Abstract
A surface directly connects the bulk of a material to the surrounding. The ability to regulate dynamically the surface without affecting the bulk of a material holds great potential for new applications. The purpose of this work was to demonstrate that the surface can be dynamically changed using nanoparticles and oligonucleotides (ODNs) in a reversible and reiterative manner. A dual-functional nanogel has been synthesized as the model of nanoparticles using miniemulsion polymerization and click chemistry. The nanogel can not only adsorb drugs for sustained drug release, but also bind a surface functionalized with complementary ODNs. Importantly, hybridization reaction and ODN degradation can drive reversible and reiterative nanogel binding to the surface for dynamic change, which in principle is unlimited. Moreover, nanogel-mediated dynamic change offers the surface with the drug-releasing function for inhibiting the growth of surrounding cells. Since nanogels can be replaced by any functional nanoparticles with a diverse array of properties, nanoparticle-programmed surface change constitutes a promising platform for various applications such as drug delivery and stent implantation.
Keywords: nanoparticles, DNAs, surfaces, hydrogels, cell regulation
Graphical abstract
1. Introduction
Synthesis of dynamic materials is important for variouos applications such as human healthcare, military camouflage, industrial catalysis, and sensor development.1–6 Their properties can be facily regulated to change under the stimulation of light, temperature, pH, mechanical loads, or electrical potentials.7–11 However, most of these materials are designed to change their bulk entirely once stimulated. Since a surface directly connects the bulk to the surrounding, it is possible to change dynamically the function of a surface rather than the underlying bulk.
A surface can be passively changed through non-specific molecular interactions.12–14 For instance, proteins and lipids can be rapidly adsorbed to a synthetic surface,15,16 which subsequently changes the initial properties of the materials. However, these passive non-specific changes are often harmful in applications.17–19 Thus, efforts have been made for surface functionalization to achieve specific changes.20,21 Both synthetic and natural molecules have been studied for this purpose. For instance, temperature-responsive poly (N-isopropylacrylamide) (PNIPAM) has been successfully used to produce dynamic surfaces for hydrophilic-hydrophobic conformational change under the stimulation of temperature variation; and peptide-functionalized surfaces have been studied for dynamic regulation of cell attachment under the stimulation of light.22,23 These elegant studies focused on the functionalization of the surfaces with molecules and the dynamic changes of the surfaces were only regulated at the molecular levels. Few studies have been carried out to develop a dynamic surface at the nanoscale level using both biomolecules and nanoparticles.
Nanoparticles can be made of polymers, metals, semiconductors, or oxides with a diverse array of mechanical, electrical, optical, magnetic, chemical, and biological properties.24–27 Moreover, they can be physically or chemically functionalized with biomolecules such as peptides, antibodies, and nucleic acids.28–30 Based on the functionalization, nanoparticles can acquire the function of molecular recognition. Notably, while nanoparticles often exhibit unique properties and functions different from bulk materials, they still possess certain key features of bulk materials. For instance, nanoparticles retain the ability of loading and releasing drugs.31,32 Thus, nanoparticles can act as a bridge between molecules and surfaces, which would be of great interest to various fields.
The purpose of this study was to demonstrate that nanoparticles and DNAs can be used to develop a novel platform for reversibly controlling the dynamic change of a surface, whcih has not been investigated. Nanoparticles and the surface are both functionalized with ODNs that form a complementary duplex. When DNA-functionalized nanoparticles meet a complementary ODN-grafted surface, they will be adsorbed onto the surface through robust hybridization and therefore change the function of the surface. Importantly, the nanoparticles can be desorbed from the surface for dynamic changes since nucleic acid hybridization can be reversed using numerous methods such as strand displacement, temperature variation and nuclease degradation.33–35 Therefore, de novo designed ODNs can precisely and actively regulate specific adsorption and desorption of nanoparticles at the solid-solution interface for dynamic changes. To demonstrate the concept and its potential applications, we synthesized a DNA-functionalized dual-functional nanogel and applied it to program a hydrogel surface for drug release and cell inhibition.
2. Results and Discussion
2.1. Synthesis and characterization of DNA-functionalized nanogels
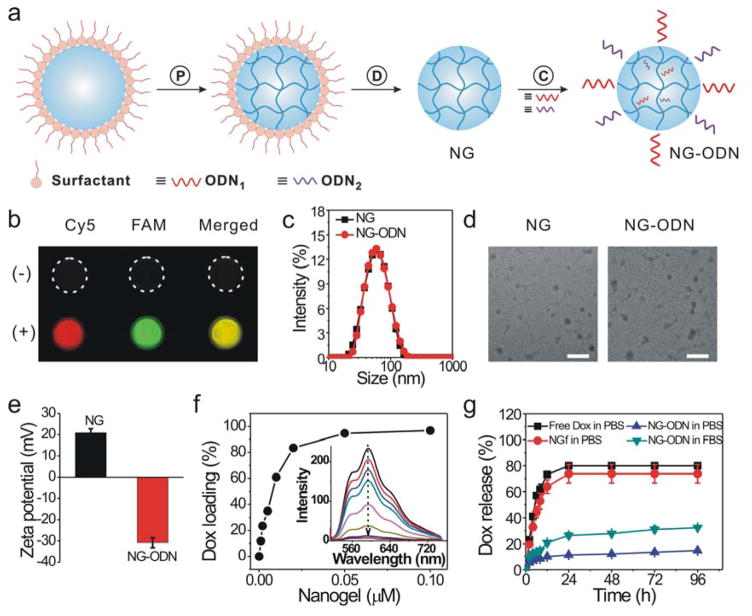
The nanogel was synthesized via the inverse miniemulsion polymerization method (Figure 1a). The aqueous phase containing acrylamide, N-(3-azidopropyl)methacrylamide and bisacrylamide was dispersed in a continuous organic phase for APS-initiated free radical polymerization. After the removal of the surfactants, the nanogels with azide groups were obtained for conjugation. To functionalize the nanogels with DNA, the 5′ ends of ODN1 and ODN2 were modified with the alkyne group for alkyne-azide cycloaddition. If these two ODN molecules were conjugated with the nanogels, the nanogels would be able to bind their complementary ODNs. Thus, to verify the success of conjugation, the nanogels were treated with Cy5-cODN1 and FAM-cODN2 that were complementary to ODN1 and ODN2, respectively. As shown in Figure 1b, the nanogels conjugated with ODNs displayed strong red and green fluorescence signals corresponding to Cy5-cODN1 and FAM-cODN2, respectively. However, the nanogels without DNA conjugation displayed very weak fluorescence signals similar to that of the background. These data show that ODN1 and ODN2 were successfully conjugated to the nanogels via click chemistry.
Figure 1.
Synthesis and characterization of the dual-functional nanogel. (a) Schematic illustration. P: polymerization of nanodroplet in emulsion; D: dialysis for the removal of surfactant from the nanogel; C: click chemistry for conjugation of oligonucleotide (ODN) to nanogel. (b) Fluorescence images of nanogels functionalized with DNA via click reaction (+). Nanogels physically mixed with DNA was used as control (−). The nanogels were treated with Cy5-cODN1 and FAM-cODN2 for fluorescence imaging. (c) Dynamic light scattering analysis. NG: native nanogel; NG-ODN: nanogel conjugated with ODN1 and ODN2. (d) TEM images of nanogels. Scale bar: 100 nm. (e) Zeta potential of nanogels. (f) Quantification of Dox loading into nanogel. Dox of 5 μM was incubated with a varied amount of nanogels. The inset figure shows the fluorescence spectra of Dox sequestration by ODN-functionalized nanogels. (g) Dox release from ODN-functionalized nanogels. The release test was performed in a dialysis tube. Free Dox: release of free Dox from the Dox solution. NGf: release of Dox after Dox was mixed with nanogels that were free of ODN. NG-ODN: release of Dox from the ODN-functionalized nanogels.
The nanogels were subsequently characterized in size, morphology and surface charge. The native nanogels had an average hydrodynamic diameter of 54 nm (Figure 1c). After the ODN conjugation, the size of the nanogels slightly increased to 56 nm. The TEM images show that the nanogels before and after DNA conjugation were similar in size and were ~40 nm on average in diameter (Figure 1d). This difference between the DLS and TEM tests may be due to the different states of the nanogels. In the DLS test, the nanogels were dispersed in the aqueous solution whereas they were air-dried before the TEM test. After water evaporation, the nanogels would shrink with a decreased diameter. The data also show that the native nanogels were positively charged with a value of 21 mV (Figure 1e). After conjugation with DNA, the nanogels became negatively charged as their value of zeta potential was −31 mV. This result is consistent with the fluorescence imaging analysis (Figure 1b), showing that ODN could be conjugated to the nanogels.
2.2. Drug loading into and release from DNA-functionalized nanogels
The nanogels had two functions. One of them was to load drugs for drug release and cell regulation. The function of loading and releasing drugs can in principle be realized by using a number of nanoparticles such as polymeric nanoparticles, polymeric micelles, and lipid-based nanoparticles.36–38 We synthesized the nanogels for drug release since nanogels can benefit potential applications for their hydrophilicity, high water absorptivity, versatility and biocompatibility.39,40 However, nanogels are highly permeable in nature like bulk hydrogels, which may not allow for sufficient drug loading within the nanogel matrix. To solve this problem, we functionalized the nanogels with double-stranded DNAs.
Small molecules or large biomolecules can strongly bind natural nucleic acids through electrostatic interactions, minor or major groove-binding, or intercalation.41–44 Synthetic ODNs could also be designed and selected from DNA or RNA libraries to bind drugs with high binding affinity and specificity.45,46 For example, short ODNs have been selected from the libraries against small molecules such as tobramycin and streptomycin.47,48 Thus, ODN-functionalized nanogels would provide a great diversity for loading drugs that can be either a small molecule, a large biomolecule or even both.
Previous studies have shown that doxorubicin (Dox) can be intercalated into double-stranded 5′-GC-3′ and 5′-CG-3′ sites effectively.49,50 Thus, we designed a DNA duplex with multiple GC pairs (Table S1) as a Dox-binding effector for the functionalization of the nanogels. Since Dox intercalation into double-stranded DNA is accompanied by the decrease of Dox fluorescence intensity, the efficiency of Dox loading into nanogels was measured by monitoring the fluorescence intensity of the Dox solution (Figure 1f). The loading efficiency virtually increased linearly when the concentration of the nanogels increased from 0 to 0.01 μM. At 0.02 μM, the loading efficiency was 83.5%. After 0.02 μM, the loading efficiency increased slowly. Thus, the concentration at 0.02 μM was used in the following experiments unless otherwise stated.
We further examined the kinetics of Dox release from the nanogels using a dialysis setup. The two control groups including the free Dox solution and the native nanogel solution both exhibited a rapid burst release (Figure 1g). The percentage of Dox release reached 80% and 74% in these two groups within the first day, respectively. By contrast, the release of Dox from the ODN-functionalized nanogels was 11% in PBS by day 1. The release results also show that the release in FBS was faster than that in PBS since FBS contained nuclease that could hydrolyze ODNs for faster release. The release of Dox in FBS was 26% in the first day. After the first-day release, Dox was gradually released from the nanogels in the FBS medium. These results suggest that ODN-functionalized nanogels could be used as an effective nanoparticle system for loading and releasing Dox.
2.3. Nanogel adsorption on the surface
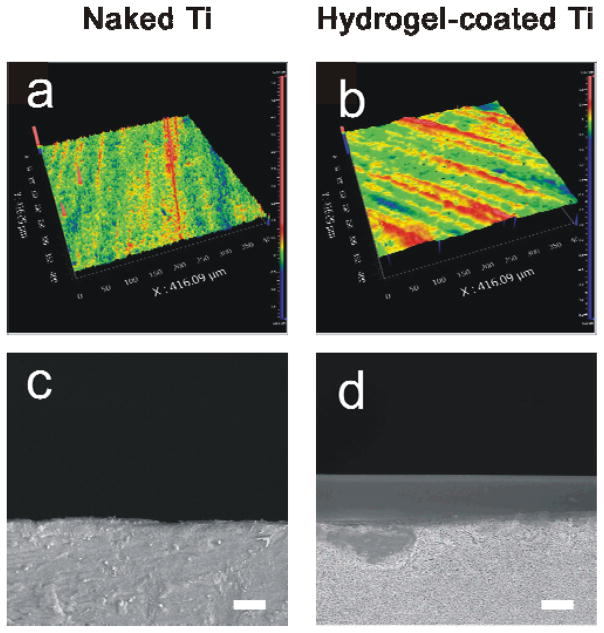
The other function of the nanogels was to bind the hydrogel surface for dynamic change. This binding function was also realized using intermolecular hybridization reaction. Two complementary ODNs, ODN1 and cODN1, were used to functionalize the nanogels and the hydrogel surface, respectively. Hydrogel has been studied as a coating material on the surfaces of different substrates.51–53 Moreover, it is easy to incorporate chemically acrydite-functionalized ODNs into the hydrogel network via free radical polymerization.54 Thus, we used hydrogel and Ti as a coating material and a bulk support, respectively, as the model system in this work. The reason for using Ti as the bulk support is that Ti is an important material for a variety of applications.55–59 Its surface functionalization has been an important task for those applications. For instance, the surface of Ti has been functionalized with biomolecules to promote cell proliferation or with thin polymeric films for drug release in inhibiting cell proliferation,60,61 which depends on the requirements of the applications. Thus, the use of hydrogel and Ti as the model system will not only demonstrate the concept, but may make a direct impact on related potential applications such as stent implantation for treatment of diseased peripheral or coronary arteris. The hydrogel surface was characterized using optical profilometry and SEM. The hydrogel layer appeared smooth with a thickness of 10 μm (Figure 2). To verify the incorporation of cODN1 into the hydrogel coating, the hydrogel was treated with FAM-ODN1, which was complementary to cODN1. As shown in Figure S2, the hydrogel surface with cODN1 displayed a much stronger fluorescence signal than that of the coating without cODN1. These data clearly showed that the acrydite-modified cODN1 was incorporated into the hydrogel coating via free radical polymerization, and also indicated that a nanogel with ODN1 would attach to the cODN1-functionalized hydrogel surface.
Figure 2.
Characterization of the surfaces. Optical (a, b) and SEM (c,d) images of Ti surface and hydrogel-coated Ti surface. Scale bar: 10 μm.
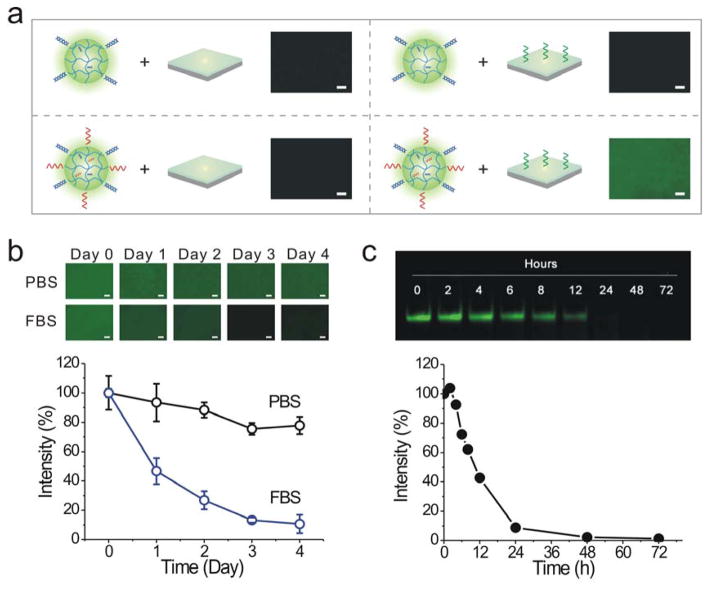
To demonstrate directly whether the nanogels could bind the hydrogel surface via DNA hybridization, fluorescein o-methacrylate was incorporated into the nanogels for clear observation. The strong green fluorescence could only be observed in the group that involved both the cODN1-functionalized hydrogel surface and the ODN1-functionalized nanogels (Figure 3a). By contrast, very weak fluorescence signals were exhibited in the other three control groups. These results clearly demonstrated that the ODN1-functionalized nanogels could specifically bind the cODN1-functionalized hydrogel surface via hybridization reaction. The comparison with the control groups also demonstrated that the dominant mechanism for the dynamic change of the surface resulted from hybridization-mediated nanogel binding rather than the passive diffusion and penetration of nanogels into the hydrogel surface. Taken together, the results showed that the hydrogel surface could be dynamically changed from a nanogel-free state to the nanogel-binding state.
Figure 3.
Nanogel adsorption and release on the surface. (a) Binding of ODN1-functionalized nanogels on the cODN1-functionalized surface. The first three samples are controls. Scale bar: 100 μm. (b) Kinetics of nanogel release from the hydrogel surface. Fluorescein o-methacrylate was incorporated into nanogels during polymerization for fluorescence examination. The nanogel release is indicated by the change of fluorescence intensity. Scale bar: 100 μm. (c) Electrophoretic gel images of free ODN1 incubated in the FBS solution. The degradation profile was generated from the analysis of the electrophoretic gel images.
2.4. Reversible nanogel adsorption and release for dynamic surface change
After examining specific nanogel adsorption on the hydrogel surface, we investigated whether the bound nanogels could be released from the surface. We incubated the nanogel-bound surface in the FBS solution. The result showed that the fluorescence intensity decreased with time (Figure 3b). The fluorescence intensity of the surface decreased to 47% after 1 day of incubation in FBS. Afterwards, the fluorescence intensity further decreased to 27%, 13% and 11% by day 2, day 3 and day 4, respectively. The daily decrease in the first three days was more than 10% whereas the daily decrease was less than 3% after day 3. These results are consistent with the degradation of free ODN1 in FBS (Figure 3c). Therefore, these observations showed that the nanogels could be released from the hydrogel surface via the nuclease degradation of ODN1.
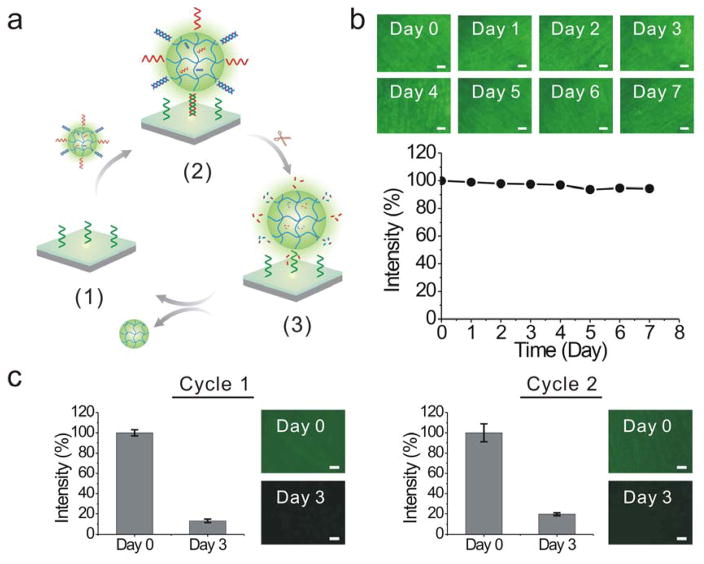
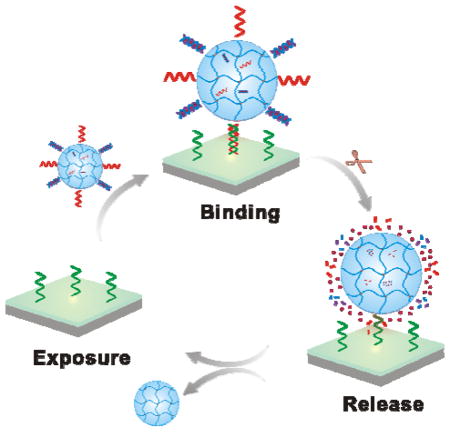
We further studied the reiterative nanogel adsorption and release (Figure 4a). The reversible and reiterative nanogel adsorption and release require the degradation of ODN1 but the stability of cODN1. The cODN1 used in this work was a chemically modified DNA sequence. Its backbone was functionalized with phosphorothioate modifications. The phosphorothioate analog is the most commonly employed synthetic modification against nuclease degradation. This modification significantly improved the stability of DNA against nuclease degradation as shown in the literature62,63 and this study (Figure 4b). In addition to the modification with phosphorothioate bond, a variety of strategies have been developed over the past two decades in the field of nucleic acids research.64–66 For instance, DNAs can be internally modified with amino, fluoro, and o-methyl groups at the 2′-position of sugar, and capped at the 3′ and 5′ ends.65,66 Recently, spiegelmers were synthesized with L-nucleotides that are unnatural to nucleases.67,68 All of these modifications have been demonstrated to improve the stability of ODNs in biological fluids and would be useful for optimization of the dynamic surface if nanoparticle-programmed surfaces would be applied to solve biological or biomedical problems in the future. As most nanogels were released from the surface by day 3, we treated the surface with the fresh nanogel solution at the end of day 3. The procedures for nanogel adsorption and release were the same as the initial treatment. The same trend as observed in the first cycle of test occurred in the second cycle of test (Figure 4c). The fluorescence intensity of the hydrogel surface recovered after the nanogel treatment; and the fluorescence intensity significantly decreased within 3 days. Therefore, these results suggested that the dynamic change of the hydrogel surface was reversible and reiterative.
Figure 4.
Evaluation of reversible and reiterative nanogel adsorption on the surface. (a) Illustration of programmable change of the surface via reiterative nanogel attachment and release. (1): cODN1 exposure; (2) nanogel adsorption; (3) nanogel release. (b) Fluorescence images of cODN1-functionalized hydrogel surface immersed in the FBS solution. The samples were treated with FAM-ODN1 for imaging. Scale bar: 100 μm. The degradation profile was acquired from the analysis of the fluorescence intensity of the surface. (c) Examination of programmable surface change in a two-cycle test. One cycle of test last for three days. Scale bar: 100 μm.
2.5. Nanogel-programmed surface for sustained drug release
After the demonstration of nanoparticle-programmed surface change, we used drug release and cell inhibition as an example to demonstrate the potential applications of this concept. We treated the hydrogel surface with the nanogel solution for two cycles and examined the Dox release kinetics from the hydrogel surface in FBS solution (Figure 5). Each cycle took three days. Dox was released from the hydrogel surface in a sustained manner. The release reached 68% and 77% by 24 h and 72 h in the first cycle of test, respectively. After 72 h, the hydrogel surface was treated with the Dox-loaded nanogels again. While the release rate was slightly higher, the overall release kinetics in the second cycle was very similar to that observed in the first cycle. The Dox release reached 72% and 81% by 24 h and 72 h in the second cycle of test, respectively. These data demonstrated that using nanogels to change the surface programmably and reiteratively could lead to continuous drug release, which is different from current bulk drug delivery systems that will not be able to release drugs once drug release is completed.
Figure 5.
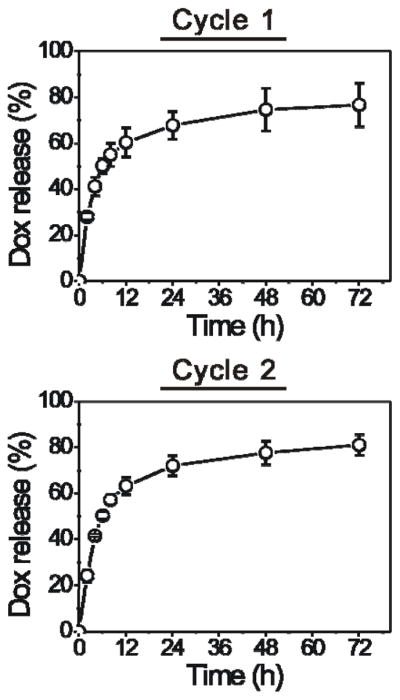
Nanogel-programmed surface change for sustained Dox release in a two-cycle test.
2.6. Drug release for cell inhibition
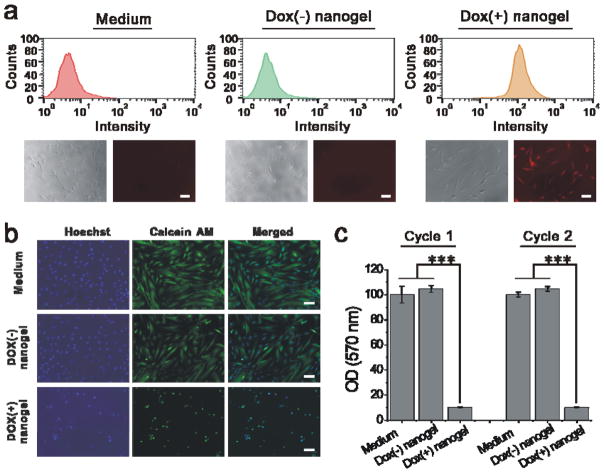
We further evaluated the cell response to Dox release. The nanogel-bound hydrogel surface was co-incubated with SMCs using a transwell cell culture insert. The Dox uptake by the SMCs was analyzed by flow cytometry and fluorescence microscopy. As shown in the flow cytometry histograms (Figure 6a), the cells that were co-incubated with the hydrogel surface treated with the Dox-loaded nanogels has a clear right shift. By contrast, the Dox signal was barely detected in the two control groups. These results were further confirmed by the fluorescence image analysis. A strong Dox signal was observed from the cells that were co-incubated with the hydrogel surface treated with the Dox-loaded nanogels.(Figure 6a). Thus, the data showed that the released Dox could be taken up by the surrounding SMCs. We further observed the effect of Dox uptake on cell morphology under a fluorescence microscope (Figure 6b). In the two control groups, the cells were stretched and elongated on the tissue culture plate. By contrast, the cells that were co-incubated with the hydrogel surface treated with the Dox-loaded nanogels displayed circular shapes or changed into cell debris. We also quantitatively studied the reiterative cell response to the dynamic change of the hydrogel surface. The similar results were observed in the first and second cycles of test (Figure 6c). The cells showed similar viability in the two control groups. However, the cell viability was decreased to 10% in the Dox-loaded nanogel group. These results clearly demonstrated that the dynamic change of the hydrogel surface programmed by the Dox-loaded nanogels could lead to effective inhibition of SMC growth.
Figure 6.
Nanogel-programmed surface for cell inhibition. (a) Upper panel: flow cytometry histograms of smooth muscle cells (SMCs). Lower panel: bright field and fluorescence images of SMCs that were co-incubated with the dynamic surfaces. The surface was treated with nanogels with (+) or without (−) Dox. Scale bar: 100 μm. (b) Fluorescence images of SMCs that were stained with Hoechst 33342 and Calcein AM after 72-h incubation with the dynamic surface. Scale bar: 100 μm. (c) Analysis of cell viability using MTT assay. Error bar: standard deviation (n = 3). *** indicates p < 0.001.
3. Conclusion
In summary, we have synthesized a programmable surface using DNAs and a dual-functional nanogel via reversible hybridization reaction. The surface can display the nanogel reiteratively, which leads to dynamic and active changes of the surface. When binding to the surface, the nanogel offers the surface with the drug-releasing function that can be applied to inhibit the growth of the surrounding cells. While cell inhibition was applied as an example to illustrate the dynamic function of the nanoparticle-programmed surface, it is worth noting that the released drugs can be applied to promote cell growth or induce cell differentiation when a cell-activating drug is used. Importantly, in principle, the nanogel can be replaced with any ODN-functionalized nanoparticles to realize a diverse array of dynamic changes on the surface. Therefore, the nanoparticle-programmed surface change may constitute a general platform for broad applications.
4. Experimental Section
4.1. Reagents
The acrylamide/bis-acrylamide solution (40% w/v; 37.5:1), ammonium persulfate (APS), N,N,N′,N′-tetramethylenediamine (TEMED), dioctyl sulfosuccinate sodium salt (AOT), Brij 30, phosphate buffered saline (PBS) and sodium hydroxide were purchased from Fisher Scientific (GA, USA). Calcein AM, Hoechst 33342, Medium 231 and smooth muscle growth supplement (SMGS) were purchased from Thermo Fisher Scientific (MA, USA). Oligonucleotides (Table S1) were synthesized by Integrated DNA Technologies (IA, USA).
4.2. Preparation of nanogels
Synthesis of N-(3-azidopropyl)methacrylamide69
3-chloropropylamine hydrochloride (6.9 g) and sodium azide (10.2 g) were dissolved and incubated in 80 mL of water at 80 °C. After the reaction for 16 h, the pH of the solution was adjusted to ~11 with NaOH pellets. Ethyl acetate was added to extract the 3-azidopropan-1-amine that was further washed by saturated NaCl solution. The organic layer was dried over Na2SO4 and concentrated under reduced pressure to obtain 3-azidopropan-1-amine, which was used for next step without further purification.
3-azidopropan-1-amine (3 g) was added into the mixture of N,N-Diisopropylethylamine (12 mL) and anhydrous dichloromethane (10 mL) at room temperature and the reaction mixture was incubated in an ice bath for 0.5 h. Then 10 mL of methacryloyl and dichloromethane (1:1) was added into the 3-azidopropan-1-amine solution. After reaction for 16 h in an ice bath, a solution of saturated NaHCO3 was added into the reaction mixture and the organic phase was further extracted by dichloromethane. The extracts were sequentially treated by an HCl solution (0.5 M) and a saturated NaCl solution. After the extracts were dried over Na2SO4, N-(3-azidopropyl)methacrylamide was purified using silica column chromatography. 1H NMR (400 MHz, CDCl3): δ 6.18 (brs, 1H), 5.67 (s, 1H), 5.31 (s, 1H), 3.40–3.35 (m, 4H), 1.94 (s, 3H), 1.81 (quintet, J = 6.4 Hz, 2H) ppm.
Preparation of nanogels
The nanogels were synthesized according to a reported protocol.70 Briefly, 0.9 g acrylamide, 0.12 g N-(3-azidopropyl)methacrylamide and 0.13 g bis-acrylamide were dissolved in 1.1 mL of water. The aqueous phase was emulsified in 25 mL of hexanes containing 0.9304 g AOT with 1.791 g Brij 30. After the emulsion was purged with argon for 30 min, 100 μL of 10% APS and 50 μL of 100% TEMED were added dropwise into the emulsion for the polymerization. The nanogels were loaded into a dialysis bag for dialysis to remove Brij 30. Finally, the nanogels were lyophilized and dispersed in the water. The same procedure was also used to prepare fluorescent-labeled nanogels except that 6 μL of 40% fluorescein o-methacrylate in DMSO solution were added to the aqueous phase.
4.3. Characterization of nanogels
The size and zeta potential of nanogels were measured with Zetasizer Nano ZS (Malvern, USA). For TEM measurement, the nanogel solution was also dropped onto a carbon-coated copper grid and then stained with 1% phosphotungstic acid. After air dry, the sample was measured on a transmission electron microscope (Talos F200X, FEI, Japan).
4.4. Conjugation of ODNs to nanogels via click reaction
Copper-catalyzed Azide-Alkyne cycloaddition was used to conjugate ODNs to nanogels. 50 μL of nanogels in water (7.5 mg/mL) were mixed with 10 nmol hexynyl-modified DNA sequences (ODN1:ODN2 = 3:7). Sodium ascorbate was added into the reaction mixture to a concentration of 10 mM and copper sulfate coupled with THPTA ligand was added to a concentration of 1 mM. The reaction was performed at 30 °C on a shaker for 2 h.
To confirm DNA conjugation to nanogels, Cy5-cODN1 and FAM-cODN2 (i.e., the complementary sequences of ODN1 and ODN2) were added to the solution. After 1 h incubation, centrifugation (14,000 × g) was applied to remove free ODNs using a filter unit (100 k MWCO, Millipore) and the samples were washed for totally four times. The nanogel solution was washed and imaged with a CRI Maestro System (MA, USA). To load DOX, cODN2 was added to the nanogel solution for hybridization with ODN2.
4.5. Evaluation of Dox loading into and release from nanogels
To determine Dox loading into nanogels, Dox of 5 μM was incubated with nanogels at room temperature for 1 h. The fluorescence intensity of the solution was monitored by Nanodrop 3300 fluorospectrometer. The loading efficiency was calculated by normalization of the intensity of the sequestered solutions to that of the initial free Dox solution.
To determine Dox release, 200 μL of Dox-loaded ODN-functionalized nanogels were transferred into a mini dialysis unit (20 k MWCO, Thermo Scientific). An equivalent amount of free Dox with or without native nanogels was prepared as control. The dialysis unit was immersed in 5% FBS solution (1.45 mL) at 37 °C with a shaking speed of 70 rpm. At the predetermined time points, 250 μL of dialysis solution was collected for fluorescence analysis.
4.6. Coating of the hydrogel on Ti surface
A Ti foil (Alfa Aesar, USA) was cut into Ti plates (4.5×4.5 mm). The Ti plates were treated in 5 M NaOH solution at 60 °C for 12 h and then with 1% TMSPMA in the mixture of ethanol and acetic acid. To coat the hydrogel film on the silanized titanium plates, a pregel solution was prepared by mixing 0.1 μL of 10% APS, 0.1 μL of 5% TEMED and 1.2 μL of the mixture of acrylamide/bis-acrylamide (37.5:1) and cODN1 (850 pmol). 1.3 μL of pregel solution was dropped onto a supporting glass and covered by the silanized Ti plate. After polymerization, the Ti plate was gently removed from the glass and washed in PBS. The surfaces of Ti samples with or without the hydrogel film were examined under an Optical Profilometer (Zygo NexView, USA).
4.7. Examination of nanogel-mediated dynamic change of the hydrogel surface
The dynamic change of the hydrogel surface was mediated by nanogel attachment and release on the hydrogel surface. To study nanogel attachment onto the hydrogel surface, 6.5 μL of nanogel solution was dropped onto the parafilm in 24-well plate and covered by the hydrogel-coated Ti plate. After 2 h of incubation, the hydrogel-coated Ti plate was gently washed trice using PBS. To examine the release of nanogels from the surface, the samples were incubated in 700 μL of cell culture medium containing 5% FBS at 37 °C. At the predetermined time points, the samples were removed from the solution and rinsed with PBS. The hydrogel surface was imaged under the fluorescence microscope (Olympus IX73, Japan). To examine the reiterative change of the hydrogel surface, the whole procedure of nanogel loading and release was repeated after the first cycle of nanogel attachment and release.
4.8. Gel electrophoresis
Gel electrophoresis was used to examine the degradation kinetics. The ODN solution was incubated in the culture medium containing 5% FBS at 37 °C. The solutions were collected at the predetermined time points and stored at −20 °C before gel electrophoresis. The medium of ODN1 was mixed with fresh cODN1 at a molar ratio of 1:1.2 for the electrophoresis analysis. The samples were loaded into the wells of 10% polyacrylamide and the electrophoresis was run at 120 V for 40 min. The hybridized duplex was stained with SYBR-safe and imaged with a CRI Maestro System (MA, USA).
4.9. Examination of stability of immobilized cODN1 on the hydrogel surface
To examine the stability of cODN1 after incorporated into the hydrogel surface, the hydrogel samples were immersed in 700 μL of cell culture medium containing 5% FBS at 37 °C. At the predetermined time points, the samples were removed from the medium and thoroughly washed. The samples were incubated with FAM-ODN1 for 1 h, washed thoroughly with PBS and imaged under the fluorescence microscope (Olympus IX73, Japan). The intensity of the samples was analyzed using ImageJ software.
4.10. Examination of Dox release from the hydrogel surface
To examine Dox release from the nanogel-programmed hydrogel surface, the hydrogel-Ti samples were put into a mini dialysis unit (20 k MWCO, Thermo Scientific) with 200 μL of 5% FBS solution. The unit was incubated in 550 μL of 5% FBS solution at 37 °C and shaken at a speed of 70 rpm. At the predetermined time points, the dialysis solution was collected for analysis and replaced with an equivalent volume of fresh solution.
4.11. Examination of Dox uptake by cells
Human Aortic Smooth Muscle Cells (SMCs) were cultured in Medium 231 supplemented with SMGS containing 5% FBS at 37 °C in a humid atmosphere with 5% CO2. The cells were seeded into a 24-well plate with a density of 1 × 104 cells/well and cultured overnight. Then, the cells were co-incubated with the hydrogel-functionalized Ti plates that were put into the transwell inserts. After 12-h incubation, the cells were washed with PBS, treated with 0.05% trypsin and resuspended in PBS for the flow cytometry analysis (Guava easyCyte, Germany). A total of 5000 events were collected and the data was analyzed with FlowJo V10. To further confirm the Dox uptake, the cells were also imaged under the fluorescence microscope (Olympus IX73, Japan).
4.12. Examination of cell morphology and viability
The cells were stained with Hoechst 33342 (2.5 μg/mL) and Calcein AM (1 μM) and imaged under the fluorescence microscope (Olympus IX73, Japan). The cells were also examined using the MTT assay. The MTT solution (5 mg/mL) was added into each well and incubated with the cells for 4 h. The absorbance at 570 nm was measured via a microplate reader (Infinite M200, Tecan, Switzerland). The values of absorbance were normalized to that of the control without Dox treatment.
4.13. Statistical analysis
The data were showed as a mean ± standard deviation. Statistical comparisons were performed by using student’s t-test. P-value < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
This work was supported in part by the U.S. NSF (DMR-1322332), and the National Heart, Lung, and Blood Institute of the NIH (R01HL122311). We thank the Husk Institute Microscopy Facility (University Park, PA) for technical support. P. J. also acknowledges the China Scholarship Council for support.
Footnotes
H NMR spectrum, fluorescence images of hydrogel surfaces and a table of DNA sequences.
Supporting Information is available on the ACS Publications website.
References
- 1.Huebsch N, Mooney DJ. Inspiration and application in the evolution of biomaterials. Nature. 2009;462:426–432. doi: 10.1038/nature08601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpi F, Frediani G, Turco S, Rossi DD. Bioinspired Tunable Lens with Muscle-Like Electroactive Elastomers. Adv Funct Mater. 2011;21:4152–4158. [Google Scholar]
- 3.Yoshida M, Lahann J. Smart Nanomaterials. ACS Nano. 2008;2:1101–1107. doi: 10.1021/nn800332g. [DOI] [PubMed] [Google Scholar]
- 4.Schurig D, Mock JJ, Justice BJ, Cummer SA, Pendry JB, Starr AF, Smith DR. Metamaterial Electromagnetic Cloak at Microwave Frequencies. Science. 2006;314:977–980. doi: 10.1126/science.1133628. [DOI] [PubMed] [Google Scholar]
- 5.Bergbreiter DE, Mariagnanam VM, Zhang L. Polymer ligands that can regulate reaction temperature in “smart” catalysts. Adv Mater. 1995;7:69–71. [Google Scholar]
- 6.Su B, Guo W, Jiang L. Learning from Nature: Binary Cooperative Complementary Nanomaterials. Small. 2015;11:1072–1096. doi: 10.1002/smll.201401307. [DOI] [PubMed] [Google Scholar]
- 7.Kim SH, Kang EB, Jeong CJ, Sharker SM, In I, Park SY. Light Controllable Surface Coating for Effective Photothermal Killing of Bacteria. ACS Appl Mater Interfaces. 2015;7:15600–15606. doi: 10.1021/acsami.5b04321. [DOI] [PubMed] [Google Scholar]
- 8.Roy D, Brooks WLA, Sumerlin BS. New directions in thermoresponsive polymers. Chem Soc Rev. 2013;42:7214–7243. doi: 10.1039/c3cs35499g. [DOI] [PubMed] [Google Scholar]
- 9.Leblond J, Gao H, Petitjean A, Leroux JC. pH-Responsive Molecular Tweezers. J Am Chem Soc. 2010;132:8544–8545. doi: 10.1021/ja103153t. [DOI] [PubMed] [Google Scholar]
- 10.Di J, Yao S, Ye Y, Cui Z, Yu J, Ghosh TK, Zhu Y, Gu Z. Stretch-Triggered Drug Delivery from Wearable Elastomer Films Containing Therapeutic Depots. ACS Nano. 2015;9:9407–9415. doi: 10.1021/acsnano.5b03975. [DOI] [PubMed] [Google Scholar]
- 11.Lahann J, Mitragotri S, Tran TN, Kaido H, Sundaram J, Choi IS, Hoffer S, Somorjai GA, Langer R. A Reversibly Switching Surface. Science. 2003;299:371–374. doi: 10.1126/science.1078933. [DOI] [PubMed] [Google Scholar]
- 12.Wisniewski N, Reichert M. Methods for reducing biosensor membrane biofouling. Colloids Surf B: Biointerfaces. 2000;18:197–219. doi: 10.1016/s0927-7765(99)00148-4. [DOI] [PubMed] [Google Scholar]
- 13.Castner DG, Ratner BD. Biomedical surface science: Foundations to frontiers. Surface Sci. 2002;500:28–60. [Google Scholar]
- 14.Banerjee I, Pangule RC, Kane RS. Antifouling Coatings: Recent Developments in the Design of Surfaces That Prevent Fouling by Proteins, Bacteria, and Marine Organisms. Adv Mater. 2011;23:690–718. doi: 10.1002/adma.201001215. [DOI] [PubMed] [Google Scholar]
- 15.Wei Q, Becherer T, Angioletti-Uberti S, Dzubiella J, Wischke C, Neffe AT, Lendlein A, Ballauff M, Haag R. Protein Interactions with Polymer Coatings and Biomaterials. Angew Chem Int Ed. 2014;53:8004–8031. doi: 10.1002/anie.201400546. [DOI] [PubMed] [Google Scholar]
- 16.Cho NJ, Frank CW, Kasemo B, Hook F. Quartz crystal microbalance with dissipation monitoring of supported lipid bilayers on various substrates. Nat Protocols. 2010;5:1096–1106. doi: 10.1038/nprot.2010.65. [DOI] [PubMed] [Google Scholar]
- 17.Gorbet MB, Sefton MV. Biomaterial-associated thrombosis: roles of coagulation factors, complement, platelets and leukocytes. Biomaterials. 2004;25:5681–5703. doi: 10.1016/j.biomaterials.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 18.Thissen H, Gengenbach T, du Toit R, Sweeney DF, Kingshott P, Griesser HJ, Meagher L. Clinical observations of biofouling on PEO coated silicone hydrogel contact lenses. Biomaterials. 2010;31:5510–5519. doi: 10.1016/j.biomaterials.2010.03.040. [DOI] [PubMed] [Google Scholar]
- 19.Harding JL, Reynolds MM. Combating medical device fouling. Trends Biotechnol. 2014;32:140–146. doi: 10.1016/j.tibtech.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Cole MA, Voelcker NH, Thissen H, Griesser HJ. Stimuli-responsive interfaces and systems for the control of protein–surface and cell–surface interactions. Biomaterials. 2009;30:1827–1850. doi: 10.1016/j.biomaterials.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 21.Mas-Moruno C, Fraioli R, Rechenmacher F, Neubauer S, Kapp TG, Kessler H. αvβ3- or α5β1-Integrin-Selective Peptidomimetics for Surface Coating. Angew Chem Int Ed. 2016;55:7048–7067. doi: 10.1002/anie.201509782. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt S, Zeiser M, Hellweg T, Duschl C, Fery A, Möhwald H. Adhesion and Mechanical Properties of PNIPAM Microgel Films and Their Potential Use as Switchable Cell Culture Substrates. Adv Funct Mater. 2010;20:3235–3243. [Google Scholar]
- 23.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable Hydrogels for Dynamic Tuning of Physical and Chemical Properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhihong N, Petukhova A, Kumacheva E. Properties and emerging applications of self-assembled structures made from inorganic nanoparticles. Nat Nanotechnol. 2010;5:15–25. doi: 10.1038/nnano.2009.453. [DOI] [PubMed] [Google Scholar]
- 25.Albanese A, Tang PS, Chan WCW. The Effect of Nanoparticle Size, Shape, and Surface Chemistry on Biological Systems. Annu Rev Biomed Eng. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 26.Kao J, Thorkelsson K, Bai P, Rancatore BJ, Xu T. Toward functional nanocomposites: taking the best of nanoparticles, polymers, and small molecules. Chem Soc Rev. 2013;42:2654–2678. doi: 10.1039/c2cs35375j. [DOI] [PubMed] [Google Scholar]
- 27.Merino S, Martín C, Kostarelos K, Prato M, Vázquez E. Nanocomposite Hydrogels: 3D Polymer–Nanoparticle Synergies for On-Demand Drug Delivery. ACS Nano. 2015;9:4686–4697. doi: 10.1021/acsnano.5b01433. [DOI] [PubMed] [Google Scholar]
- 28.Tkachenko AG, Xie H, Coleman D, Glomm W, Ryan J, Anderson MF, Franzen S, Feldheim DL. Multifunctional Gold Nanoparticle–Peptide Complexes for Nuclear Targeting. J Am Chem Soc. 2003;125:4700–4701. doi: 10.1021/ja0296935. [DOI] [PubMed] [Google Scholar]
- 29.Ning ST, Lee SY, Wei MF, Peng CL, Lin SYF, Tsai MH, Lee PC, Shih YH, Lin CY, Luo TY, Shieh MJ. Targeting Colorectal Cancer Stem-Like Cells with Anti-CD133 Antibody-Conjugated SN-38 Nanoparticles. ACS Appl Mater Interfaces. 2016;8:17793–17804. doi: 10.1021/acsami.6b04403. [DOI] [PubMed] [Google Scholar]
- 30.Hong BJ, Eryazici I, Bleher R, Thaner RV, Mirkin CA, Nguyen ST. Directed Assembly of Nucleic Acid-Based Polymeric Nanoparticles from Molecular Tetravalent Cores. J Am Chem Soc. 2015;137:8184–8191. doi: 10.1021/jacs.5b03485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tibbitt MW, Dahlman JE, Langer R. Emerging Frontiers in Drug Delivery. J Am Chem Soc. 2016;138:704–717. doi: 10.1021/jacs.5b09974. [DOI] [PubMed] [Google Scholar]
- 32.Doane T, Burda C. Nanoparticle mediated non-covalent drug delivery. Adv Drug Deliery Rev. 2013;65:607–621. doi: 10.1016/j.addr.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Gaddes ER, Chen N, Wang Y. Molecular Encryption and Reconfiguration for Remodeling of Dynamic Hydrogels. Angew Chem Int Ed. 2015;54:5957–5961. doi: 10.1002/anie.201500397. [DOI] [PubMed] [Google Scholar]
- 34.Randeria PS, Jones MR, Kohlstedt KL, Banga RJ, Olvera de la Cruz M, Schatz GC, Mirkin CA. What Controls the Hybridization Thermodynamics of Spherical Nucleic Acids? J Am Chem Soc. 2015;137:3486–3489. doi: 10.1021/jacs.5b00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grieves JL, Fye JM, Harvey S, Grayson JM, Hollis T, Perrino FW. Exonuclease TREX1 degrades double-stranded DNA to prevent spontaneous lupus-like inflammatory disease. Proc Natl Acad Sci USA. 2015;112:5117–5122. doi: 10.1073/pnas.1423804112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Win KY, Teng CP, Ye E, Low M, Han MY. Evaluation of Polymeric Nanoparticle Formulations by Effective Imaging and Quantitation of Cellular Uptake for Controlled Delivery of Doxorubicin. Small. 2015;11:1197–1204. doi: 10.1002/smll.201402073. [DOI] [PubMed] [Google Scholar]
- 37.Yang G, Wang J, Wang Y, Li L, Guo X, Zhou S. An Implantable Active-Targeting Micelle-in-Nanofiber Device for Efficient and Safe Cancer Therapy. ACS Nano. 2015;9:1161–1174. doi: 10.1021/nn504573u. [DOI] [PubMed] [Google Scholar]
- 38.Kauffman KJ, Dorkin JR, Yang JH, Heartlein MW, DeRosa F, Mir FF, Fenton OS, Anderson DG. Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 39.Hamidi M, Azadi A, Rafiei P. Hydrogel nanoparticles in drug delivery. Adv Drug Deliery Rev. 2008;60:1638–1649. doi: 10.1016/j.addr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Chacko RT, Ventura J, Zhuang J, Thayumanavan S. Polymer nanogels: A versatile nanoscopic drug delivery platform. Adv Drug Deliery Rev. 2012;64:836–851. doi: 10.1016/j.addr.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermann T, Westhof E. Aminoglycoside binding to the hammerhead ribozyme: a general model for the interaction of cationic antibiotics with RNA1. J Mol Biol. 1998;276:903–912. doi: 10.1006/jmbi.1997.1590. [DOI] [PubMed] [Google Scholar]
- 42.Edwards KJ, Jenkins TC, Neidle S. Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry. 1992;31:7104–7109. doi: 10.1021/bi00146a011. [DOI] [PubMed] [Google Scholar]
- 43.Hermann T, Westhof E. Non-Watson-Crick base pairs in RNA-protein recognition. Chem Biol. 1999;6:R335–R343. doi: 10.1016/s1074-5521(00)80003-4. [DOI] [PubMed] [Google Scholar]
- 44.Wilson WD, Ratmeyer L, Zhao M, Strekowski L, Boykin D. The search for structure-specific nucleic acid interactive drugs: Effects of compound structure on RNA versus DNA interaction strength. Biochemistry. 1993;32:4098–4104. doi: 10.1021/bi00066a035. [DOI] [PubMed] [Google Scholar]
- 45.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 46.Tan W, Wang H, Chen Y, Zhang X, Zhu H, Yang C, Yang R, Liu C. Molecular aptamers for drug delivery. Trends Biotechnol. 2011;29:634–640. doi: 10.1016/j.tibtech.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Rando RR. Specific binding of aminoglycoside antibiotics to RNA. Chem Biol. 1995;2:281–290. doi: 10.1016/1074-5521(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 48.Wallace ST, Schroeder R. In vitro selection and characterization of streptomycin-binding RNAs: recognition discrimination between antibiotics. RNA. 1998;4:112–123. [PMC free article] [PubMed] [Google Scholar]
- 49.Chaires JB, Herrera JE, Waring MJ. Preferential binding of daunomycin to 5′TACG and 5′TAGC sequences revealed by footprinting titration experiments. Biochemistry. 1990;29:6145–6153. doi: 10.1021/bi00478a006. [DOI] [PubMed] [Google Scholar]
- 50.Bagalkot V, Farokhzad OC, Langer R, Jon S. An Aptamer–Doxorubicin Physical Conjugate as a Novel Targeted Drug-Delivery Platform. Angew Chem Int Ed. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 51.Neubert A, Sternberg K, Nagel S, Harder C, Schmitz KP, Kroemer HK, Weitschies W. Development of a vessel-simulating flow-through cell method for the in vitro evaluation of release and distribution from drug-eluting stents. J Control Release. 2008;130:2–8. doi: 10.1016/j.jconrel.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 52.Yoshikawa HY, Rossetti FF, Kaufmann S, Kaindl T, Madsen J, Engel U, Lewis AL, Armes SP, Tanaka M. Quantitative Evaluation of Mechanosensing of Cells on Dynamically Tunable Hydrogels. J Am Chem Soc. 2011;133:1367–1374. doi: 10.1021/ja1060615. [DOI] [PubMed] [Google Scholar]
- 53.Cha C, Antoniadou E, Lee M, Jeong JH, Ahmed WW, Saif TA, Boppart SA, Kong H. Tailoring Hydrogel Adhesion to Polydimethylsiloxane Substrates Using Polysaccharide Glue. Angew Chem Int Ed. 2013;52:6949–6952. doi: 10.1002/anie.201302925. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Z, Chen N, Li S, Battig MR, Wang Y. Programmable Hydrogels for Controlled Cell Catch and Release using Hybridized Aptamers and Complementary Sequences. J Am Chem Soc. 2012;134:15716–15719. doi: 10.1021/ja307717w. [DOI] [PubMed] [Google Scholar]
- 55.Liu X, Chu PK, Ding C. Surface modification of titanium, titanium alloys, and related materials for biomedical applications. Mater Sci Eng : R: Rep. 2004;47:49–121. [Google Scholar]
- 56.Liu P, Zhang H, Liu H, Wang Y, Yao X, Zhu G, Zhang S, Zhao H. A Facile Vapor-Phase Hydrothermal Method for Direct Growth of Titanate Nanotubes on a Titanium Substrate via a Distinctive Nanosheet Roll-Up Mechanism. J Am Chem Soc. 2011;133:19032–19035. doi: 10.1021/ja207530e. [DOI] [PubMed] [Google Scholar]
- 57.Lee K, Mazare A, Schmuki P. One-Dimensional Titanium Dioxide Nanomaterials: Nanotubes. Chem Rev. 2014;114:9385–9454. doi: 10.1021/cr500061m. [DOI] [PubMed] [Google Scholar]
- 58.Jiang P, Liang J, Song R, Zhang Y, Ren L, Zhang L, Tang P, Lin C. Effect of Octacalcium-Phosphate-Modified Micro/Nanostructured Titania Surfaces on Osteoblast Response. ACS Appl Mater Interfaces. 2015;7:14384–14396. doi: 10.1021/acsami.5b03172. [DOI] [PubMed] [Google Scholar]
- 59.Zhang C, Miyatake H, Wang Y, Inaba T, Wang Y, Zhang P, Ito Y. A Bioorthogonal Approach for the Preparation of a Titanium-Binding Insulin-like Growth-Factor-1 Derivative by Using Tyrosinase. Angew Chem Int Ed. 2016;55:11447–11451. doi: 10.1002/anie.201603155. [DOI] [PubMed] [Google Scholar]
- 60.Lee JS, Kim K, Lee K, Park JP, Yang K, Cho SW, Lee H. Surface Chemistry of Vitamin: Pyridoxal 5′-Phosphate (Vitamin B6) as a Multifunctional Compound for Surface Functionalization. Adv Funct Mater. 2015;25:4754–4760. [Google Scholar]
- 61.Choi J, Konno T, Takai M, Ishihara K. Regulation of cell proliferation by multi-layered phospholipid polymer hydrogel coatings through controlled release of paclitaxel. Biomaterials. 2012;33:954–961. doi: 10.1016/j.biomaterials.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 62.Monia BP, Johnston JF, Sasmor H, Cummins LL. Nuclease Resistance and Antisense Activity of Modified Oligonucleotides Targeted to Ha-ras. J Biol Chem. 1996;271:14533–14540. doi: 10.1074/jbc.271.24.14533. [DOI] [PubMed] [Google Scholar]
- 63.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA Interference in Mammalian Cells by Chemically-Modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 64.Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr Opin Chem Biol. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 65.Mayer G. The Chemical Biology of Aptamers. Angew Chem Int Ed. 2009;48:2672–2689. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 66.Keefe AD, Pai S, Ellington AD. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eulberg D, Klussmann S. Spiegelmers: Biostable Aptamers. ChemBioChem. 2003;4:979–983. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]
- 68.Yatime L, Maasch C, Hoehlig K, Klussmann S, Andersen GR, Vater A. Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer. Nat Commun. 2015;6:6481. doi: 10.1038/ncomms7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hannant J, Hedley JH, Pate J, Walli A, Farha Al-Said SA, Galindo MA, Connolly BA, Horrocks BR, Houlton A, Pike AR. Modification of DNA-templated conductive polymer nanowires via click chemistry. Chem Commun. 2010;46:5870–5872. doi: 10.1039/c0cc00693a. [DOI] [PubMed] [Google Scholar]
- 70.Zeng Z, Hoshino Y, Rodriguez A, Yoo H, Shea KJ. Synthetic Polymer Nanoparticles with Antibody-like Affinity for a Hydrophilic Peptide. ACS Nano. 2010;4:199–204. doi: 10.1021/nn901256s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.