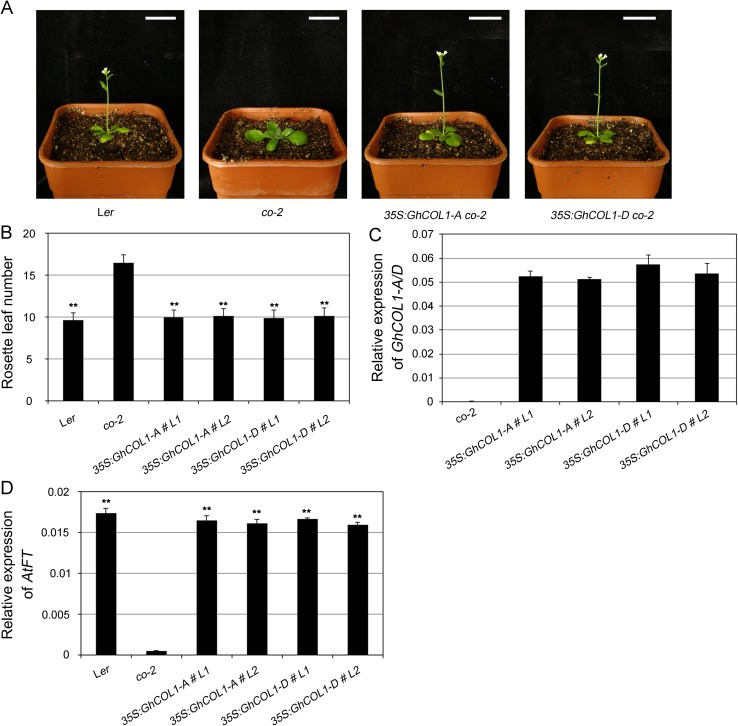
Ectopic expression of GhCOL1-A and GhCOL1-D promotes flowering in Arabidopsis
To further explore possible roles in flowering control of cotton homoeologous genes derived from past whole-genome duplication events, we cloned the coding sequences of the seven homoeologous COL gene pairs in Group I from G. hirsutum cv. XLZ42. Multiple alignments of amino acid sequences among 14 cotton GhCOLs and Arabidopsis AtCO and rice Hd1 are shown in S3 Fig. We then expressed them under the CaMV 35S promoter in co-2 mutant Arabidopsis. While co-2 mutant Arabidopsis exhibited obvious late flowering compared to wild-type (Ler) under LD, the transgenic plants expressing GhCOL1 homoeologs flowered significantly earlier than co-2 mutants (Fig 6A), with similar rosette leaf numbers to Ler (Fig 6B), and GhCOL genes were confirmed to be overexpressed in the transgenic co-2 plants by qRT-PCR (Fig 6C and Figure C in S4 Fig). In the co-2 mutant, expression of endogenous AtFT was hardly detectable compared with basal levels in Ler. However, AtFT transcripts in transgenic plants approached the level detected in the Ler background (Fig 6D). These results showed that ectopic expression of GhCOL1 homoeologs complemented the late-flowering effect of co-2. We also found that the flowering times in transgenic plants expressing GhCOL3 and GhCOL7 homoeologs were also slightly earlier than co-2, but still later than wild-type under the same conditions (Figures A and B in S4 Fig). The endogenous AtFT transcripts exceeded levels in the co-2 mutant, but were far below the levels in Ler (Figure D in S4 Fig), suggesting that GhCOL3 and GhCOL7 homoeologs partially complemented the later flowering phenotype of co-2. However, overexpression of GhCOL4, GhCOL5, GhCOL6, and GhCOL8 homoeologous gene pairs had no influence on flowering time of co-2 (S4 Fig).
Fig 6. Overexpression of GhCOL1-A and GhCOL1-D rescued the late-flowering phenotype of the Arabidopsis co-2 mutant.
(A) Representative phenotype of 20 d Ler, co-2, 35S:GhCOL1-A and 35S:GhCOL1-D transgenic line in phytotron under LD conditions. Scale bar, 1 cm. (B) Flowering time was measured as the rosette leaves number per plant. Data represent a minimum of 10 plants for each line ± SE. (C) Detection of GhCOL1-A/D expression by qRT-PCR in 35S:GhCOL1s transgenic lines and co-2 under LD conditions. (D) The expression level of endogenous AtFT (AT1G65480) was determined by qRT-PCR. Data represent the mean ± SE (n = 3) obtained from three independent biological repeats in (C) and (D), and AtACT 2 (AT3G18780) was used as internal control. ** indicate significant differences compared with co-2 at P < 0.01 according to the Student’s t-test.
Discussion
Functional conservation and divergence of COL gene family in cotton
Many studies have shown that the CO/FT regulon in photoperiod-responsive plant species plays an important role in regulating flowering transition, but our understanding of the molecular mechanism is still limited, especially in polyploid species. Upland cotton is a domesticated allotetraploid and is cultivated worldwide, and has gradually lost their photoperiod sensitivity. The CO/FT module in cultivated cotton remains unclear. In total, 42 GhCOLs family genes from the G. hirsutum genome (acc. TM-1) were identified and characterized in the present study. They were distributed unevenly along 18 different chromosomes (Fig 1), and phylogenetic analysis clustered them into three groups (Fig 2 and Fig 3A). Both gene structures and the conserved protein motifs of GhCOLs shared high similarity with known COL homologs involved in photoperiod-responsive plant species, suggesting that the function of COL family genes was highly conserved during evolution of a wide range of plant species (Fig 3B and 3C).
Fourteen COL proteins in Group I in upland cotton had two B-box, one VP motif, and one CCT domain (Fig 3C and S2 Fig). Zhang et al [43] analyzed the expression levels of eight COL genes derived from TM-1 in Group I, and found that they all had diurnal expression patterns. In Zhang’s study, however, qRT-PCR primers used for gene expression detection did not discriminate between A- or D-subgenomes, and so the expression patterns of homoeologous genes were not clear. Due to polyploidization, expression levels of many homoeologous genes are unequal in allotetraploid cotton [45]. To understand their possible roles, we performed a detailed transcript-level characterization of seven homoeologous COL genes pairs in Group I. We analyzed the diurnal expression patterns of the A- and D-homoeologs in detail by designing gene-specific primers based on their single nucleotide polymorphism, showing a clear expression of diurnal rhythm for all 14 genes in cv. XLZ 42, consistent with published data [43].
COL1, COL3, and COL5 homoeologs showed similar diurnal expression patterns under both light conditions, with more consistent expression rhythm in SD (Fig 5B), and their expression peaked at dawn and declined rapidly to minima at dusk. Under LD, COL6, COL7, and COL8 homoeologs had clear cyclic expression patterns, and their expression peaks occurred at dawn 4 h later; whereas under SD, their expression patterns were similar to COL1, COL3, and COL5 homoeologs. Under LD, the peak times for COL4 homoeologs differed from each other; whereas the expression rhythms in SD were also similar to COL1, COL3, and COL5 homoeologs. Among seven COL homoeologs in upland cotton, there were slightly more genes with expression bias toward Dt than toward At homoeologs, consistent with published data [45]. In summary, the diurnal expression analyses indicated that in photoperiodic flowering, cotton COL family genes in Group I had similar or conserved functions. Unequal expression of COL homoeologs between At and Dt subgenomic loci may lead to subfunctionalization or neofunctionalization in allotetraploid cotton, but detailed functional analyses of cotton COL family genes are still needed.
In addition to regulating flower times, the COL gene family is involved in a wide range of events in plant development in response to photoperiodic signaling, including seedling growth [54,55], dormancy [7], tuberization [56], and cell growth [38]. Functional divergence of the COL gene family in Arabidopsis has been frequently reported. For example, AtCO, AtCOL1, and AtCOL2 share high sequence similarity. However, altered expression of AtCOL1 and AtCOL2 in transgenic plants accelerated the circadian clock, but had little effect on flowering time [57]. Unlike AtCO, AtCOL3 represses flowering and influences root growth and lateral root formation [54]. The expression of AtCOL9 is also regulated by the circadian clock in the photoperiod pathway. Unexpectedly, AtCOL9 overexpression repressed flowering through repression of AtCO as well as AtFT [58]. Diverse diurnal expression patterns of the GhCOL family genes strongly suggested functional divergence of cotton COL homoeologs in multiple aspects of photoperiodic response, including flowering.
Furthermore, tissue-specific expression patterns also strongly indicated that multiple functions of GhCOLs were not necessarily related to flowering. Although 42 COL genes were expressed in all examined tissues, the average expression levels and numbers of expressed genes varied among the seven different tissues. GhCOL1 homoeologs and GhCOL3-A were solely highly expressed in the SAM. GhCOL5-A was predominantly expressed in roots. GhCOL9-D, GhCOL10 homoeologs, and GhCOL20-A were solely highly expressed in leaves. GhCOL6, GhCOL7 homoeologs, and GhCOL16-A were solely highly expressed in flowers. These data suggest specific functions in root, leaf, flower, and SAM for specific COL genes, whereas the similar expression patterns suggest functional redundancy, and biased-expressed homoeologous COLs genes may lead to diverse functionalization. In addition, GhCOL17 homoeologs, GhCOL18-A and GhCOL22-D, were highly expressed in fibers, suggesting involvement in fiber development. Their functional divergence and exact roles in cotton growth require further study.
GhCOL1-A and ChCOL1-D are potential flowering inducers and activators of GhFT1 in G. hirsutum
Of the 42 cotton COLs, we explored which COL homoeologs were the flowering inducers in cotton. We gathered evidence indicating that the GhCOL1-A and GhCOL1-D homoeologs were the flowering inducers in G. hirsutum. First, GhCOL1-A was shown to have 55.1 and 43.5% amino acid sequence similarity with the Arabidopsis CO and rice Hd1, which both function as flowering inducers, while correspondingly, GhCOL1-D had 55.6 and 45.3% similarity (S3 Fig). Second, phylogenetic analysis indicated that GhCOL1-A and GhCOL1-D clustered together with AtCO and Hd1 (Fig 2). Third, GhCOL1-A and GhCOL1-D mRNA abundance showed similar oscillations under both LD and SD conditions, and the highest levels of mRNA were at dawn (Fig 5), showing similarity with Arabidopsis CO [11]. The GhCOL1-A and GhCOL1-D mRNA levels continued to oscillate for a period of 24 h, indicating that they were regulated by the circadian clock. Last, our transgenic study showed that overexpression of GhCOL1-A and GhCOL1-D rescued the late-flowering phenotype of the Arabidopsis loss-of-function co-2 mutant, thereby demonstrating their crucial role in flowering (Fig 6). Moreover, by over-expressing GhCOL1-A, or GhCOL1-D, endogenous AtFT transcription was almost fully restored to the normal levels (Fig 6D).
Our previous study showed that under LD or SD conditions, the expression pattern of GhFT1 (FT ortholog of G. hirsutum) was rhythmic with an expression peak 4 h into the light period [52]. The 4-h time lag between the expression peak of GhCOL1 homoeologs and GhFT1 suggest a putative novel mechanism in cotton CO/FT regulation.
Taken together, the results show that GhCOL1-A and GhCOL1-D were the potential activator of GhFT1, and the CO/FT module reported in Arabidopsis, rice, and other plants was conserved in G. hirsutum. We suggest that GhCOL1 homoeologs play important roles in flowering regulation of cotton in response to changing photoperiod. Further experiments will clarify the molecular mechanism and explore the functions of other GhCOL homoeologs.
Supporting information
Left and right black arrows indicated the locations of forward and reverse primers, respectively. Red frames indicated the differences of nucleotides between GhCOL-A and GhCOL-D homoeologs in Group I.
(TIF)
Multiple alignments of amino acid sequences of 42 GhCOLs were performed using ClustalW [46]. (A) 14 COLs in Group I. (B) six COLs in Group II. (C) 22 COLs in group III. Conserved amino acids were highlighted in black and the similar in grey. The B-box1, B-box2, VP motif, zinc finger and CCT conserved sequences were marked with horizontal lines.
(TIF)
AtCO (NP_1978088.1) and Hd1 (BAB17627.1) were retrieved from GenBank. Conserved amino acids are highlighted in black and the similar in grey. The gaps indicated by dashes are attributed to the lack of amino acids.
(TIF)
(A) Representative phenotype of 20 days Ler, co-2 and transgenic plants grown in phytotron under LD conditions. Scale bar, 1 cm. (B) Flowering time was measured as the rosette leaves number per plant. Data represent a minimum of 10 plants scored for each line ± SE. (C) Detection of GhCOLs expression by qRT-PCR in 35S:GhCOLs transgenic lines and co-2 under LD conditions. (D) The expression level of Arabidopsis FT was determined by qRT-PCR. Data represent the mean ± SE from three biological replicates in (C) and (D), and AtACT2 (AT3G18780) was used as internal control. ** and * indicate significant differences in comparison with co-2 mutant at P < 0.01 and P < 0.05 according to the Student’s t-test compared to mutant, respectively.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability
GenBank accession numbers for upland cotton COL genes in Group I are as follows: GhCOL1-A (KY769104), GhCOL1-D (KY769111), GhCOL3-A (KY769105), GhCOL3-D (KY769112), GhCOL4-A (KY769106), GhCOL4-D (KY769113), GhCOL5-A (KY769107), GhCOL5-D (KY769114), GhCOL6-A (KY769108), GhCOL6-D (KY769115), GhCOL7-A (KY769109), GhCOL7-D (KY769116), GhCOL8-A (KY769110), GhCOL8-D (KY769117).
Funding Statement
This work was financially supported by the National Natural Science Foundation of China (31360366) to XH; the Program for New Century Excellent Talents in University (grant no. NCET-12-1072) to XH; the Scientific and Technological Innovation Leading Talents of Xinjiang Production and Construction Corps (2006BC001) to XH; the Innovation Team Project for Xinjiang Production and Construction Corps (2014CC005) to XH.
References
- 1.Thomas B, Vince-Prue D. Photoperiodism in plants. Academic Press; 1997. [Google Scholar]
- 2.Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000; 288: 1613–1616. doi: 10.1126/science.288.5471.1613 [DOI] [PubMed] [Google Scholar]
- 3.Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, et al. Activation tagging of the floral inducer FT. Science. 1999; 286(5446): 1962–1965. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999; 286(5446):1960–1962. [DOI] [PubMed] [Google Scholar]
- 5.Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003; 422(6933): 719–722. doi: 10.1038/nature01549 [DOI] [PubMed] [Google Scholar]
- 6.Hayama R, Coupland G. The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 2004; 135(2): 677–684. doi: 10.1104/pp.104.042614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006; 312(5776): 1040–1043. doi: 10.1126/science.1126038 [DOI] [PubMed] [Google Scholar]
- 8.Ballerini ES, Kramer EM. In the light of evolution: a reevaluation of conservation in the CO-FT regulon and its role in photoperiodic regulation of flowering time. Front Plant Sci. 2011; 2: 81 doi: 10.3389/fpls.2011.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Putterill J, Robson F, Lee K, Coupland G. Chromosome walking with YAC clones in Arabidopsis: isolation of 1700 kb of contiguous DNA on chromosome 5, including a 300 kb region containing the flowering-time gene CO. Mol Gen Genet. 1993; 239(1–2): 145–157. [DOI] [PubMed] [Google Scholar]
- 10.Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995; 80(6): 847–857. [DOI] [PubMed] [Google Scholar]
- 11.Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001; 410(6832): 1116–1120. doi: 10.1038/35074138 [DOI] [PubMed] [Google Scholar]
- 12.Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007; 316(5827): 1030–1033. doi: 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- 13.Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, et al. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-panning domains. EMBO J. 1999; 18(17): 4679–4688. doi: 10.1093/emboj/18.17.4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huq E, Tepperman JM, Quail PH. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2000; 97(17): 9789–9794. doi: 10.1073/pnas.170283997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005; 309(5732): 293–297. doi: 10.1126/science.1110586 [DOI] [PubMed] [Google Scholar]
- 16.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007; 318(5848): 261–265. doi: 10.1126/science.1146994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA,et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009; 17(1): 75–86. doi: 10.1016/j.devcel.2009.06.015 [DOI] [PubMed] [Google Scholar]
- 18.Song YH, Ito S, Imaizumi T. Flowering time regulation: photoperiod-and temperature-sensing in leaves. Trends Plant Sci. 2013; 18(10): 575–583. doi: 10.1016/j.tplants.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, et al. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008; 27(8): 1277–1288. doi: 10.1038/emboj.2008.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srikanth A, Schmid M. Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci. 2011; 68(12): 2013–2037. doi: 10.1007/s00018-011-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazaro A, Valverde F, Piñeiro M, Jarillo JA. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell. 2012; 24(3): 982–999. doi: 10.1105/tpc.110.081885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazaro A, Mouriz A, Piñeiro M, Jarillo JA. Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis. Plant Cell. 2015; 27(9): 2437–2454. doi: 10.1105/tpc.15.00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, et al. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell. 2008; 20(2): 292–2306. doi: 10.1105/tpc.107.057281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell. 2008; 32(5): 617–630. doi: 10.1016/j.molcel.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, et al. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011; 25(10): 1023–1028. doi: 10.1101/gad.2025111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H, Liu B, Zhao C, Pepper M, Lin C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011; 16(12): 684–691. doi: 10.1016/j.tplants.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou Z, Liu H, Liu B, Liu X, Lin C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol. 2011; 21(10): 841–847. doi: 10.1016/j.cub.2011.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagercrantz U, Axelsson T. Rapid evolution of the family of CONSTANS LIKE genes in plants. Mol Biol Evol. 2000; 17(10): 1499–1507. [DOI] [PubMed] [Google Scholar]
- 29.Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000; 12(12): 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffiths S, Dunford RP, Coupland G, Laurie DA. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003; 131(4): 1855–1867. doi: 10.1104/pp.102.016188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chia T, Müller A, Jung C, Mutasa-Göttgens E. Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J Exp Bot. 2008; 59(10): 2735–2748. doi: 10.1093/jxb/ern129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong AC, Hecht VF, Picard K, Diwadkar P, Laurie RE, Wen J, et al. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front Plant Sci. 2014; 5: 486 doi: 10.3389/fpls.2014.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu F, Price BW, Haider W, Seufferheld G, Nelson R, Hanzawa Y. Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. PLoS One. 2014;9(1): e85754 doi: 10.1371/journal.pone.0085754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song X, Duan W, Huang Z, Liu G, Wu P, Liu T, et al. Comprehensive analysis of the flowering genes in Chinese cabbage and examination of evolutionary pattern of CO-like genes in plant kingdom. Sci Rep. 2015; 5: 14631 doi: 10.1038/srep14631 PMCID: PMC4586889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu J, Yang L, Dai S. Identification and characterization of the CONSTANS-like gene family in the short-day plant Chrysanthemum lavandulifolium. Mol Genet Genomics. 2015; 290(3): 1039–1054. doi: 10.1007/s00438-014-0977-3 [DOI] [PubMed] [Google Scholar]
- 36.Liu T, Zhu S, Tang Q, Tang S. Identification of a CONSTANS homologous gene with distinct diurnal expression patterns in varied photoperiods in ramie (Boehmeria nivea L. Gaud). Gene. 2015; 560(1): 63–70. doi: 10.1016/j.gene.2015.01.045 25623329 [Google Scholar]
- 37.Chaurasia AK, Patil HB, Azeez A, Subramaniam VR, Krishna B, Sane AP, et al. Molecular characterization of CONSTANS-Like (COL) genes in banana (Musa acuminata L. AAA Group, cv. Grand Nain). Physiol Mol Biol Plants. 2016; 22(1): 1–15. doi: 10.1007/s12298-016-0345-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serrano G, Herrera-Palau R, Romero JM, Serrano A, Coupland G, Valverde F. Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr Biol. 2009; 19(5): 359–368. doi: 10.1016/j.cub.2009.01.044 [DOI] [PubMed] [Google Scholar]
- 39.Gangappa SN, Botto JF. The BBX family of plant transcription factors. Trends Plant Sci. 2014; 19(7): 460–470. doi: 10.1016/j.tplants.2014.01.010 [DOI] [PubMed] [Google Scholar]
- 40.Wendel JF, Albert VA. Phylogenetics of the Cotton genus (Gossypium): Character-State weighted parsimony analysis of chloroplast-DNA restriction site ata and its systematic and biogeographic implications. Syst Bot. 1992; 17: 115–143 [Google Scholar]
- 41.Saha S, Jenkins JN, Wu J, McCarty JC, Gutiérrez OA, Percy RG, et al. Effects of hromosome-specific introgression in upland cotton on fiber and agronomic traits. Genetics. 2006; 172: 1927–1938. doi: 10.1534/genetics.105.053371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang T, Guo W, et al. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2007; 145: 1303–1310. doi: 10.1104/pp.107.107672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang R, Ding J, Liu C, Cai C, Zhou B, Zhang T, et al. Molecular evolution and phylogenetic analysis of eight COL superfamily genes in group I related to photoperiodic regulation of flowering time in wild and domesticated cotton (Gossypium) species. PloS One. 2015; 10(2): e0118669 doi: 10.1371/journal.pone.0118669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015; 33(5): 524–530. doi: 10.1038/nbt.3208 [DOI] [PubMed] [Google Scholar]
- 45.Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015; 33(5): 531–537. doi: 10.1038/nbt.3207 [DOI] [PubMed] [Google Scholar]
- 46.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22(22): 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28(10): 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo A, Zhu Q, Chen X, Luo J. GSDS: a gene structure display server. Yi chuan. 2007; 29(8): 1023–1026. [PubMed] [Google Scholar]
- 49.Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. 2001; 25(4): 402–408 doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 50.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: a toolkit for illustrating heatmaps. PLoS One. 2014; 9(11): e111988 doi: 10.1371/journal.pone.0111988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994; 25(6): 989–994. [DOI] [PubMed] [Google Scholar]
- 52.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998; 16(6): 735–743. [DOI] [PubMed] [Google Scholar]
- 53.Guo D, Li C, Dong R, Li X, Xiao X, Huang X. Molecular cloning and functional analysis of the FLOWERING LOCUS T (FT) homolog GhFT1 from Gossypium hirsutum L. J Integr Plant Biol. 2015; 57(6):522–533. doi: 10.1111/jipb.12316 [DOI] [PubMed] [Google Scholar]
- 54.Datta S, Hettiarachchi GH, Deng XW, Holm M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell. 2006; 18(1): 70–84. doi: 10.1105/tpc.105.038182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datta S, Hettiarachchi C, Johansson H, Holm M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell. 2007; 19(10): 3242–3255. doi: 10.1105/tpc.107.054791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.González-Schain ND, Suárez-López P. CONSTANS delays flowering and affects tuber yield in potato. Biol Plantarum. 2008; 52(2): 251–8 [Google Scholar]
- 57.Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 2001; 26(1): 15–22. [DOI] [PubMed] [Google Scholar]
- 58.Cheng XF, Wang ZY. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005; 43(43): 758–768. doi: 10.1111/j.1365-313X.2005.02491.x . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Left and right black arrows indicated the locations of forward and reverse primers, respectively. Red frames indicated the differences of nucleotides between GhCOL-A and GhCOL-D homoeologs in Group I.
(TIF)
Multiple alignments of amino acid sequences of 42 GhCOLs were performed using ClustalW [46]. (A) 14 COLs in Group I. (B) six COLs in Group II. (C) 22 COLs in group III. Conserved amino acids were highlighted in black and the similar in grey. The B-box1, B-box2, VP motif, zinc finger and CCT conserved sequences were marked with horizontal lines.
(TIF)
AtCO (NP_1978088.1) and Hd1 (BAB17627.1) were retrieved from GenBank. Conserved amino acids are highlighted in black and the similar in grey. The gaps indicated by dashes are attributed to the lack of amino acids.
(TIF)
(A) Representative phenotype of 20 days Ler, co-2 and transgenic plants grown in phytotron under LD conditions. Scale bar, 1 cm. (B) Flowering time was measured as the rosette leaves number per plant. Data represent a minimum of 10 plants scored for each line ± SE. (C) Detection of GhCOLs expression by qRT-PCR in 35S:GhCOLs transgenic lines and co-2 under LD conditions. (D) The expression level of Arabidopsis FT was determined by qRT-PCR. Data represent the mean ± SE from three biological replicates in (C) and (D), and AtACT2 (AT3G18780) was used as internal control. ** and * indicate significant differences in comparison with co-2 mutant at P < 0.01 and P < 0.05 according to the Student’s t-test compared to mutant, respectively.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
GenBank accession numbers for upland cotton COL genes in Group I are as follows: GhCOL1-A (KY769104), GhCOL1-D (KY769111), GhCOL3-A (KY769105), GhCOL3-D (KY769112), GhCOL4-A (KY769106), GhCOL4-D (KY769113), GhCOL5-A (KY769107), GhCOL5-D (KY769114), GhCOL6-A (KY769108), GhCOL6-D (KY769115), GhCOL7-A (KY769109), GhCOL7-D (KY769116), GhCOL8-A (KY769110), GhCOL8-D (KY769117).