Abstract
Background
Studies have found that c-Abl oncogenic kinases may regulate gene transcription by RNA polymerase II phosphorylation or by direct regulation of specific transcription factors or coactivators. However, the global regulation of differential gene expression by c-Abl/Arg is largely unknown. In this study, differentially expressed genes (DEGs) regulated by c-Abl/Arg were identified, and related cellular functions and associated pathways were investigated.
Material/Methods
RNA obtained from wild-type and c-Abl/Arg gene-silenced MCF-7 cells was analyzed by RNA-Seq. DEGs were identified using edgeR software and partially validated by qRT-PCR. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were used to explore the potential functions of these DEGs.
Results
A total of 1,034 DEGs were significantly regulated by c-Abl/Arg (399 were up-regulated and 635 were down-regulated after c-Abl/Arg double knockdown). GO and KEGG analyses showed that the DEGs were primarily involved in cellular metabolic processes, neurodegenerative disease, the metabolic process and signaling pathway of cAMP, angiogenesis, and cell proliferation.
Conclusions
Our data collectively support the hypothesis that c-Abl/Arg regulate differential gene expression, providing new insights into the biological functions of c-Abl and Arg.
MeSH Keywords: Proto-Oncogene Proteins c-abl; Sequence Analysis, RNA; Transcription, Genetic
Background
c-Abl was initially discovered as a homologous oncogene of the Abelson murine leukemia virus and implicated in chromosomal translocations in human leukemia [1–3]. The N-terminal structure of c-Abl contains Src homology 2 (SH2), SH3, and kinase domains [1,4], and the C-terminus has a DNA binding motif and a nuclear localization signal (NLS) for the nuclear localization of c-Abl [5,6]. Arg (a c-Abl-related gene) shares ~90% homology with c-Abl at the N-terminus, implying that they share overlapping functions in cell growth [7], apoptosis [8,9], response to DNA damage [10,11], and tumorigenesis [12,13]. Activation of c-Abl/Arg has been detected in many solid tumors, including breast, lung, prostate, and colon carcinoma [14–19].
In contrast to other tyrosine kinases, c-Abl binds to DNA. Although early studies determined that c-Abl preferentially interacts with A/T-rich DNA or AAC motif-containing sequences, the specific DNA sequence and the related function of the c-Abl-DNA interaction were not clearly identified until recently. The high mobility group-like boxes (HLBs) of c-Abl prefer to bind bent or bendable DNA [20], and c-Abl mainly contacts the minor groove of the double helix or strongly interacts with distorted DNA structures [21]. Kipreos et al. also reported that the hyperphosphorylation of the c-Abl DNA binding domain inactivates its DNA binding activity when cells enter mitosis, implying that c-Abl may play a role in cell cycle progression by transcriptional regulation [6,22]. The C-terminal repeated domain (CTD) of RNA polymerase II (RNAPII) has been identified as the nuclear substrate of c-Abl [23]. Tyrosine phosphorylation of the consensus sequence YSPTSPS disrupts the recruitment of CTD-binding proteins [24], by which c-Abl is likely to regulate RNA processing and chromatin modification globally. Moreover, c-Abl is also involved in specific gene transcription by targeting many transcription factors or regulators, such as p53, p73, NF-κB, c-Myc, C/EBPβ, or Yap [25–30], during numerous biological processes. These findings collectively indicate that c-Abl/Arg plays an important role in gene transcription regulation.
To comprehensively study c-Abl/Arg-related transcription regulation, RNA was prepared from MCF-7/Scr and MCF-7/c-Abl/Arg-knockdown cells and was subjected to RNA-Seq analysis. The differentially expressed genes (DEGs) were identified by comparing the RNA from MCF-7/Scr cells with that from MCF-7/c-Abl/Arg-knockdown cells, and GO and KEGG analyses of the DEGs suggested that c-Abl/Arg may play critical roles in multiple cellular processes by regulating gene transcription.
Material and Methods
Cell culture and c-Abl/Arg-targeting gene silencing
All cells were grown in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% heat-inactivated fetal bovine serum (HyClone), 2 mM L-glutamine, 100 mg/ml streptomycin and 100 units/ml penicillin at 37°C. The siRNA sequence targeting the c-Abl/Arg mRNA was 5′-GGGAAATTGCTACCTATGG-3′ (100% homology between c-Abl and Arg). These primers were cloned into the psiSTRIKETM-Hygromycin vector (Promega, Madison, WI, USA). To obtain MCF-7/c-Abl/Arg-knockdown cell line, wild-type MCF-7 cells were transfected with the c-Abl/Arg-targeted siRNA plasmids using Lipofectamine 2000 DNA Transfection Reagent (Invitrogen) when the cells reached 50–70% confluence. At 36 h after transfection, the cells were maintained in DMEM with 50 mg/ml hygromycin B until individual clones were obtained. To evaluate siRNA efficacy, qRT-PCR and western blotting were performed to detect the mRNA and protein levels of c-Abl and Arg, respectively.
The scrambling gene silencing
The scrambling gene silencing cell line was established and used as a control of c-Abl/Arg-targeting gene silencing cell line. The scrambled siRNA sequence was 5′-GTGACATAGCAGGAACTAC-3′, corresponding to a negative siRNA control with no match with any Homo sapiens mRNA sequences. The primers were cloned into the psiSTRIKETM-Hygromycin Vector (Promega, Madison, WI, USA). The MCF-7/Scramble cell line was established by the same method used in the MCF-7/c-Abl/Arg-knockdown cell line.
Quantitative real-time polymerase chain reaction analysis
Total RNA was extracted from the MCF-7/Scr and MCF-7/c-Abl/Arg-knockdown cells using the RNeasy Mini Kit (QIAGEN). The total RNA was then used for cDNA synthesis with the GoScriptTM Reverse Transcription System (Promega) according to the manufacturer’s protocol. qRT-PCR was performed using PowerUpTM SYBRTM Green Master Mix (Thermo Fisher), and the primer sequences are shown in Table 1. The relative expression of the target genes was calculated according to the 2−ΔΔCT method and was normalized to the actin gene.
Table 1.
Primers used for quantitative real time polymerase chain reaction.
| Gene | Gene ID | Primer sequence | |
|---|---|---|---|
| c-Abl | 25 | Forward | AGCTCTACGTCTCCTCCGAG |
| Reverse | CAGCTTGTGCTTCATGGTGA | ||
|
| |||
| Arg | 27 | Forward | ACAGCACCAGAGAGTCTTGC |
| Reverse | AGCAAAAGAGGGCCTATCGG | ||
|
| |||
| Actin | 60 | Forward | TGGCACCCAGCACAATGAA |
| Reverse | CTAAGTCATAGTCCGCCTAGAAGCA | ||
|
| |||
| DUT | 1854 | Forward | CTGAAGAGACACCCGCCATT |
| Reverse | AAGTGTTTTGCAGCCAAGCC | ||
|
| |||
| SERP1 | 27230 | Forward | GAGAAGGCGTCTGTAGGACC |
| Reverse | TGACTGAATAAGTAGGGTCCACT | ||
|
| |||
| TSR3 | 115939 | Forward | CAGATTCGGCGGTCTGGTG |
| Reverse | TTCCGCAGCAAAATGACAGC | ||
|
| |||
| ELF3 | 1999 | Forward | GTACTGACCCTGAGCAACCC |
| Reverse | CATGCCATCCTTCTCCAGCA | ||
|
| |||
| HOXA7 | 3204 | Forward | TACGACCAAAACATCCCCGG |
| Reverse | TTAATCTGGCGCTCGGTGAG | ||
|
| |||
| NRP2 | 8828 | Forward | ATCATCTCCTCGGGCTCCAT |
| Reverse | AGGGTGTTTTGGTCCCACAG | ||
|
| |||
| PDGFA | 5154 | Forward | ATTCCTCGGAGTCAGGTCGA |
| Reverse | GGAGGAGAACAAAGACCGCA | ||
|
| |||
| PDE4D | 5144 | Forward | TGCACAGCTCTAGTCTGACT |
| Reverse | ACTGGACAACATCTGCAGCA | ||
Western blot analysis
Cell lysates were prepared in lysis buffer (50 mM Tris-HCl [pH 7.5], 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 10 mM sodium fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 10 μg/ml pepstatin A) containing 1% Nonidet P-40 and were then subjected to SDS-PAGE and blotted onto a transfer membrane (Millipore). Anti-c-Abl (Santa Cruz SC-131), anti-Arg (Santa Cruz SC-20708), and anti-β-actin (Santa Cruz SC-1616) antibodies were used for the immunoblotting analysis. The antigen-antibody complexes were visualized by chemiluminescence (PerkinElmer Life Sciences).
Preparation of libraries and RNA sequencing
Total RNA was extracted from the 2 cell lines, and the quality and quantity of the RNA were determined by measuring the absorbance at 260 nm and 280 nm (A260/A280) using a SmartSpec Plus (Bio-Rad). The RNA integrity was further determined by 1.5% agarose gel electrophoresis. For each sample, 10 μg of total RNA was used to prepare the RNA-Seq libraries. Polyadenylated mRNAs were purified and concentrated with oligo(dT)-conjugated magnetic beads (Thermo Fisher), and reverse transcription was performed with primers harboring a 3′ adaptor sequence and randomized hexamers. PCR products of cDNA corresponding to 200–500 bp were purified, quantified and stored at −80°C until sequencing.
High-throughput sequencing was performed on the Illumina HiSeq 2000 system by ABLife lnc. (Wuhan, China). A FASTX-Toolkit (Version 0.0.13) was used to obtain clean reads, and the raw sequencing data were also evaluated using FAST-QC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Parameters such as quality distribution of nucleotides, position-specific sequencing quality, GC content, proportion of PCR duplication, and k-mer frequency were evaluated to obtain a deeper understanding of the data prior to variant evaluation.
Analysis of DEGs
According to the genome annotation, uniquely localized reads were used for the DEG analysis. The DEG-Seq Algorithm and edgeR software were used to analyze the differentially expressed genes between MCF-7/Scr and MCF-7/c-Abl/Arg-knockdown cells (fold change of case/control ≥2 or ≤0.5, false discovery rate (FDR) <0.05)
Gene ontology and pathway analysis
Gene Ontology (GO) analysis was performed to identify the potential functions of the DEGs. By analyzing the genes with DAVID version 6.8 (https://david.ncifcrf.gov/), the biological process, molecular function, and cellular component functions were classified, and P-values were computed. The Kyoto Encyclopedia of Genes and Genomes (KEGG) was employed to determine the significant pathways in which the genes were involved. Fisher’s exact test was used to identify significantly enriched pathways, and the P-value was set to define the threshold of significance. Significant pathway terms were indicated by a P-value ≤0.05, whereas non-significant pathway terms were indicated by a P-value >0.05. The smaller the P-value, the more specific the gene’s function. This analysis can aid the identification of significant pathways involving DEGs.
Results
c-Abl/Arg gene silencing and RNA sequence analysis
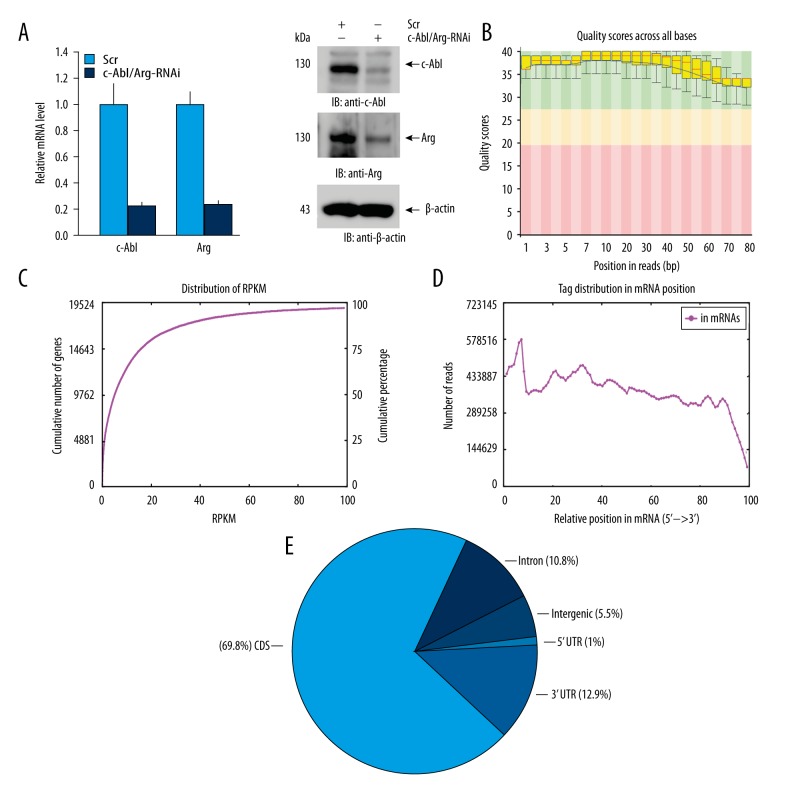
To comprehensively investigate the regulation of gene transcription by c-Abl/Arg, the expression of both c-Abl and Arg in MCF-7 cells was knocked down by transfection with targeted siRNAs, and the MCF-7/c-Abl/Arg-knockdown stable cell line was established. qRT-PCR analysis showed that the mRNA levels of c-Abl/Arg were down-regulated by ~80% in the MCF-7/c-Abl/Arg-knockdown cells compared with the scrambled siRNA control cells (Figure 1A, left). Accordingly, the expression levels of c-Abl and Arg in the c-Abl/Arg gene-silenced cells were also significantly decreased as measured by immunoblotting (Figure 1A, right). To construct cDNA libraries for RNA-Seq analysis, total RNA was extracted from the MCF-7/c-Abl/Arg-knockdown or scrambled control cell lines and was subjected to poly(A) enrichment, RNA fragmentation, random hexamer-primed cDNA synthesis, linker ligation, size selection, and PCR amplification. Finally, a total of 69 million and 82 million clean RNA-Seq reads were obtained by Illumina HiSeq 2000 sequencing of the c-Abl/Arg gene-silenced and scrambled control cDNA libraries, respectively.
Figure 1.
Gene silencing and RNA-Seq quality control. (A) Relative c-Abl and Arg mRNA levels were measured by qRT-PCR in MCF-7 cells stably expressing c-Abl/Arg-siRNA or scrambled-siRNA. Data are shown as the average ±SD of three independent experiments (left). The expression of c-Abl or Arg was also analyzed by immunoblotting with the indicated antibodies (right). (B) The range of the quality values across all bases at each position was analyzed by FastQ and is shown as a box-and-whisker plot. The central red line represents the median value; the yellow box represents the inter-quartile range (25–75%); the upper and lower whiskers represent the 10% and 90% points; and the black line represents the mean quality. According to the values on the y-axis, the background of the graph was divided into very-good-quality calls (green), reasonable-quality calls (orange), and poor-quality calls. Quality scores >20 imply that the accuracy of the mapping is greater than 99%. (C) mRNA expression profile of the MCF-7/c-Abl/Arg-knockdown sample is reflected by RPKM (reads per kilobase of a gene per million reads). The ordinate of each point on the red line represents the number and proportion of genes when the RPKM is less than or equal to the abscissa of this point. (D) mRNA coverage (from 5′ to 3′) analysis per hundred bins. The x-axis represents the relative position of the sequencing reads in the mRNA sequences, and the y-axis indicates the number of reads. (E) Read distribution across the genomic regions. The data shown in (B–E) were acquired from the MCF-7/c-Abl/Arg-knockdown samples.
Quality control analysis by Fast-QC showed that the Q scores (from 1 to 100) of all reads were >28, indicating that high-quality RNA-Seq data had been obtained (Figure 1B). The reads per kilobase million reads (RPKM) values were less than 20 for ~75% of the reads that were mapped to genes, indicating that most of the genes in the libraries were expressed at low levels (Figure 1C). The read distribution analysis showed that the mapped reads were evenly distributed in the mRNA from the 5′ to 3′ ends (Figure 1D). Approximately 90% of the 69 million mapped reads mapped to exon coding sequences, and ~69.8% of the reads mapped to protein coding sequences (Figure 1E). Similar RNA-Seq data were obtained from the scrambled control cells (Figure 2). These results demonstrated that the RNA-Seq data were high quality and reliable, and therefore, these data were used for the following differential expression analysis.
Figure 2.
(A–D) Quality analysis of the control sample (MCF-7 cells stably expressing the scrambled RNAi).
c-Abl/Arg regulates gene transcription
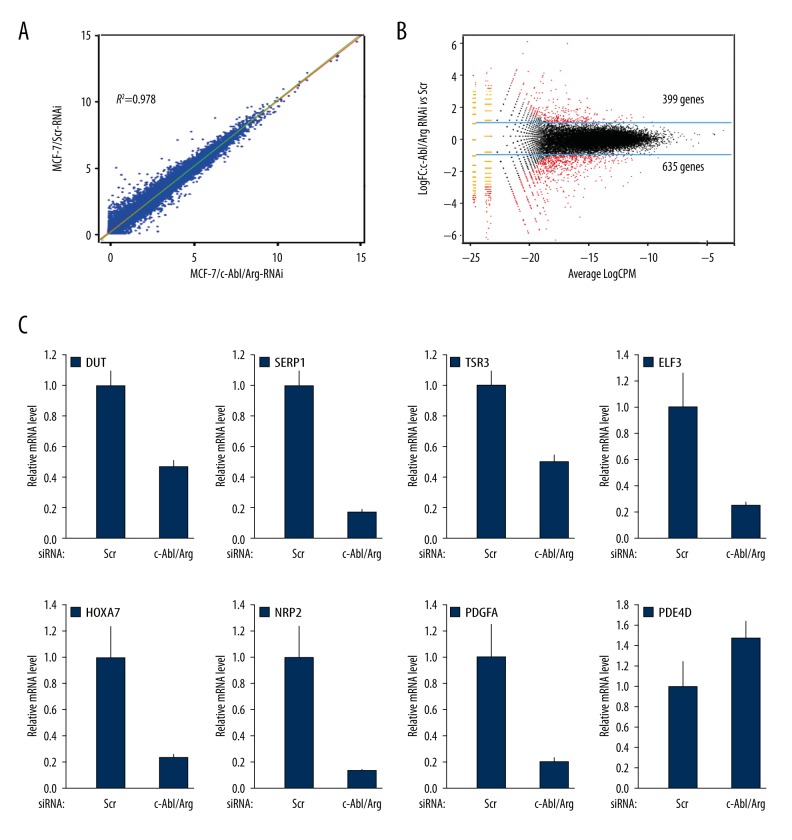
c-Abl has been reported to be involved in the regulation of gene transcription. To investigate transcription regulation by Abl kinases, the DEGs between the MCF-7/Scr and MCF-7/c-Abl/Arg-knockdown cells were determined based on the RPKM values of all mapped genes. The correlation analysis showed that the expression levels of specific genes in the 2 samples from the MCF-7/Scr and MCF-7/c-Abl/Arg-knockdown cells were well correlated (R2=0.978) (Figure 3A), suggesting that the RNA-Seq data were highly repeatable. Using edgeR software, the DEGs were filtered using the following criteria: P≤0.01 and fold change ≥2 or ≤0.5. The results showed that 1,034 DEGs were significantly regulated by the Abl kinases, of which 635 were down-regulated and 399 were up-regulated in the c-Abl/Arg gene-silenced cells (Figure 3B). Furthermore, the significant regulation of mRNA transcription levels by c-Abl/Arg was verified by qRT-PCR (Figure 3C). The data for 60 genes significantly down- or up-regulated by c-Abl/Arg gene silencing are presented in Table 2.
Figure 3.
Abl kinases regulate differential gene expression. (A) Analysis of the differential gene expression patterns between the MCF-7/c-Abl/Arg-knockdown and MCF-7/Scr samples. (B) DEGs between the 2 samples were identified by edgeR analysis. DEGs were defined as genes with a P-value ≤0.01 and a fold change ≥2 or ≤0.5. Log10CPM shows the overall concentration of each compared gene in the 2 groups. (C) qRT-PCR validation of the DEGs using RNA extracted from the MCF-7/c-Abl/Arg-knockdown and MCF-7/Scr cells.
Table 2.
30 down/up regulated genes by c-Abl/Arg knockdown.
| Down-regualted | Up-regualted | ||||
|---|---|---|---|---|---|
| Gene | Log2FC | FDR | Gene | Log2FC | FDR |
| INA | −20 | 0 | MCF2 | 6.00 | 0 |
| VCAN | −20 | 0 | KLHL13 | 5.75 | 0 |
| GPX3 | −20 | 2.61E-13 | ROR2 | 5.55 | 2.22E-16 |
| TRIB2 | −20 | 1.97E-12 | MAPK4 | 5.17 | 0 |
| NAP1L2 | −20 | 3.91E-12 | FZD3 | 3.19 | 2.31E-14 |
| MCTP1 | −20 | 0 | CYFIP2 | 3.17 | 1.98E-11 |
| OLFM1 | −6.73 | 0 | RNF182 | 3.13 | 0 |
| TP53I11 | −6.32 | 0 | ST8SIA4 | 3.09 | 0 |
| LOC100506465 | −5.65 | 0 | MCTP2 | 3.03 | 0 |
| SLC9A2 | −5.60 | 3.11E-11 | IL31RA | 2.99 | 7.27E-11 |
| CSF2RA | −5.42 | 0 | TMEM47 | 2.82 | 3.08E-08 |
| LXN | −5.03 | 0 | KMO | 2.69 | 1.03E-06 |
| NLRP1 | −4.98 | 4.15E-13 | ZFHX4 | 2.62 | 0 |
| MECOM | −4.76 | 2.77E-11 | NEGR1 | 2.59 | 0 |
| IFI44 | −4.74 | 3.55E-15 | SYDE2 | 2.37 | 2.61E-12 |
| MNS1 | −4.54 | 0 | CIDEB | 2.35 | 1.48E-09 |
| MAGEB2 | −4.27 | 0 | OAS1 | 2.26 | 1.24E-06 |
| STARD8 | −4.21 | 1.51E-10 | BACE2 | 2.21 | 0 |
| KRT16 | −3.97 | 7.82E-09 | SCUBE1 | 2.21 | 5.16E-05 |
| PDE3B | −3.97 | 4.77E-11 | ENPP1 | 2.20 | 1.56E-10 |
| SLCO2A1 | −3.93 | 1.12E-12 | SGK196 | 2.14 | 5.88E-11 |
| MIR675 | −3.92 | 0 | ATP8 | 2.10 | 0 |
| MAP1B | −3.83 | 0 | DGKH | 2.05 | 1.28E-12 |
| NRP2 | −3.01 | 0 | PDE3A | 2.04 | 0 |
| PDGFA | −2.81 | 0 | PTGS2 | 2.03 | 1.05E-07 |
| ELF3 | −2.21 | 2.22E-15 | NR4A3 | 2.03 | 1.21E-08 |
| HOXA7 | −2.07 | 5.74E-09 | FAM91A3P | 1.99 | 1.40E-04 |
| SERP1 | −1.82 | 1.09E-12 | RN7SL2 | 1.97 | 0 |
| TSR3 | −1.72 | 1.31E-11 | PRKAA2 | 1.97 | 0 |
| DUT | −1.54 | 2.43E-09 | PDE4D | 1.66 | 2.43E-12 |
Functional analysis of the c-Abl/Arg-regulated DEGs
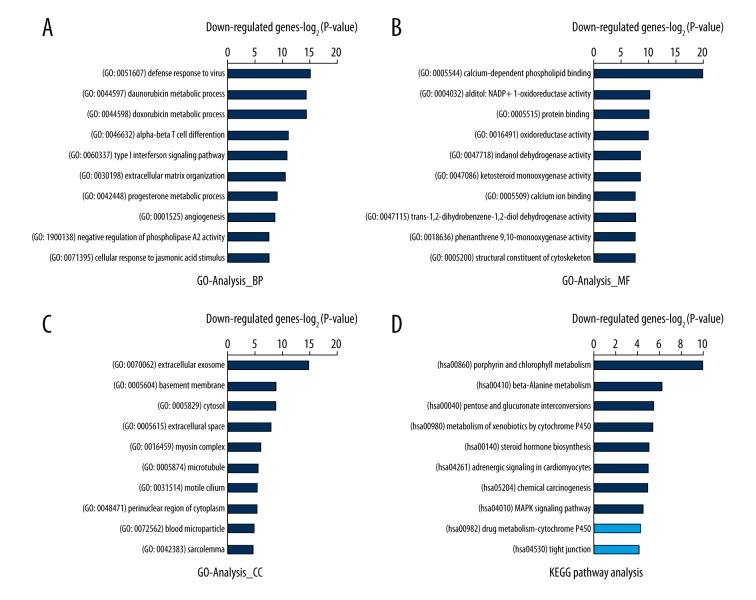
To further understand the potential cellular function and associated pathways of these DEGs, we performed GO term (including biological process, molecular function and cellular component) and KEGG pathway analyses. As shown by the biological process analysis, the functions of the genes that were down-regulated by c-Abl/Arg gene silencing (i.e., genes positively regulated by the Abl kinase) were mainly involved in virus defense response (GO: 0051607, GO: 0060337 and GO: 0045071) and oxidoreductase-involved metabolic processes, including chemotherapeutic drugs (GO: 0044597 and GO: 0044598), progesterone (GO: 0042448), steroid (GO: 0008202), and fatty acid (GO: 0006631) metabolic processes (Figure 4A). Proteins with calcium-dependent phospholipid binding activity (GO: 0005544), oxidoreductase activity (GO: 0004032, GO: 0016491, GO: 0047718, GO0047086, etc.), and protein binding capacity (GO: 0005515) were the most enriched molecular functions (Figure 4B). Extracellular exosome (GO: 0070062), basement membrane (GO: 0005604), and cytosol (GO: 0005829) were the most enriched cellular components (Figure 4C). The KEGG analysis revealed that the greatest enrichments also occurred in metabolism regulation, such as the metabolism of porphyrin (hsa00860) and beta-alanine (hsa00410), pentose and glucuronate interconversions (hsa00040), the metabolism of xenobiotics by cytochrome P450 (hsa00980), and steroid hormone biosynthesis (hsa00140), etc. (Figure 4D).
Figure 4.
Top 10 GO and KEGG pathways enriched in DEGs that were down-regulated by c-Abl/Arg double knockdown. (A–C) The significantly enriched GO terms of c-Abl/Arg up-regulated genes are shown. (A) BP – biological process; (B) MF – molecular functions; (C) CC – cellular components. (D) The KEGG pathway analysis of c-Abl/Arg up-regulated genes is shown. All data are shown as −Log2P-value. Dark blue: significant pathway term (P-value ≤0.05); Light blue: non-significant pathway term (P-value >0.05).
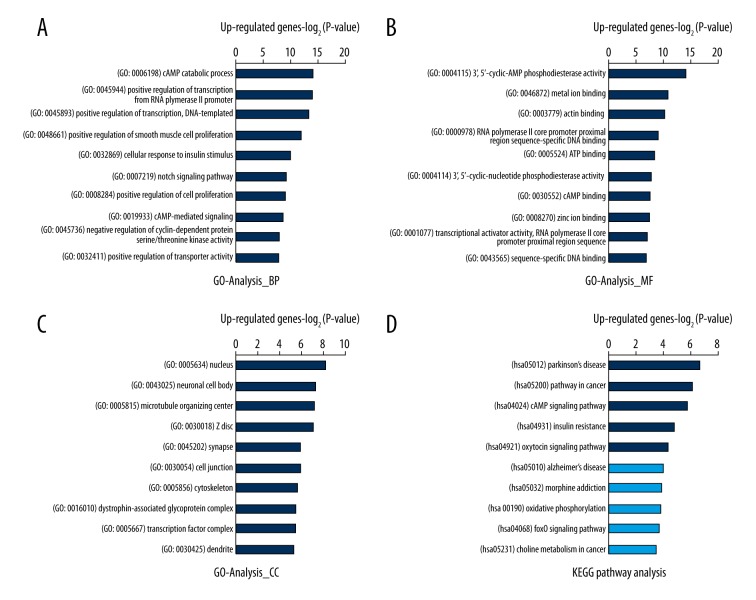
According to the biological process analysis, the genes that were up-regulated by c-Abl/Arg gene silencing (i.e., genes negatively regulated by the Abl kinase) were enriched in cAMP catabolic and signaling process (GO: 0006198 and GO: 0019933), positive regulation of transcription (GO: 0045944 and GO: 0045893), and positive regulation of cell proliferation (GO: 0048661 and GO: 0008284) (Figure 5A). cAMP phosphodiesterase activity (GO: 0004115, GO: 0004114, and GO: 0030552), metal ion binding (GO: 0046872 and GO: 0008270), actin binding (GO: 0003779), and DNA or chromatin binding (GO: 0000978, GO: 0001077, GO: 0043565, and GO: 0003682) were the most enriched molecular functions (Figure 5B). Moreover, the nucleus (GO: 0005634), neuronal cell body (GO: 0043025), and microtubule organizing center (GO: 0005815) were the most enriched cellular components (Figure 5C). The KEGG pathway analysis showed that signaling pathways in Parkinson’s disease (hsa05012), signaling pathways in cancer (hsa05200), the cAMP signaling pathway (hsa04024), signaling pathways in insulin resistance (hsa04931), and the oxytocin signaling pathway (hsa04921) were the most enriched (Figure 5D). These data collectively demonstrate that c-Abl/Arg can positively or negatively regulate gene transcription to participate in metabolism, virus defense responses, cell proliferation, nervous system disease and tumorigenesis.
Figure 5.
Top 10 GO and KEGG pathways enriched in DEGs that were up-regulated by c-Abl/Arg double knockdown. (A–C) The significantly enriched GO terms of c-Abl/Arg down-regulated genes are shown. (A) BP – biological process; (B) MF – molecular functions; (C) CC – cellular components. (D) The KEGG pathway analysis of c-Abl/Arg down-regulated genes is shown. All data are shown as –Log2P-value. Dark blue: significant pathway term (P-value ≤0.05); Light blue: non-significant pathway term (P-value >0.05).
Discussion
The Abl family of protein tyrosine kinases, c-Abl and Arg, functions to link diverse stimuli, such as DNA damage, oxidative stress, Src tyrosine kinases, and growth factors, to signaling pathways controlling cell growth, survival, stress, invasion, adhesion, and migration. Mice with targeted disruption of the c-abl gene are born runted, exhibit head and eye abnormalities, and succumb as neonates with defective lymphopoiesis [31,32]. Moreover, embryos deficient in both c-abl and arg exhibit defects in neurulation and die by 11 days postcoitus [33]. These findings demonstrate that c-Abl and Arg play essential and overlapping roles in biological processes. In contrast to Arg, the C-terminus of c-Abl has a DNA binding domain and NLS, which allow c-Abl kinase to enter the nucleus and regulate gene transcription. Although c-Abl is involved in gene transcription by directly mediating the tyrosine phosphorylation of specific transcription factors or regulators, the global regulation of gene transcription by Abl kinases is unknown.
Because c-Abl and Arg have common and unique domains and thus exhibit overlapping as well as unique functions, the expression of both genes was simultaneously silenced by RNAi, and c-Abl/Arg-mediated transcription regulation was investigated by RNA-Seq. By comparing the differentially expressed genes between scrambled cells and MCF-7/c-Abl/Arg-knockdown cells, we revealed that 1,034 DEGs were regulated by c-Abl/Arg, and several of these genes were validated by qRT-PCR (Figure 3C). To further understand their potential cellular functions, we employed GO and KEGG to analyze these DEGs. The genes down-regulated by c-Abl/Arg gene silencing were mainly enriched in oxidoreductase-involved metabolic processes (including chemotherapeutic drugs, progesterone, steroid, retinoic acid, and fatty acid) and the anti-virus response (including type I interferon signaling pathway and virus replication), whereas the up-regulated genes were mainly enriched in cAMP metabolic processes and signaling pathways, metal ion binding, DNA or chromatin binding, cell proliferation, Parkinson’s disease, and signaling pathways in cancer (Figures 4, 5). Most of these biological pathways have been related to the cellular functions of c-Abl/Arg in previous reports. For example, c-Abl phosphorylates Parkin, an E3 ubiquitin ligase involved in Parkinson’s disease, to contribute to the degeneration of dopaminergic neurons [34]. Our findings suggest that the negatively regulated transcription of mitochondrial membrane respiration chain complex subunits, such as ND1, ND4L, COX2, and ATP8, by c-Abl/Arg expression might participate in neurodegenerative disease. Notably, c-Abl/Arg regulates the transcription of not only nucleus-encoded but also mitochondrion-encoded genes (as mentioned above). DEGs regulated by c-Abl/Arg were also significantly enriched in chemotherapeutic drug metabolic processes (GO: 0044597 and GO: 0044598), consistent with several studies that have reported activation of c-Abl/Arg in cells in response to doxorubicin (DOX) treatment. For example, the data suggest that c-Abl kinase plays a vital role in DOX-induced NF-κB signaling, and inhibition of kinase activity leads to increased cellular sensitivity to the induction of apoptosis [35]. In response to DOX-induced DNA damage, c-Abl also acts upstream of DNA damage-activated signaling cascades via activation of the PI-3 kinase-like kinases Atm and Atr and thus has been implicated in controlling apoptosis, DNA repair and cell cycle progression [36]. Our finding identified that Abl kinase-regulated DEGs may be involved in the metabolic of doxorubicin, providing a potential mechanism by which Abl kinase responses to doxorubicin or other chemotherapeutic drug treatment.
Genes that were positively transcriptionally by c-Abl/Arg expression were also significantly enriched in metabolic pathways, which have seldom been studied in Abl kinase functions. A recent study found that c-Abl was activated in fumarate hydratase-deficient kidney tumors (hereditary leiomyomatosis and renal cell carcinoma, HLRCC), which are characterized by high glycolysis and accumulate high levels of fumarate, lactate and hypoxia-stimulated transcription factor (HIF1α) [37]. Activation of c-Abl in HLRCC functions to promote aerobic glycolysis through activation of mTOR-HIF1α. Consistent with this finding, our study demonstrated that aldo-keto reductase family 1 members (including AKR1C3, AKR1C2, AKR1B10, AKR1B1, and AKR1C1), UDP glucuronosyltransferase family 1 (UGT1A7 and UGT1A8), and the aldehyde dehydrogenase family (ALDH1A3, ALDH2 and ALDH3A1) were positively regulated by c-Abl/Arg, which may contribute to Abl kinase-dependent energy metabolism and cytochrome P450-involved drug metabolism, implying that c-Abl/Arg play critical roles in metabolic processes via transcription regulation.
Conclusions
In this study, the global regulation of gene transcription by c-Abl/Arg was investigated using RNA-Seq, and the potential cellular functions and associated pathways of these DEGs were analyzed in depth. The DEGs were mainly enriched in cellular metabolic processes, neurodegenerative disease, cAMP metabolic processes, signaling pathways, angiogenesis, and cell proliferation. Our data collectively support the hypothesis that c-Abl/Arg regulate differential gene expression, which may provide novel insights into the discovery and annotation of signaling pathways that are related to c-Abl/Arg.
Footnotes
Conflicts of interest
The authors have no conflicts of interest.
Source of support: This work was supported by the National Natural Science Foundation of China [31070674]
References
- 1.Goff SP, Gilboa E, Witte ON, Baltimore D. Structure of the Abelson murine leukemia virus genome and the homologous cellular gene: Studies with cloned viral DNA. Cell. 1980;22(3):777–85. doi: 10.1016/0092-8674(80)90554-1. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Neriah Y, Daley GQ, Mes-Masson AM, et al. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science. 1986;233(4760):212–14. doi: 10.1126/science.3460176. [DOI] [PubMed] [Google Scholar]
- 3.Abelson HT, Rabstein LS. Lymphosarcoma: Virus-induced thymic-independent disease in mice. Cancer Res. 1970;30(8):2213–22. [PubMed] [Google Scholar]
- 4.Kruh GD, Perego R, Miki T, Aaronson SA. The complete coding sequence of arg defines the Abelson subfamily of cytoplasmic tyrosine kinases. Proc Natl Acad Sci USA. 1990;87(15):5802–6. doi: 10.1073/pnas.87.15.5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Etten RA, Jackson PK, Baltimore D, et al. The COOH terminus of the c-Abl tyrosine kinase contains distinct F- and G-actin binding domains with bundling activity. J Cell Biol. 1994;124(3):325–40. doi: 10.1083/jcb.124.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kipreos ET, Wang JY. Cell cycle-regulated binding of c-Abl tyrosine kinase to DNA. Science. 1992;256(5055):382–85. doi: 10.1126/science.256.5055.382. [DOI] [PubMed] [Google Scholar]
- 7.Goga A, Liu X, Hambuch TM, et al. p53 dependent growth suppression by the c-Abl nuclear tyrosine kinase. Oncogene. 1995;11(4):791–99. [PubMed] [Google Scholar]
- 8.Gong JG, Costanzo A, Yang HQ, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399(6738):806–9. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 9.Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 1999;399(6738):809–13. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 10.Yuan ZM, Huang Y, Whang Y, et al. Role for c-Abl tyrosine kinase in growth arrest response to DNA damage. Nature. 1996;382(6588):272–74. doi: 10.1038/382272a0. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Zhang J, Lee J, et al. A kinase-independent function of c-Abl in promoting proteolytic destruction of damaged DNA binding proteins. Mol Cell. 2006;22(4):489–99. doi: 10.1016/j.molcel.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 12.Wong S, Witte ON. The BCR-ABL story: Bench to bedside and back. Annu Rev Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- 13.Lin J, Arlinghaus R. Activated c-Abl tyrosine kinase in malignant solid tumors. Oncogene. 2008;27(32):4385–91. doi: 10.1038/onc.2008.86. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan D, Plattner R. Activation of Abl tyrosine kinases promotes invasion of aggressive breast cancer cells. Cancer Res. 2006;66(11):5648–55. doi: 10.1158/0008-5472.CAN-06-0734. [DOI] [PubMed] [Google Scholar]
- 15.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Sun T, Ji L, et al. Oncogenic activation of c-Abl in non-small cell lung cancer cells lacking FUS1 expression: Inhibition of c-Abl by the tumor suppressor gene product Fus1. Oncogene. 2007;26(49):6989–96. doi: 10.1038/sj.onc.1210500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganguly SS, Fiore LS, Sims JT, et al. c-Abl and Arg are activated in human primary melanomas, promote melanoma cell invasion via distinct pathways, and drive metastatic progression. Oncogene. 2012;31(14):1804–16. doi: 10.1038/onc.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furlan A, Stagni V, Hussain A, et al. Abl interconnects oncogenic Met and p53 core pathways in cancer cells. Cell Death Differ. 2011;18(10):1608–16. doi: 10.1038/cdd.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drake JM, Graham NA, Stoyanova T, et al. Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc Natl Acad Sci USA. 2012;109(5):1643–48. doi: 10.1073/pnas.1120985109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao YJ, Wang JY. Binding of A/T-rich DNA by three high mobility group-like domains in c-Abl tyrosine kinase. J Biol Chem. 1996;271(37):22823–30. doi: 10.1074/jbc.271.37.22823. [DOI] [PubMed] [Google Scholar]
- 21.David-Cordonnier MH, Hamdane M, Bailly C, D’Halluin JC. The DNA binding domain of the human c-Abl tyrosine kinase preferentially binds to DNA sequences containing an AAC motif and to distorted DNA structures. Biochemistry. 1998;37(17):6065–76. doi: 10.1021/bi973030w. [DOI] [PubMed] [Google Scholar]
- 22.Kipreos ET, Wang JY. Differential phosphorylation of c-Abl in cell cycle determined by cdc2 kinase and phosphatase activity. Science. 1990;248(4952):217–20. doi: 10.1126/science.2183353. [DOI] [PubMed] [Google Scholar]
- 23.Baskaran R, Dahmus ME, Wang JY. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc Natl Acad Sci USA. 1993;90(23):11167–71. doi: 10.1073/pnas.90.23.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayer A, Heidemann M, Lidschreiber M, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336(6089):1723–25. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 25.Levav-Cohen Y, Goldberg Z, Zuckerman V, et al. C-Abl as a modulator of p53. Biochem Biophys Res Commun. 2005;331(3):737–49. doi: 10.1016/j.bbrc.2005.03.152. [DOI] [PubMed] [Google Scholar]
- 26.Yuan ZM, Shioya H, Ishiko T, et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399(6738):814–17. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 27.Kawai H, Nie L, Yuan ZM. Inactivation of NF-kappaB-dependent cell survival, a novel mechanism for the proapoptotic function of c-Abl. Mol Cell Biol. 2002;22(17):6079–88. doi: 10.1128/MCB.22.17.6079-6088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong KK, Hardin JD, Boast S, et al. A role for c-Abl in c-myc regulation. Oncogene. 1995;10(4):705–11. [PubMed] [Google Scholar]
- 29.Li X, Liu X, Wang G, et al. Non-receptor tyrosine kinases c-Abl and Arg regulate the activity of C/EBPbeta. J Mol Biol. 2009;391(4):729–43. doi: 10.1016/j.jmb.2009.06.055. [DOI] [PubMed] [Google Scholar]
- 30.Keshet R, Adler J, Ricardo Lax I, et al. c-Abl antagonizes the YAP oncogenic function. Cell Death Differ. 2015;22(6):935–45. doi: 10.1038/cdd.2014.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tybulewicz VL, Crawford CE, Jackson PK, et al. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65(7):1153–63. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 32.Schwartzberg PL, Stall AM, Hardin JD, et al. Mice homozygous for the ablm1 mutation show poor viability and depletion of selected B and T cell populations. Cell. 1991;65(7):1165–75. doi: 10.1016/0092-8674(91)90012-n. [DOI] [PubMed] [Google Scholar]
- 33.Koleske AJ, Gifford AM, Scott ML, et al. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron. 1998;21(6):1259–72. doi: 10.1016/s0896-6273(00)80646-7. [DOI] [PubMed] [Google Scholar]
- 34.Ko HS, Lee Y, Shin JH, et al. Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin’s ubiquitination and protective function. Proc Natl Acad Sci USA. 2010;107(38):16691–96. doi: 10.1073/pnas.1006083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esparza-López J, Medina-Franco H, Escobar-Arriaga E, et al. Doxorubicin induces atypical NF-κB activation through c-Abl kinase activity in breast cancer cells. J Cancer Res Clin Oncol. 2013;139(10):1625–35. doi: 10.1007/s00432-013-1476-3. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Zeng L, Wang J, et al. A positive role for c-Abl in Atm and Atr activation in DNA damage response. Cell Death Differ. 2011;18(1):5–15. doi: 10.1038/cdd.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sourbier C, Ricketts CJ, Matsumoto S, et al. Targeting ABL1-mediated oxidative stress adaptation in fumarate hydratase-deficient cancer. Cancer Cell. 2014;26(6):840–50. doi: 10.1016/j.ccell.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]