Abstract
Purpose
To propose and validate a respiratory motion resolved, self‐gated (SG) 4D‐MRI technique to assess patient‐specific breathing motion of abdominal organs for radiation treatment planning.
Methods
The proposed 4D‐MRI technique was based on the balanced steady‐state free‐precession (bSSFP) technique and 3D k‐space encoding. A novel rotating cartesian k‐space (ROCK) reordering method was designed which incorporates repeatedly sampled k‐space centerline as the SG motion surrogate and allows for retrospective k‐space data binning into different respiratory positions based on the amplitude of the surrogate. The multiple respiratory‐resolved 3D k‐space data were subsequently reconstructed using a joint parallel imaging and compressed sensing method with spatial and temporal regularization. The proposed 4D‐MRI technique was validated using a custom‐made dynamic motion phantom and was tested in six healthy volunteers, in whom quantitative diaphragm and kidney motion measurements based on 4D‐MRI images were compared with those based on 2D‐CINE images.
Results
The 5‐minute 4D‐MRI scan offers high‐quality volumetric images in 1.2 × 1.2 × 1.6 mm3 and eight respiratory positions, with good soft‐tissue contrast. In phantom experiments with triangular motion waveform, the motion amplitude measurements based on 4D‐MRI were 11.89% smaller than the ground truth, whereas a −12.5% difference was expected due to data binning effects. In healthy volunteers, the difference between the measurements based on 4D‐MRI and the ones based on 2D‐CINE were 6.2 ± 4.5% for the diaphragm, 8.2 ± 4.9% and 8.9 ± 5.1% for the right and left kidney.
Conclusion
The proposed 4D‐MRI technique could provide high‐resolution, high‐quality, respiratory motion‐resolved 4D images with good soft‐tissue contrast and are free of the “stitching” artifacts usually seen on 4D‐CT and 4D‐MRI based on resorting 2D‐CINE. It could be used to visualize and quantify abdominal organ motion for MRI‐based radiation treatment planning.
Keywords: 4D‐MRI , k‐space binning, respiratory motion management, self‐gating
1. Introduction
In radiation therapy of abdominal tumors, it is necessary to assess patient‐specific breathing pattern to determine individual margins for optimal radiation beam delivery.1 As the current clinical standard, four‐dimensional computed tomography (4D‐CT) acquires a series of oversampled 2D slices, which are subsequently sorted into multiple respiratory motion‐resolved 3D datasets using a motion surrogate.2, 3, 4 However, 4D‐CT delivers excessive ionizing radiation to the subject due to its relatively long scan time, and its soft‐tissue contrast is often insufficient for tumor delineation in the abdomen. In addition, the 2D image sorting may result in “stitching” artifacts which potentially undermines abdominal organ visualization as well as the accuracy of motion quantification.5, 6, 7
As an alternative modality, four‐dimensional magnetic resonance imaging (4D‐MRI) offers excellent soft‐tissue contrast and is free of ionizing radiation. Following the concept of 4D‐CT, the majority of existing 4D‐MRI approaches acquire multiple 2D‐CINE images in consecutive slices and retrospectively sort them based on recorded motion surrogates.5, 6, 7, 8, 9, 10 As a result, these 2D‐based methods have similar “stitching” artifacts as 4D‐CT, and their > 2‐mm inplane resolution and 5–10 mm slice thickness may be inadequate for accurate motion quantification.11 Deng et al. recently proposed a novel 4D‐MRI technique with high spatial resolution and free of the “stitching” artifacts because 2D image sorting is replaced by 3D k‐space binning.11 In their approach, golden‐ratio 3D radial k‐space trajectory12, 13 with respiratory motion self‐gating (SG)14, 15 is used. However, the mandatory cubic fields‐of‐view (FOV) and computationally demanding image reconstruction associated with 3D radial trajectory potentially limit the scan efficiency and overall clinical utility.
In this work, we propose a 4D‐MRI technique based on the balanced steady‐state free‐precession (bSSFP) technique and a 3D Cartesian k‐space trajectory. The use of bSSFP contrast improves the scan efficiency due to its signal‐to‐noise ratio (SNR) advantage over gradient recalled echo (GRE). In our technique, we used a rotating cartesian k‐space (ROCK) reordering method which incorporates repeatedly sampled k‐space centerline as respiratory motion surrogate and allows retrospective k‐space data binning into multiple respiratory positions. The proposed technique was validated in imaging studies on a motion phantom and healthy volunteers, where quantitative motion measurements based on 4D‐MRI were compared with those based on 2D‐CINE images.
2. Method
2.A. Imaging sequence
A standard bSSFP MRI pulse sequence was modified using ROCK,16 a 3D Cartesian k‐space reordering method that provides: (a) variable density k‐space sampling for improved scan efficiency17; (b) repetitively sampled k‐space centerline18 as respiratory motion surrogate; (c) flexibility of retrospective data binning into multiple respiratory positions.
Figure 1(a) shows the logical ky−kz plane of the Cartesian k‐space grid, where each point represents a read‐out line in the kx direction. First, all points in the plane are divided into N concentric rings. A spiral path with azimuthal angle is generated and subsequently mapped to N Cartesian grid points along the spiral trajectory, one point from each ring. Details on the generation and mapping of the spiral path have been previously described.16 The entire process is then repeated for the next spiral path with incremented using the golden angle (), until the desired number of samples is reached. The golden angle rotation ensures near uniform k‐space sampling pattern for each respiratory position after retrospective data binning while allowing flexible sampling rates.12, 19
Figure 1.
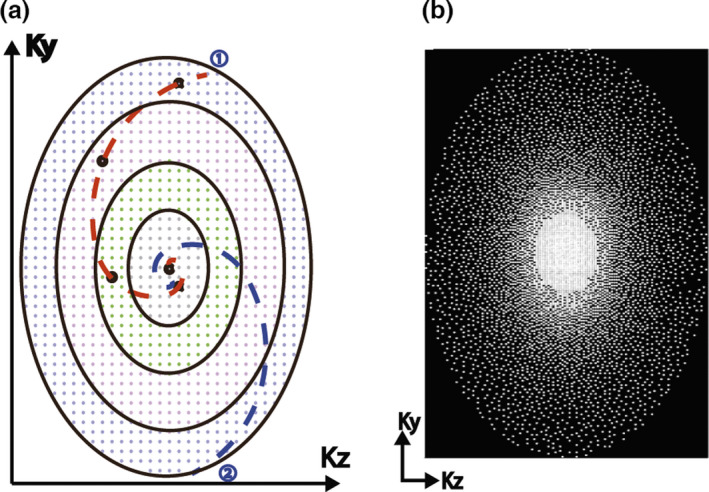
(a) Illustration of the ROCK sampling pattern; (b) The undersampling rate grow exponentially along radial direction. This variable density undersampling is more efficient than uniform sampling when compressed sensing image reconstruction is used.
In this study, the Cartesian grid was divided into N = 20 rings, which was empirically chosen considering the matrix size in the ky‐kz plane and the sampling frequency of the self‐gating lines. The number of Cartesian grid points within each ring grew exponentially from the innermost ring to the outer rings. In this way, the final sampling pattern had variable sampling density20 where the center k‐space was sampled more often than the peripheral (Fig. 1(b)), because an equal number of sampling points were chosen from each ring. Moreover, the innermost ring had only one point in the center, which was therefore sampled once in every quasispiral path and subsequently used for SG.
2.B. Motion estimation and data binning
The scan was performed in coronal view with k‐space read‐out in the superior–inferior (SI) direction. The k‐space centerlines in the ROCK pattern were used for calculating the respiratory motion surrogate signal. They were first Fourier transformed into projections and then interpolated eightfold in order to improve the spatial definition of the estimated motion and offer smoother motion weights in the soft‐gated image reconstruction. The shifts relative to the reference (i.e., first SI projection), which were estimated by maximizing the cross‐correlation, were subsequently used as the respiratory motion surrogate.16 Compared with using cross‐correlation coefficient alone,11, 21 this method calculates the respiratory motion in millimeters and is insensitive to the choice of reference.
All acquired k‐space data were then binned into Nresp respiratory positions based on the amplitude of the motion surrogate. Instead of a conventional binary data binning decision, a soft‐gating approach was used based on a Gaussian weighting function with its Full‐Width Half maximum (FWHM) set to 1/Nresp of the dynamic range (i.e., highest peak to lowest valley) of the respiratory surrogate signal, as shown in Fig. 2. The Gaussian weightings were used as the soft‐gating term in the ESPIRiT22 image reconstruction, which has been shown to effectively improve the signal‐to‐noise‐ratio (SNR) without introducing noticeable motion blurring artifacts.23, 24, 25
Figure 2.
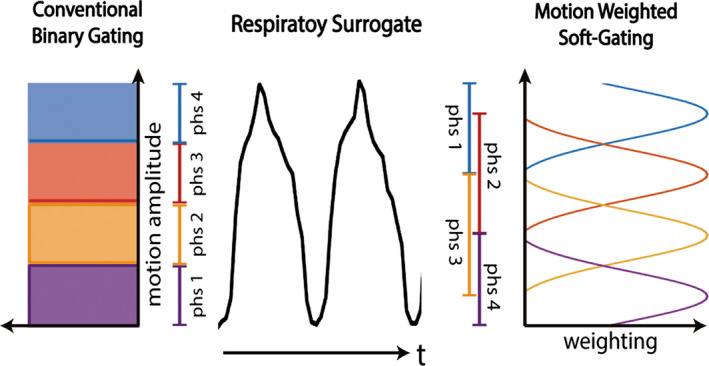
Conventional respiratory gating methods are based on binary gating decisions. The proposed 4D‐MRI uses the soft‐gating approach with Gaussian‐shaped motion weighting.
2.C. Image reconstruction
Two different image reconstruction strategies were used in this study. In motion‐independent reconstruction (strategy 1), 3D k‐space from each respiratory position was reconstructed independently using:
| (1) |
where is the reconstructed volumetric image; and represent undersampling and Fourier transform operations, respectively; is the coil sensitivity map estimated by ESPIRiT;22 is the Gaussian‐shaped weightings for the respiratory motion soft‐gating; is the k‐space measurements; is the spatial regularization term using the randomized shifting Daubechies wavelets.26 The motion‐independent reconstruction (strategy 1) was used in phantom experiment since each k‐space bin has approximately the same number of samples due to the relative homogeneous motion profile.
In the motion‐regularized image reconstruction (strategy 2), images from all respiratory positions were reconstructed together using:
| (2) |
where i = {1…Nresp} represents different respiratory positions and is a sparsifying transform applied along the respiratory dimension. In this work, the temporal finite differences (i.e., total variation) were used as .27 The additional regularization along the respiratory dimension potentially enforces the similarity among images of different respiratory positions. We expected that it is helpful in compensating for the image quality inconsistency throughout the different respiratory positions.
Data processing and image reconstruction algorithms for both strategies were implemented in a previously described customized inline image reconstruction system.28 K‐space data were transmitted to the server (3.5 GHz 6‐core CPU, 32 GB RAM, NVIDIA GTX970) as it was acquired. The motion estimation, data binning and image reconstruction programs were started automatically upon the finish of the scan. The reconstructed pixel data were transmitted back to the scanner console to generate the DICOM files. The total processing time was 10 min with GPU and CPU acceleration using the Berkley Advanced Reconstruction Toolbox (bart‐0.2.09).29 A web‐based graphic interface was employed for users to configure and monitor the reconstruction process.
2.D. Phantom experiment
A custom‐made, MRI‐compatible motion phantom was constructed to validate the proposed 4D‐MRI techniques. As shown in Fig. 3, a computer‐controlled step motor drives a hydraulic cylinder that transmits the reciprocating motion to a second cylinder connected to a moving dolly inside the scanner room. A linear potentiometer was used to measure the displacement of the cylinders for feedback control and system calibration.
Figure 3.
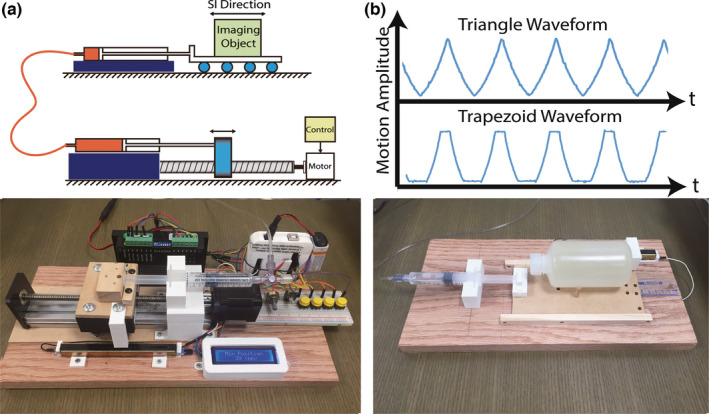
(a) The MRI‐compatible motion phantom is able to generate periodic motion waveforms with triangular and trapezoid shape (b). The reciprocating motion of a computer‐controlled step motor (c) is transmitted to a motion dolly placed inside the scanner (d) hydraulic cylinders and silicon tubes. The dolly is made with wood and 3D‐printed PLA materials for MRI compatibility.
A 300‐ml plastic bottle filled with water was placed on the dolly as the moving object, which is surrounded by two 1000‐ml static water bottles on both sides. The motion was programmed with variable motion parameters for a periodic motion (displacement range: 28 and 14 mm; period: 8 s, 12 s, 16 s, and 20 s; triangle and trapezoid waveform). All phantom imaging experiments were performed on a 1.5T MRI scanner (MAGNETOM TIM Avanto, Siemens Medical Solutions, Erlangen, Germany). The proposed 4D‐MRI technique was performed and repeated for each motion configuration. Important pulse sequence parameters included: field‐of‐view = 500 × 300 × 200 mm; resolution = 1.2 × 1.2 × 1.6 mm; coronal view with read‐out in SI direction; TE/TR = 2.0/4.0 ms; flip angle = 65; scan time = 5min; fat saturation pulses every 200 ms.
Data were retrospectively binned into Nresp = 8 respiratory positions and reconstructed using strategy 1. The locations of the moving object in each respiratory position were measured on 4D‐MRI images and compared with the ground truth motion parameters.
2.E. In vivo experiments
The proposed 4D‐MRI technique was tested in six healthy volunteers using the same 1.5T scanner used in the phantom experiment. The study was approved by our institutional review board and each volunteer provided a written informed consent. No specific breathing instructions were given to the subjects. 4D‐MRI scans were performed using identical sequence parameters as those used in our phantom experiment. After the 5‐min 4D‐MRI sequence, additional 2D‐CINE (bSSFP, resolution = 1.8 × 1.8 × 6 mm; 5fps; scan time = 30 s) scans were repeated six times with different slice orientations (two sagittal, two coronal, and two axial). Total scan time on each volunteer was less than 20 min including the set‐up and the localizer. 4D‐MRI data were retrospectively binned into Nresp = 8 respiratory positions and reconstructed using both strategies 1 and 2.
A peak detection algorithm was used to identify the peak and valley of each breathing cycle in the derived SG surrogate. The breathing cycle variability was quantitatively calculated as the normalized standard deviation (NSD) of the magnitude and position of all breathing cycles as shown in Eqs. (3) and (4).
| (3) |
| (4) |
In order to compare the motion measurements based on 4D‐MRI and 2D‐CINE, the reconstructed 4D images (strategy 2) were reformatted to match the slice orientations of the 2D‐CINE images. Inplane motion amplitudes of three regions of interest (ROI), including the diaphragm dome; left and right kidney, were measured based on reformatted 4D and 2D‐CINE images. The 30 s 2D‐CINE images contain more than one breathing cycle and therefore the motion amplitudes of all cycles were averaged as the final measurements.
3. Result
Figure 4 shows the 4D‐MRI of the motion phantom using the following motion configuration: displacement range = 28 mm; period = 16 s; triangle waveform. The motion surrogate derived from k‐space centerlines was in accordance with the ground truth motion waveform. The images of the first and last respiratory positions are color‐overlaid for motion amplitude measurement in Fig. 4(c). The measured motion amplitude based on our 4D MRI was 24.82 mm, 11.36% smaller than the ground truth peak amplitude (28 mm), which is expected due to the averaging effect of data binning. For a triangular motion waveform where the motion is uniform within each bin, we expect a 1/Nresp (i.e., 12.5%) difference between the measured displacement and the ground truth, which is from the topmost point of the triangle to the very bottom (Fig. 4(d)). Table 1 lists all motion amplitude measurements under different configurations and they were 11.89% (expected: 12.5%) smaller than the ground truth when triangular waveform was used, and 6.22% smaller than the ground truth when trapezoid waveform was used.
Figure 4.
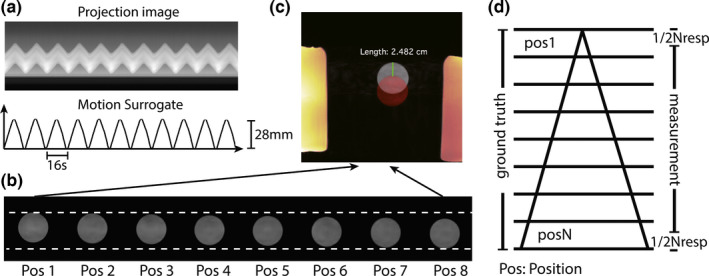
(a) the time series of projections and the derived motion surrogate; (b) the object in the 4D images (selected slice) has different locations; (c) images of the first and the last positions are color‐overlaid for motion amplitude measurement; (d) the motion amplitude measurement based on 4d‐mri has a system deviation of −1/Nresp with the ground truth for triangular‐shaped motion.
Table 1.
Motion amplitude measurements on phantom using 4D MRI in comparison with the ground truth
| Amplitude (mm) | Cycle (s) | Waveform | Measurement (mm) | Difference (%) |
|---|---|---|---|---|
| 28 | 20 | Trapezoid | 26.61 | −4.96 |
| 14 | 20 | Trapezoid | 12.92 | −7.71 |
| 14 | 16 | Trapezoid | 13.16 | −6.00 |
| 28 | 16 | Triangle | 24.82 | −11.36 |
| 14 | 16 | Triangle | 12.24 | −12.57 |
| 14 | 12 | Triangle | 12.13 | −13.35 |
| 14 | 8 | Triangle | 12.56 | −10.28 |
All volunteer scans were successful. The averaged NSD of the motion amplitude and position were 15.87% and 11.68%, respectively. Figure 5(a) shows selected pieces of respiratory motion surrogates from a single 4D‐MRI on a healthy volunteer. The amplitude, position, and frequency of respiratory motion were highly inconsistent among different breathing cycles. Several inspiration peaks in this example (marked in red) had different amplitudes, which would be binned together and result in motion artifacts if conventional respiratory phase based data binning was used.30 In the proposed method, however, data binning was based on the respiratory amplitude and these inspiration peaks were binned into different respiratory positions. The 3D K‐space data samples after retrospective respiratory binning were roughly uniform in the azimuthal direction (Fig. 5(c)) due to the golden angle rotation of quasispiral paths in our ROCK k‐space sampling pattern. Figure 6 shows the reconstructed 4D‐MRI images (4 out of 8 respiratory positions) reformatted into sagittal, coronal, and axial slice orientations. See online supplemental material Video‐S1 for a movie on the same dataset. Major abdominal organs, including the liver, kidneys, and pancreas are clearly visualized in 3D and in different respiratory positions.
Figure 5.
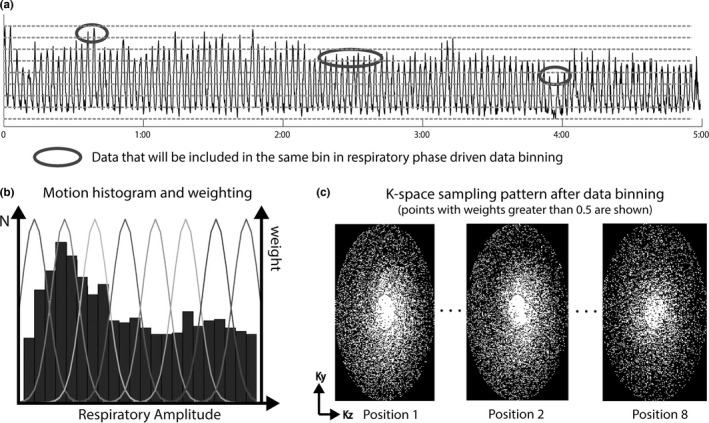
The amplitude of the derived self‐gating motion surrogate on a healthy volunteer (a) is highly inconsistent in different temporal windows. The respiratory amplitude driven data binning used in the proposed technique could address intrabin motion issues when using conventional respiratory phase driven data binning. The heterogeneity of respiratory motion, as shown in the motion histogram in (b), will result in different k‐space undersampling factor for each respiratory position. But the golden‐ratio rotation in ROCK sampling pattern ensures near uniform angular sampling in all k‐space bins.
Figure 6.
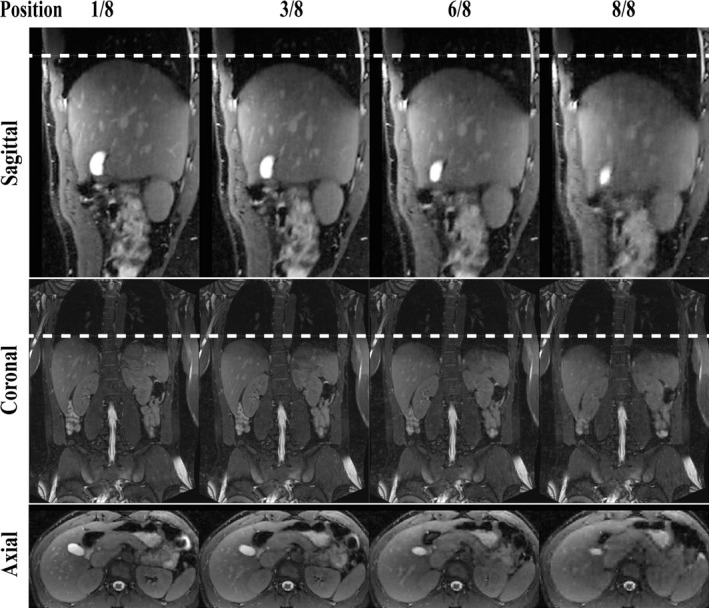
4D‐MRI images on a healthy volunteer reformatted in sagittal, coronal, and axial orientations. Four of nine respiratory positions are shown. See online supplemental material Video‐S1 for a movie on the same dataset.
In our study, the averaged undersampling rates of the 3D k‐space over all volunteers and in all respiratory positions were 7.5 ± 1.7. The different respiratory positions have different undersampling rates due to the heterogeneity of each individual's breathing pattern. This results in image quality variations across different respiratory positions when motion‐independent image reconstruction (strategy 1) is used. In the example shown in Fig. 7, the k‐spaces of the first and last respiratory positions had undersampling rates of 5.4x and 9.5x, respectively, and image of phase 8 using strategy 1 reconstruction had severe artifacts that potentially limit the delineations of abdomen organs or tumors. In motion‐regularized image reconstruction (strategy 2), the images for all the respiratory positions are reconstructed in a joint optimization process and the image quality is more consistent throughout the respiratory cycle.
Figure 7.
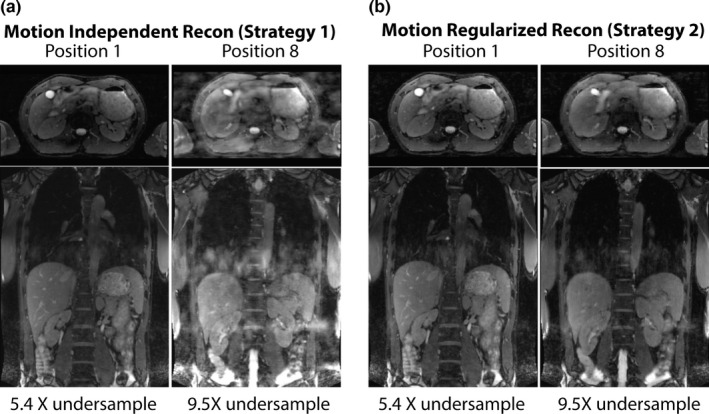
Comparison of the images reconstructed using two strategies on the same volunteer dataset. The image quality is inconsistent in motion‐independent image reconstruction (strategy 1) because the undersampling factors of the k‐space bins are different due to the heterogeneity of respiratory motion. In motion regularized image reconstruction (strategy 2), the image quality is more consistent in different respiratory positions.
Figure 8 shows a comparison of the reformatted 4D‐MRI and 2D‐CINE images from the same subject and in the same slice orientation. The 4D‐MRI image has comparable image quality with the 2D‐CINE, although it offers much higher resolution and 3D coverage. Quantitative in vivo motion amplitude measurements over three ROIs are listed in Table 2. The difference between the measurements based on 4D‐MRI and the ones based on 2D‐CINE were 6.2 ± 4.5% for the diaphragm, 8.2 ± 4.9% and 8.9 ± 5.1% for the right and left kidney. The difference between the two measurements may be due to the intrascan and interscan variation in the breathing pattern and the relatively low spatial resolution of 2D‐CINE images.
Figure 8.
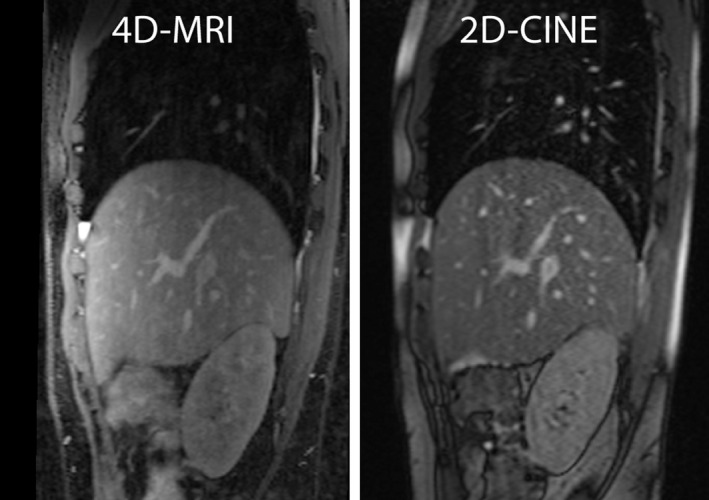
A Comparison of the 4D‐MRI and 2D‐CINE images in the same slice of the same subject.
Table 2.
Quantitative motion amplitude measurement volunteers
| Diaphragm (mm) Coronal slice | Right Kidney (mm) Coronal slice | Error % | Left Kidney (mm) Sagittal Slice | Error % | |||||
|---|---|---|---|---|---|---|---|---|---|
| 4D‐MRI | 2D‐CINEa | Error | 4D‐MRI | 2D‐CINEa | 4D‐MRI | 2D‐CINEa | |||
| Subject 1 | 29.58 | 25.46 | 13.93% | 23.72 | 20.21 | 14.80% | 22.98 | 19.68 | 14.36% |
| Subject 2 | 16.14 | 15.01 | 6.00% | 12.40 | 11.62 | 6.29% | 11.01 | 12.61 | −14.53% |
| Subject 3 | 20.54 | 20.02 | 2.53% | 14.91 | 13.13 | 11.94% | 12.45 | 11.87 | 4.66% |
| Subject 4 | 13.21 | 13.38 | −1.29% | 8.77 | 9.40 | −7.18% | 7.38 | 8.17 | −10.70% |
| Subject 5 | 17.53 | 16.66 | 4.96% | 14.00 | 12.80 | 8.57% | 12.54 | 11.70 | 6.70% |
| Subject 6 | 24.19 | 22.20 | 8.23% | 18.91 | 18.78 | 0.69% | 18.44 | 18.85 | −2.22% |
Measurements averaged over 30 s (4–6 cycles).
4. Discussion
In this work, we proposed a 4D‐MRI method based on 3D ROCK Cartesian sampling and retrospective k‐space data binning based on a respiratory motion self‐gating signal. The preliminary results on phantom and healthy volunteers demonstrated that the proposed method could be used to quantitatively assess the respiratory motion in abdominal organs. This approach may represent a significant improvement over conventional multislice 2D dynamic MRI approaches that have been developed over the past decade due to its intrinsic 3D encoding, higher SNR and absence of miss‐registration issues between slices. These features could translate to significant benefits for MRI‐based radiotherapy treatment planning.
The custom‐made programmable motion phantom was able to generate parametric and reproducible motion to quantitatively validate the proposed 4D‐MRI technique. The motion configurations used in the study were analogous to the respiratory motion of human subjects in terms of amplitude and cycle. The triangular waveform offered uniform spatial distribution over entire motion range so that the difference in motion amplitude measurements due to data binning could be predicted. The 4D‐MRI images were reconstructed into eight respiratory positions and therefore the amplitude measurement based on those positions was expected to be 12.5% less than the ground truth. Our result shows that those measurements were 11.89% less than the ground truth, a good match with the theoretical analysis. Furthermore, the trapezoid waveform used in the study has nonuniform spatial distribution in the first and last positions with bias toward the peak. Accordingly, the motion amplitude measurements in these configurations were 6.22% less than the ground truth. These results demonstrated the accuracy of motion measurements based on the proposed 4D‐MRI technique.
Our volunteer study results showed a relatively large difference between motion amplitude measurements based on 4D‐MRI and those based on 2D‐CINE. As demonstrated in Fig. 5 and our breathing cycle analysis results, we speculate that the larger breathing cycle and breathing motion amplitude variabilities associated with the longer 5‐min 4D‐MRI scan are the main reason for this difference. In addition, the relative large voxel size (1.8 × 1.8 × 6 mm3) of 2D‐CINE is also a potential source of error for the measurement based on 2D‐CINE. On the other hand, the aforementioned 4D‐MRI binning effect, as demonstrated in the motion phantom experiment, could contribute to the measurement error of 4D‐MRI. Such an error can be reduced by increasing the number of respiratory bins during the reconstruction.
One of the key components in the proposed 4D‐MRI method is the Cartesian ROCK sampling pattern. Several Cartesian‐based, motion‐imaging methods have been proposed recently and applied to abdominal31 and neuro imaging32 applications. Similar to these methods, the ROCK used golden angle rotation of k‐space sampling trajectory to ensure near‐uniform angular distribution after retrospective data binning. However, our ROCK approach differs from these previous approaches. First, the Cartesian k‐space is reordered using quasispiral instead of quasiradial trajectories. We speculate that the quasispiral trajectories may provide k‐space sampling that is more incoherent, especially when an anisotropic ky−kz matrix is used. Second, the Cartesian grid was divided into several rings and the size of each ring controls the sampling density in the radial direction. This design enables a more efficient variable density k‐space sampling where the center region is more densely sampled than the outer region. We showed several examples of the k‐space sampling pattern after data binning that demonstrate the benefits of these designs.
In the proposed 4D‐MRI method, the self‐gated respiratory motion surrogate is derived from the k‐space centerline of the imaging volume. Compared with motion surrogates based on external devices (e.g., respiratory bellows), the k‐space centerlines provide information directly from the imaging object and therefore may be theoretically more accurate and reliable. In addition, the acquisition of k‐space centerline is fully integrated within the ROCK sampling pattern so that the magnetization steady state is not disrupted by the acquisition of k‐space centerlines, whereas another commonly used alternative technique, diaphragm navigator, usually creates dark bands over the liver. The accuracy of k‐space centerline as motion surrogate was demonstrated in our phantom experiments where the derived motion signal matches well with the programmed motion waveform.
It is noteworthy that the data binning used in the proposed method is based on the amplitude of the respiratory motion rather than the phase (i.e., time within the respiratory cycle). The major benefit of amplitude‐based data binning is that all acquired data can be used in the reconstruction even in the presence of varying breathing patterns. Our result shows that the amplitude and position of the breathing cycles within the 5‐min 4D‐MRI scan were highly variable even in these healthy volunteers (averaged NSD of amplitude and position were 15.78% and 11.68%, respectively). For instance, two pieces of data acquired during a deep inspiration and a shallow inspiration would have been included together using respiratory phase‐based binning methods, and resulted in blurring and ghosting artifacts in the reconstructed images. Therefore, certain data rejection strategies are usually used to exclude “outlier” breathing cycle in phase‐based binning methods. In addition to compromised scan efficiency, the 4D images after outlier data rejection may no longer accurately represent the true breathing pattern during radiotherapy. On the other hand, the amplitude‐based approach provides only position information, whereas the phase‐based approach provides both position and temporal information within a breathing cycle. In addition, amplitude‐based approaches could fail to capture the well‐known hysteresis effects, which is a limitation that warrants further investigations.
The ROCK sampling could provide k‐space bins with near‐uniform sampling in the azimuthal direction and variable density sampling in the radial direction. However, the actual sampling rate of each k‐space bin also depends on the breathing pattern. A heterogeneous breathing pattern could result in inconsistent k‐space undersampling rates and, consequently, variations in image quality among different respiratory positions. In the proposed method, we addressed this issue by using a motion regularized image reconstruction where all 4D images were reconstructed using a joint equation with a regularization term that penalizes differences among images of different respiratory positions. This concept was proposed by Feng et al.27 for accelerations in cardiovascular imaging applications. Our preliminary result shows that this method could compensate for the inconsistency in k‐space undersampling rate and results in 4D images with more consistent quality across the respiratory cycle. However, the total variation regularization used in the proposed method may not accurately model the underlying respiratory motion. Therefore, the image quality could be compromised in extreme cases of very highly undersampled k‐space.
The proposed method is based on 3D Cartesian sampling that holds several technical and practical advantages over non‐Cartesian sampling like 3D radial. First, the imaging field‐of‐view of Cartesian sampling is more flexible, whereas the 3D radial sampling is limited to isotropic field‐of‐views. For imaging application in the abdomen, an isotropic field‐of‐view may inevitably include regions outside the body and potentially compromise the scan efficiency. Moreover, Cartesian sampling is less sensitive to hardware imperfections. Additional data manipulation, including gradient delay correction and phase correction, is warranted to remove imaging artifacts in reconstructing non‐Cartesian dataset and these techniques are usually designed specifically for certain systems and are not widely applicable. In Cartesian sampling, the behaviors of these hardware imperfections are quite different and hardly noticeable in the reconstructed images. In addition, the computation power required to reconstruct Cartesian dataset is much less than those required for a non‐Cartesian dataset. This problem becomes more prominent when iterative image reconstruction algorithms like compressed sensing are used. In this study, all data binning and image reconstruction algorithms were finished in 10 min on a conventional PC, which is currently not possible if non‐Cartesian sampling was used.33, 34 Furthermore, the proposed 4D‐MRI could potentially enable adaptive motion management in MRI‐guided radiation therapy suite,35 where the low‐latency image reconstruction is highly desirable.
The scan efficiency and image quality of the proposed 4D‐MRI could be further improved by optimizing the pulse sequence and image reconstruction. First, the sequence applies fat saturation pulses before every quasi‐spiral read‐out. This is because subcutaneous fat is associated with high signal, which could undermine the motion estimation accuracy and sensitivity of the self‐gating projections. However, we acknowledge that such implementation is not optimal since only half of the scan time is spent on actual k‐space sampling. This issue could be mitigated by reducing the frequency of fat saturation pulses, or completely solved by using fat suppression techniques that does not interrupt the steady state (e.g., fat‐water separation or water only excitation). Second, the data processing and image reconstruction time, which currently takes approximately 10 min, could be further reduced by optimizing the algorithm and using more powerful computing systems with improved parallel processing capabilities. Furthermore, the performance of the image reconstruction algorithm could be improved by incorporating a respiratory motion model into the reconstruction framework. Odille et al.36 first proposed to simultaneously estimate the images and the motion model by solve two coupled inverse problems. Several recent works37, 38 combined this approach with compressed sensing and non‐Cartesian sampling, and were able to reconstruct images based on up to 16X undersampled k‐space.
Our study has limitations. The customized motion phantom can only generate simple translational motion in one dimension. Due to the lack of gold‐standard for in vivo quantitative motion measurements, it is highly desirable to further validate the proposed technique using phantoms with complex nonrigid motion.39, 40 Furthermore, our in vivo study only included six healthy volunteers. Despite the relative small number of subjects, we were unable to use the proposed techniques to characterize the motion of abdominal tumors. Future work includes clinical studies to validate the proposed technique on patients and to compare it against 4D‐CT, which is the current clinical standard.
5. Conclusion
The proposed 4D‐MRI technique could provide high‐resolution, high‐quality respiratory motion‐resolved 4D images with good soft‐tissue contrast and are free of the “stitching” artifacts usually seen on 4D‐CT and 4D MRI based on resorting 2D‐CINE. Results from motion phantom and volunteer studies demonstrated the feasibility of using proposed 4D‐MRI technique to visualize and quantify high‐resolution abdominal organ motion for MRI‐based radiation treatment planning.
Conflicts of interest
The authors have no relevant conflicts of interest to disclose.
Supporting information
Video S1. Movie of the proposed 4D‐MRI method acquired in a healthy volunteer.
Acknowledgments
The authors acknowledge funding support from the National Institutes of Health under the award numbers R01HL127153 and R01CA188300. The authors thank Yuanfei Jiang, for her help in the design and implementation of the MRI‐compatible motion phantom.
References
- 1. Nelson C, Balter P, Morice RC, et al. Evaluation of tumor position and PTV margins using image guidance and respiratory gating. Int J Radiat Oncol Biol Phys. 2010;76:1578–1585. [DOI] [PubMed] [Google Scholar]
- 2. Keall P. 4‐dimensional computed tomography imaging and treatment planning. Semin Radiat Oncol. 2004;14:81–90. [DOI] [PubMed] [Google Scholar]
- 3. Low DA, Nystrom M, Kalinin E, et al. A method for the reconstruction of four‐dimensional synchronized CT scans acquired during free breathing. Med Phys. 2003;30:1254–1263. [DOI] [PubMed] [Google Scholar]
- 4. Vedam SS, Keall PJ, Kini VR, Mostafavi H, Shukla HP, Mohan R. Acquiring a four‐dimensional computed tomography dataset using an external respiratory signal. Phys Med Biol. 2003;48:45–62. [DOI] [PubMed] [Google Scholar]
- 5. Cai J, Chang Z, Wang Z, Paul Segars W, Yin F‐F. Four‐dimensional magnetic resonance imaging (4D‐MRI) using image‐based respiratory surrogate: a feasibility study. Med Phys. 2011;38:6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu Y, Caruthers SD, Low DA, Parikh PJ, Mutic S. Respiratory amplitude guided 4‐dimensional magnetic resonance imaging. Int J Radiat Oncol Biol Phys. 2013;86:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Y, Yin F‐F, Chang Z, et al. Investigation of sagittal image acquisition for 4D‐MRI with body area as respiratory surrogate. Med Phys. 2014;41:101902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tryggestad E, Flammang A, Han‐Oh S, et al. Respiration‐based sorting of dynamic MRI to derive representative 4D‐MRI for radiotherapy planning. Med Phys. 2013;40:51909. [DOI] [PubMed] [Google Scholar]
- 9. von Siebenthal M, Székely G, Gamper U, Boesiger P, Lomax A, Cattin P. 4D MR imaging of respiratory organ motion and its variability. Phys Med Biol. 2007;52:1547–1564. [DOI] [PubMed] [Google Scholar]
- 10. Remmert G, Biederer J, Lohberger F, Fabel M, Hartmann GH. Four‐dimensional magnetic resonance imaging for the determination of tumour movement and its evaluation using a dynamic porcine lung phantom. Phys Med Biol. 2007;52:N401–N415. [DOI] [PubMed] [Google Scholar]
- 11. Deng Z, Pang J, Yang W, et al. Four‐dimensional MRI using three‐dimensional radial sampling with respiratory self‐gating to characterize temporal phase‐resolved respiratory motion in the abdomen. Magn Reson Med. 2016;75:1574–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Winkelmann S, Schaeffter T, Koehler T, Eggers H, Doessel O. An optimal radial profile order based on the golden ratio for time‐resolved MRI. IEEE Trans Med Imaging. 2007;26:68–76. [DOI] [PubMed] [Google Scholar]
- 13. Chan RW, Ramsay EA, Cheung EY, Plewes DB. The influence of radial undersampling schemes on compressed sensing reconstruction in breast MRI. Magn Reson Med. 2012;67:363–377. [DOI] [PubMed] [Google Scholar]
- 14. Larson AC, Kellman P, Arai A, et al. Preliminary investigation of respiratory self‐gating for free‐breathing segmented cine MRI. Magn Reson Med. 2005;53:159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larson AC, White RD, Laub G, McVeigh ER, Li D, Simonetti OP. Self‐gated cardiac cine MRI. Magn Reson Med. 2004;51:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han F, Zhou Z, Han E, et al. Self‐gated 4D multiphase, steady‐state imaging with contrast enhancement (MUSIC) using rotating cartesian K‐space (ROCK): validation in children with congenital heart disease. Magn Reson Med. 2016; doi: 10.1002/mrm.26376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zijlstra F, Viergever MA, Seevinck PR. Evaluation of variable density and data‐driven k‐space undersampling for compressed sensing magnetic resonance imaging. Invest Radiol. 2016;51:410–419. [DOI] [PubMed] [Google Scholar]
- 18. Crowe ME, Larson AC, Zhang Q, et al. Automated rectilinear self‐gated cardiac cine imaging. Magn Reson Med. 2004;52:782–788. [DOI] [PubMed] [Google Scholar]
- 19. Han F, Zhou Z, Rapacchi S, Nguyen KL, Finn JP, Hu P. Segmented golden ratio radial reordering with variable temporal resolution for dynamic cardiac MRI. Magn Reson Med. 2016;76:94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lustig M, Donoho D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58:1182–1195. [DOI] [PubMed] [Google Scholar]
- 21. Hu P, Hong S, Moghari MH, et al. Motion correction using coil arrays (MOCCA) for free‐breathing cardiac cine MRI. Magn Reson Med. 2011;66:467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uecker M, Lai P, Murphy MJ, et al. ESPIRiT‐an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA. Magn Reson Med. 2013;71:990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forman C, Piccini D, Grimm R, Hutter J, Hornegger J, Zenge MO. Reduction of respiratory motion artifacts for free‐breathing whole‐heart coronary MRA by weighted iterative reconstruction. Magn Reson Med. 2015;73:1885–1895. [DOI] [PubMed] [Google Scholar]
- 24. Johnson KM, Block WF, Reeder SB, Samsonov A. Improved least squares MR image reconstruction using estimates of k‐space data consistency. Magn Reson Med. 2012;67:1600–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng JY, Zhang T, Ruangwattanapaisarn N, et al. Free‐breathing pediatric MRI with nonrigid motion correction and acceleration. J Magn Reson Imaging. 2015;42:407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Donoho DL. De‐noising by soft‐thresholding. IEEE Trans Inf Theory. 1995;41:613–627. [Google Scholar]
- 27. Feng L, Axel L, Chandarana H, Block KT, Sodickson DK, Otazo R. XD‐GRASP: golden‐angle radial MRI with reconstruction of extra motion‐state dimensions using compressed sensing. Magn Reson Med. 2016;75:775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Z, Han F, Rapacchi S, et al. Accelerated ferumoxytol‐enhanced 4D multiphase, steady‐state imaging with contrast enhancement (MUSIC) cardiovascular MRI: validation in pediatric congenital heart disease. NMR Biomed. 2017; doi: 10.1002/nbm.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uecker M, Ong F, Tamir JI, et al. Berkeley advanced reconstruction toolbox. In: ISMRM 23rd Sci. Sess, Toronto, Canada, 2015: 23: 2486. [Google Scholar]
- 30. Du D, Caruthers SD, Glide‐Hurst C, et al. High quality t2‐weighted 4D magnetic resonance imaging for radiation therapy applications. Int J Radiat Oncol Biol Phys. 2015;92:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Küstner T, Würslin C, Schwartz M, et al. Self‐navigated 4D cartesian imaging of periodic motion in the body trunk using partial k‐space compressed sensing. Magn Reson Med. 2016; doi: 10.1002/mrm.26406. [DOI] [PubMed] [Google Scholar]
- 32. Zhu Y, Guo Y, Lingala SG, Lebel RM, Law M, Nayak KS. GOCART: golden‐angle cartesian randomized time‐resolved 3D MRI. Magn Reson Imaging. 2016;34:940–950. [DOI] [PubMed] [Google Scholar]
- 33. Chandarana H, Feng L, Block TK, et al. Free‐breathing contrast‐enhanced multiphase MRI of the liver using a combination of compressed sensing, parallel imaging, and golden‐angle radial sampling. Invest Radiol. 2013;48:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pang J, Bhat H, Sharif B, et al. Whole‐heart coronary MRA with 100% respiratory gating efficiency: self‐navigated three‐dimensional retrospective image‐based motion correction (TRIM). Magn Reson Med. 2014;71:67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang Y, Cao M, Sheng K, et al. Longitudinal diffusion MRI for treatment response assessment: preliminary experience using an MRI‐guided tri‐cobalt 60 radiotherapy system. Med Phys. 2016;43:1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Usman M, Atkinson D, Odille F, et al. Motion corrected compressed sensing for free‐breathing dynamic cardiac MRI. Magn Reson Med. 2013;70:504–516. [DOI] [PubMed] [Google Scholar]
- 37. Küstner T, Würslin C, Schmidt H, Yang B. Combining compressed sensing with motion correction in acquisition and reconstruction for PET/MR. In: 2015 IEEE Int. Conf. Acoust. Speech Signal Process, Brisbane, Australia; 2015: 788–792. [Google Scholar]
- 38. Rank CM, Heußer T, Buzan MTA, et al. 4D respiratory motion‐compensated image reconstruction of free‐breathing radial MR data with very high undersampling. Magn Reson Med. 2017;77:1170–1183. [DOI] [PubMed] [Google Scholar]
- 39. Würslin C, Küstner T, Bartel S, Schwenzer N, Yang B, Schmidt H. A simple‐to‐build and cost‐efficient MR‐compatible phantom for the simulation of non‐rigid motion. In: Proc. ISMRM Motion Correct. MRI Work, Tromsø, Norway, 2014. [Google Scholar]
- 40. Fieseler M, Kugel H, Gigengack F, et al. A dynamic thorax phantom for the assessment of cardiac and respiratory motion correction in PET/MRI: a preliminary evaluation. Nucl Instruments Methods Phys Res Sect A Accel Spectrometers Detect Assoc Equip. 2013;702:59–63. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Movie of the proposed 4D‐MRI method acquired in a healthy volunteer.


