Abstract
We investigate the protective effect of combined vaccination based on live attenuated influenza vaccine (LAIV) and group B streptococcus (GBS) recombinant polypeptides against potential pandemic H7N9 influenza infection followed by GBS burden. Mice were intranasally immunized using 107 50% egg infectious dose (EID50) of H7N3 LAIV, the mix of the 4 GBS peptides (group B streptococcus vaccine [GBSV]), or combined LAIV + GBSV vaccine. The LAIV raised serum hemagglutination-inhibition antibodies against H7N9 in higher titers than against H7N3. Combined vaccination provided advantageous protection against infections with A/Shanghai/2/2013(H7N9)CDC-RG influenza and serotype II GBS. Combined vaccine significantly improved bacterial clearance from the lungs after infection compared with other vaccine groups. The smallest lung lesions due to combined LAIV + GBSV vaccination were associated with a prevalence of lung interferon-γ messenger RNA expression. Thus, combined viral and bacterial intranasal immunization using H7N3 LAIV and recombinant bacterial polypeptides induced balanced adaptive immune response, providing protection against potential pandemic influenza H7N9 and bacterial complications.
Keywords: Live influenza vaccine, group B streptococcus, recombinant polypeptides, intranasal vaccination
Introduction
Influenza A(H7N9) viruses that cause severe disease and death in humans continue to circulate widely in some poultry populations. These remaining influenza viruses have pandemic potential because most humans likely have no immunity to them. According to the World Health Organization report, by August 17, 2016, human infections of H7N9 virus have reached 798 laboratory-confirmed cases since early 2013 when the virus emerged in China.1 By January 31, 2016, the case fatality rate for H7N9 has been reported as 39.5% (285 from 721).2 Due to increased binding to mammalian respiratory cells, H7N9 has a greater potential pandemic risk for further mammalian adaptation with a possible human-to-human transmission than H5N1 avian influenza viruses.3
Group B streptococcus (GBS), or Streptococcus agalactiae surface proteins have recently drawn attention to the promising vaccine candidates, playing an important role at various stages of infection and being the antigenic determinants of a number of other bacteria, such as Streptococcus pneumoniae and Streptococcus pyogenes.4
Group B streptococcus colonizing the respiratory, gastrointestinal, and urogenital tract is an important human pathogen that causes pneumonia, sepsis, and meningitis in neonates. It also poses a significant threat to immunocompromised adults. Bacterial coinfections, including H1N1 pandemic influenza, are common in influenza-infected patients and are more frequently noted in older aged patients who are associated with higher rates of complications.5 Recently, in the prevention of bacterial infection, much attention has been paid to the development of peptide vaccines based on superficial factors of bacteria pathogenicity. Thus, the P6 protein, used in this study, is based on the immunodominant bac protein containing IgA-binding motif.6 About 30% of GBS possess the bac gene which can be predominantly found in the strains of serotypes I and II.7 The ScaAB protein is the major surface lipoprotein similar to the lipoprotein receptor-associated antigen I (LraI) family..8,9 This protein is presented on the surface of gram-positive pathogens and closely related to PsaA protein of S pneumoniae which is considered a protein-based vaccine candidate against pathogenic pneumococci which are the most common cause of community-acquired pneumonia.10 The ScpB1 protein represents the portion of C5a peptidase, which is contained among 100% of strains of group A and B streptococci, thus providing a universal target for vaccines against pathogenic streptococci.11 Stv protein is a portion of the glycan-binding protein and adhesion SspB1 which can be found in about one-third of highly pathogenic GBS strains predominantly circulating throughout Russia. Immunization with this protein selectively directs the immune response against the highly virulent GBS strains and is able to stimulate the cross-reacting antibody against group A streptococci surface proteins.12 The complex of 4 streptococcal antigens included in the composition of recombinant GBS vaccine will contribute to the formation of protection against various determinants of bacterial pathogenicity, regardless of their functional state.
In a previous study, we have shown that associated intranasal immunization of outbred mice using live attenuated influenza vaccine (LAIV) of H7N3 subtype and 4 GBS recombinant polypeptides was effective against pulmonary infection with five hundred 50% mouse infectious doses (MID50) of avian A/mallard/Netherlands/12/00(H7N3) or 100 MID50 A/PR8/34(H1N1) influenza followed by GBS serotype II secondary infection.13 In this study, we evaluated whether immunity generated against H7N3 LAIV and GBS polypeptides can protect against H7N9 influenza infection followed by GBS burden. We also made an attempt to reveal the role of some causes of innate and adaptive immunity in such protection. Although avian influenza A(H7N9) is infrequent complicated by streptococcal secondary infection,10 new animal models of viral-bacterial interactions can be applied for further study of vaccines against human infections.
Methods
Viruses and vaccine preparations
The reassortant influenza virus A/17/Mallard/Netherlands/00/95(H7N3) (LAIV) was provided from the Virology Department, Institute of Experimental medicine (Saint Petersburg, Russia) collection of viruses. The A/Shanghai/2/2013(H7N9) CDC-RG influenza virus (H7N9) was provided by the Centers for Disease Control and prevention, USA. The viruses were propagated in embryonated chicken eggs (CE) and stored at −70°C.
The group B streptococcus vaccine (GBSV) contained GBS recombinant polypeptides P6 (30 kDa), ScaAB (35 kDa), ScpB1 (43-kDa), and Stv (130 kDa) which were expressed in Escherichia coli and purified as described earlier.14
Immunization
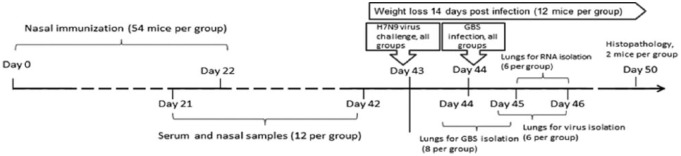
The 8- to-10-week-old female outbred mice were provided by the laboratory breeding nursery of the Russian Academy of Sciences (Rappolovo, Leningrad Region). Groups of mice (54 animals in a group) were lightly anesthetized with ether and intranasally vaccinated with 50 µL divided equally per nostril using the following preparations: (1) LAIV contained 1 × 107 50% egg infectious dose (EID50) of the H7N3 vaccine virus; (2) GBSV contained the mix of P6, ScaAB, ScpB1, and Stv recombinant polypeptides (5 µg each, 20 µg total); 3) mixed LAIV + GBSV vaccine including 1 × 107 EID50 of H7N3 virus and GBSV; and (4) control animals were inoculated with phosphate-buffered saline (PBS). The mice were immunized twice at an interval of 21 days (Figure 1).
Figure 1.
Mouse study design. GBS indicates group B streptococcus.
Three weeks after vaccination and after revaccination, sera were collected from ether-anesthetized mice via submandibular plexus. Nasal secretions were collected from mice after intraperitoneal administration of 0.1 mL of a 0.5% pilocarpine solution (Sigma-Aldrich, St. Louis, MO, USA) into the tubes containing 0.001 M of serine protease inhibitor phenylmethylsulfonyl fluoride. Sera and nasal samples were stored at −20°C.
All invasive procedures involving animals were performed under ether anesthesia, according to the “Rules Laboratory Practice” Ministry of Health of the Russian Federation No. 708 n.
Hemagglutination-inhibition assays
For hemagglutination-inhibition (HI) assay, sera were treated with receptor-destroying enzyme (Denka Seiken, Tokyo, Japan) and tested for HI antibodies against H7N3 vaccine virus and against H7N9 influenza virus as previously described.15
The enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assay (ELISA) was conducted to determine serum IgG, IgG1, IgG2a, and nasal IgA antibodies in 96-well microplates (Sarstedt AG & Co, Nümbrecht, Germany) as previously described.15 For absorption, we used 20 HAU (hemagglutination units)/0.1 mL of the whole purified H7N3 virus or 20 HAU/0.1 mL of the whole purified H7N9 virus or 0.2 mg/0.1 mL of GBSV individual components. The end point ELISA titers were expressed as the highest dilution that yielded an optical density at 450 nm (OD450) greater than the mean OD450 plus 3 SDs of negative controls at an equivalent dilution of sera.
Group B streptococcus
The S agalactiae (serotype II) was obtained from the collection of the Institute of Experimental medicine (Saint Petersburg, Russia) and grown as previously described.13
Challenge study
On day 21 after revaccination, the mice from all vaccine groups were inoculated intranasally with 300 MID50 of H7N9. Twenty-four hours post infection (p.i.), the mice were intranasally infected with 1 × 107 colony-forming units (CFUs) of GBS (type II). Weight was monitored over the course of 14 days p.i. Lung samples were collected for virus load, bacterial clearance, and transcriptional analysis and homogenized using a Tissue Lyser LT system (Qiagen, Venlo, The Netherlands), as per the manufacturer’s protocol. Infectious virus titers were detected in the lung homogenates at 48 and 72 hours p.i. in developing CE as previously described.16 Lung virus titers were expressed as log EID50. Bacterial clearance from lungs was estimated 5 and 24 hours after GBS infection. On day 7 p.i., the lungs from 2 mice per group were collected for histopathologic analysis.
Messenger RNA expression analysis
Total RNA was isolated from lung homogenates using GeneJet RNA Purification Kit (Thermo Scientific, Waltham, MA, USA). For complementary DNA (cDNA) synthesis, reverse transcription (RT) with 100 pg of total RNA was performed using oligo(dt) primer and random hexamer mix and the SuperScript III kit (Invitrogen, Waltham, MA, USA). Real-time polymerase chain reaction (RT-PCR) was performed in a CFX96 (Biorad, Hercules, CA, USA) thermocycler using SybrGreen as fluorogenic probe in 25 µL reactions containing 5 µL cDNA sample, 10 supermix (Thermo Scientific, Waltham, MA, USA), 50 pmol of forward and reverse primer, and nuclease-free water (Applied Biosystems, Waltham, MA, USA). Each RT-PCR experiment was run in triplicate with nontemplate controls. The following forward and reverse primers were used: glyceraldehyde-3-phosphate dehydrogenase (GAPDH; NM_008084, Primer Bank ID: 126012538c3)—TGGCCTTCCGTGTTCCTAC, GAGTTGCTGTTGAAGTCGCA; interferon (IFN) 1β (NM_206975, Primer Bank ID: 6754304a1)—CAGCTCCAAGAAAGGACGAAC, GGCAGTGTAACTCTTCTGCAT; IFN-γ (NM_008337, Primer Bank ID: 145966741c2)—ACAGCAAGGCGAAAAAGGATG, TGGTGGACCACTCGGATGA; tumor necrosis factor α (TNF-α; NM_013693, Primer Bank ID: 133892368c3)—CCTGTAGCCCACGTCGTAG, GGGAGTAGACAAGGTACAACCC; interleukin 6 (IL-6; NM_031168, Primer Bank ID: 13624311a1)—TAGTCCTTCCTACCCCAATTTCC, TTGGTCCTTAGCCACTTCCTTC.
Melting curve analysis at the end of the reaction was performed to confirm the specificity of amplification of each pair of primers.
Data were analyzed using the comparative Ct method, normalized to GAPDH, and presented as fold-changes in gene expression of vaccinated and infected mice, relative to control nontreated animals.
Statistics
Data were processed using the Statistica software, version 6.0 (StatSoft, Inc. Tulsa, OK, USA). Mean values and standard error of the mean (SEM) were calculated to represent virus or bacterial titers. Serum titers of IgG, IgG1, and IgG2a were analyzed using log10 transformed data, and HI serum titers and local IgA were analyzed using transformed data and presented as mean ± SEM. To compare 2 independent groups, we used Mann-Whitney U test. To compare multiple independent groups, we used Kruskal-Wallis analysis of variance (ANOVA) test. To estimate the strength of the correlation between paired data, we used Spearman correlation coefficient (rs). A value of P < .05 was considered to be statistically significant.
Results
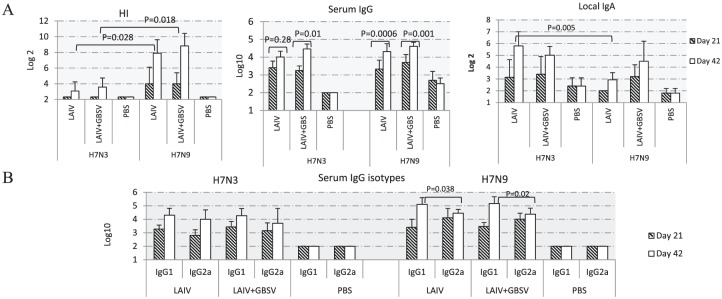
To study immunogenicity, 3 independent experiments were conducted. As the data were similar, the results of only one of them are provided in this publication. Sera and nasal washes from 10 to 12 mice were collected 3 weeks after the primary vaccination and revaccination (Figure 1). We used PBS-immunized sera as negative controls. As shown in Figure 2A, the LAIV of H7N3 subtype raised serum antibodies not only against the homologous virus but also against H7N9, which possessed the difference of 3% in the amino acid sequence of hemagglutinin (HA). In the sera from mice double-vaccinated with LAIV-containing preparations, serum HI titers against H7N9 were 20 to 40 times higher than against H7N3 (P < .05). Serum IgG levels measured by ELISA were similar against both H7N3 and H7N9 except that after immunization with LAIV only, the boost effect was more pronounced against the heterologous virus. In contrast, local IgA levels were 2.7 times higher against homologous H7N3 compared with H7N9 after vaccination with LAIV (P = .005; Figure 2A).
Figure 2.
Antibody responses against H7N3 and H7N9 influenza viruses in mice (n = 10-12) vaccinated intranasally with LAIV or LAIV + GBSV. Shown are the responses on day 21 after first vaccination and 3 weeks after the boost immunization. All data are presented as mean ± standard error of the mean. (A) Antibodies against H7N3 and H7N9 viruses in sera and nasal secrets, and (B) serum IgG subclasses after intranasal immunization with LAIV or LAIV + GBSV were detected using horseradish peroxidase–conjugated goat anti-mouse IgG1 or IgG2a. GBSV indicates group B streptococcus vaccine; HI, hemagglutination-inhibition; LAIV, live attenuated influenza vaccine; PBS, phosphate-buffered saline.
The IgG subtypes such as IgG1 and IgG2a against viral and bacterial vaccine components in each vaccine group were also evaluated by ELISA. As shown in Figure 2B, after primary vaccination with LAIV or LAIV + GBSV, the IgG2a antibody levels against H7N9 were slightly higher compared with IgG1, whereas on day 42 significantly higher IgG1 titers were seen (P = .038 and P = .02). At the same time, IgG1 and IgG2a antibody levels against H7N3 in both vaccine groups containing LAIV were detected in equal titers.
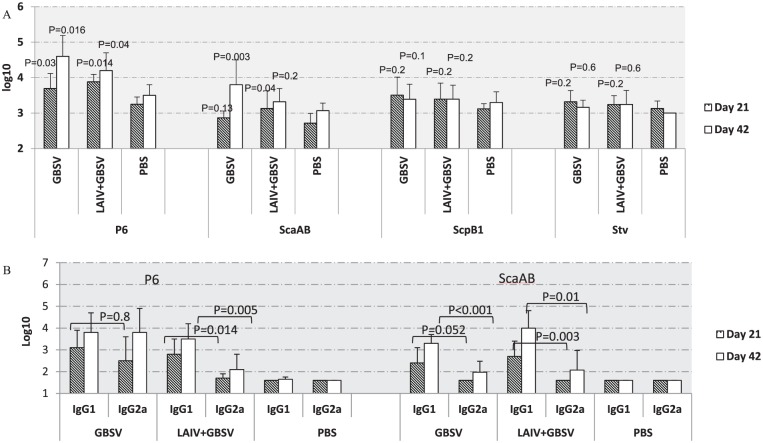
Among bacterial polypeptides, the most prominent immune response was demonstrated against the P6 polypeptide. Serum IgG titers against P6 either as a part of peptide vaccine or in the combined vaccine were significantly higher compared with mock-vaccinated animals after primary vaccination and revaccination (P < .05). Remarkably, the ScaAB polypeptide stimulated significant antibody rise (P = .028) in comparison with controls after revaccination when given only as a GBSV vaccine, unlike in a combination with LAIV (P = .2) (Figure 3A). The antibody titers against ScpB1 and Stv polypeptides did not differ from those of the control animals. Therefore, the subclasses of IgG were determined only against the P6 and ScaAB GBSV components. IgG1 titers against GBS polypeptides in almost all vaccine groups were significantly higher than the IgG2a titers (P < .05) after intranasal boost immunization. The highest IgG1/IgG2a ratio (21.1–85.1) against ScaAB peptide was seen within the LAIV + GBSV-immunized group, with a lower ratio (0–3.9) against immunodominant P6 polypeptide observed in GBSV group (Figure 3B). This finding may suggest different immunogenic properties of GBS peptides derived from various protein families.
Figure 3.
Antibody responses in vaccinated mice against the GBSV components (n = 10-12). All data are presented as mean ± standard error of the mean. (A) Serum ELISA IgG antibodies against individual GBSV peptides. P values presented compared with mock-vaccinated animals. (B) Serum IgG isotypes against P6 and ScaAB after intranasal immunization with GBSV or LAIV + GBSV. ELISA indicates enzyme-linked immunosorbent assay; GBSV, group B streptococcus vaccine; LAIV, live attenuated influenza vaccine; PBS, phosphate-buffered saline.
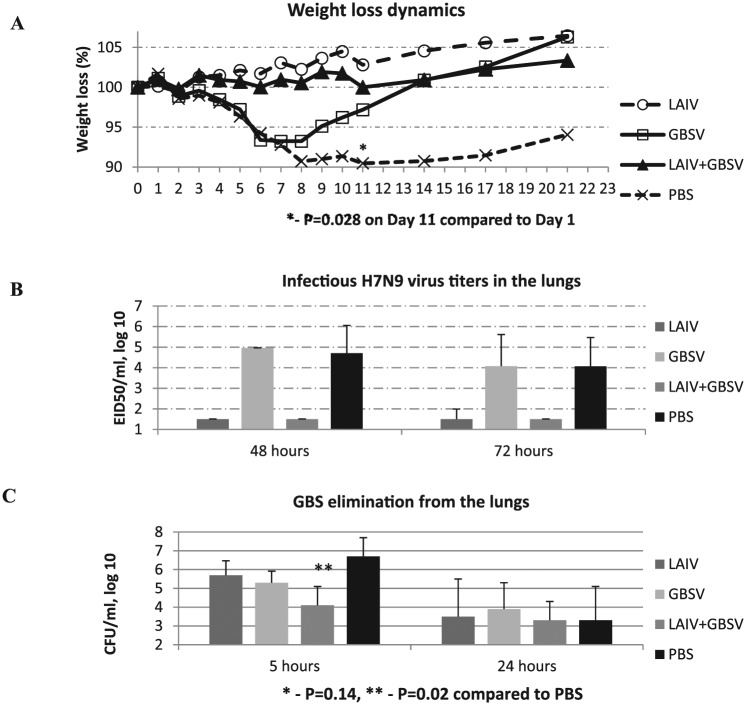
Challenge studies were conducted using various influenza viruses, including 2 studies with influenza A(H7H9) virus. In this study, data from one of the A(H7H9) experiments are provided. After primary challenges with A/Shanghai/2/2013(H7N9) CDC-RG influenza virus in an infectious dose of 300 MID50 followed by GBS infection, the maximum body weight loss (10%) was registered on days 8 to 11 in mock-vaccinated mice (Figure 4A). Vaccination with LAIV or with LAIV + GBSV completely protected mice against weight loss (0%), whereas immunization with GBSV alone only partially protected (6%). These data matched with virus isolation from the lungs, which was completely prevented in vaccine groups comprising LAIV while all of the mice were cleared (Figure 4B).
Figure 4.
Comparisons of the clinical manifestations of viral and bacterial infections in mice with the isolation of H7N9 influenza virus and group B streptococcus from the lungs. (A) Weight loss dynamics after A/Shanghai/2/2013(H7N9) infection followed by secondary GBS infection 24 hours apart; the results are expressed as the mean of 12 animals. (B) The H7N9 virus isolation from the lungs 48 and 72 hours after primary viral infection (n = 6). (C) GBS isolation from the lungs of mice 5 and 24 hours after secondary bacterial infections (n = 8). CFU indicates colony-forming unit; EID50, 50% egg infectious dose; GBS, group B streptococcus; GBSV, group B streptococcus vaccine; LAIV, live attenuated influenza vaccine; PBS, phosphate-buffered saline.
Group B streptococcus serotype II isolation from the lungs 5 hours post bacterial superinfection was an average of 400 times lower in the LAIV + GBSV group compared with nonvaccinated controls (P = .02), whereas LAIV or GBSV alone leads to 10 or 25 times reduction in bacterial titers, respectively (P = .14; Figure 4C). These results demonstrate that associated vaccination was the most effective way to reinforce bacterial elimination from the lungs after primary virus infection. The fact that GBS burden in GBSV-only immunized mice was not significantly reduced may be explained by the strengthened severity of influenza infection in the background of primary influenza. At the same time, in GBSV-alone vaccinated group—despite having high viral titers—the mice body weight is preserved in the long run. This can be explained by the enhanced clearance of infectious streptococci from the lungs, as shown in Figure 4C.
Figure 5 shows the results of pathological studies of lung slices from mice obtained on day 7 after primary virus infection. Figure 5A and B presents the lungs of the normal mice. On day 7 after the H7N9 virus infection followed by group B streptococci secondary infection, in the lungs of nonvaccinated mice, we observed severe progressive macrofocal pneumonia defined by a severe serous-hemorrhagic exudative process with massive damage of the alveolar epithelium and severe alveolar edema (Figure 5I). This lesion has occurred against the backdrop of viral damage of the small to medium bronchi (Figure 5J) and the severe vasculitis. In the case of immunization with GBS peptides only, the viral and bacterial infections lead to exudative acute bronchitis, lobular bronchopneumonia, and small focal serous pneumonia with obvious viral and bacterial damage (Figure 5E and F). In the lungs of mice vaccinated with the LAIV, only mild inflammatory changes, such as a microfocal serous pneumonia, were revealed (Figure 5C and D). The combined vaccination was the most effective in preventing the lung lesions after viral and bacterial infections (Figure 5G and H). Morphological signs of severe viral and bacterial lung injury (large or confluent pneumonia, widespread destruction of the bronchial epithelium, leukocyte infiltration of the alveoli) correlated with the levels of infectious virus isolation from the lungs (rs = 0.87, P < .01, n = 9).
Figure 5.
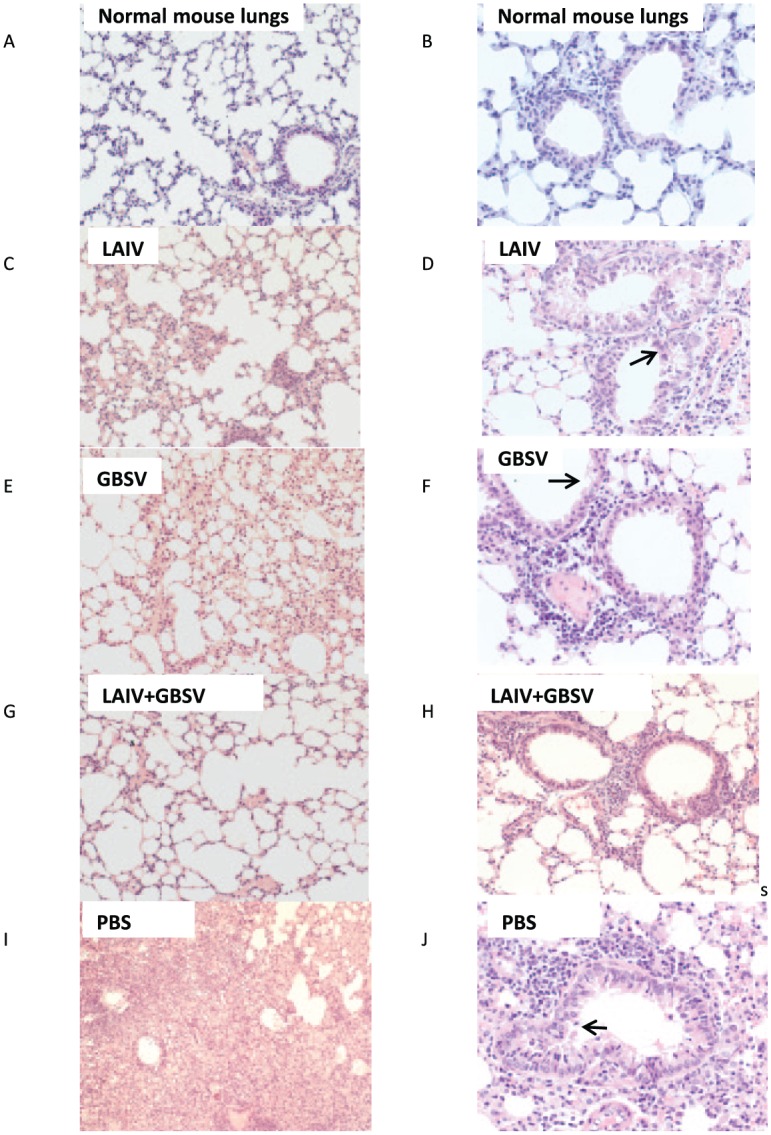
Hematoxylin-eosin stained sections from lung tissues of H7N9 and group B streptococcus–infected mice at day 7 post virus infection. (A, B) Normal mouse lungs. (A, C, E, G, I) Lung tissue pneumatization (magnification, ×200): E, G—focally slightly decreased, G—saved, I—pneumonic infiltration and alveolar edema. (B, D, F, H, J) Status of small bronchi and bronchioles (magnification, ×400): D—partial desquamation of epithelial cells (marked with an arrow), F—degenerative polymorphonuclear leukocytes in the lumen with severe damage of the epithelium (marked with an arrow), H—no any expressed pathological changes, J—desquamation of the epithelial cells (marked with an arrow), forming of pseudostratified epithelium, and local intraepithelial lymphocytic infiltration.
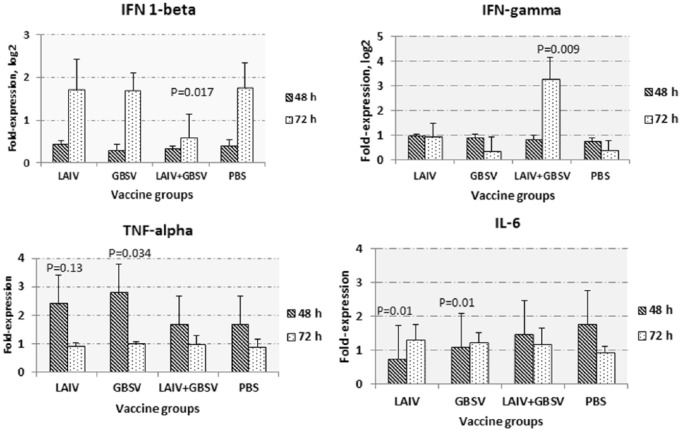
Using real-time reverse transcription polymerase chain reaction, we compared the elevation of early cytokines such as TNF-α, IL-6, and IFN 1 and 2 types, which were involved in defense against systemic viral infection by analyzing the messenger RNA (mRNA) expression in mouse lungs 48 and 72 hours post primary virus challenge. Differences between vaccine groups in the expression of IFN were detected 72 hours after viral infection. Although the standard deviations were quite high, the ANOVA test revealed statistically significant differences between groups, which were suggested by pairwise comparison between individual groups. In the LAIV + GBSV-immunized mice, the IFN-1β expression levels after the viral and bacterial challenge were significantly lower compared with nonvaccinated mice (P = .017) at this point in time (Figure 6). We did detect 2 to 8 times higher increase in IFN-γ expression in the LAIV + GBSV group compared with nontreated animals (P = .009). Statistically significant differences have also been found when comparing LAIV-immunized and PBS-vaccinated mice (P = .045). As for early cytokines, the difference in the expression of both TNF-α and IL-6 was more pronounced 48 hours after the infection. The TNF-α expression 48 hours p.i. was significantly higher in LAIV or GBSV group compared to PBS group (P = .013 and P = .034, respectively). The IL-6 expression was higher in the PBS group compared with those vaccinated with LAIV only or GBSV only. These data may indicate that in this particular case, in the development of lung injury during reinfection, an important role was played by mechanisms of adaptive immunity, rather than nonspecific inflammatory reactions.
Figure 6.
The mRNA expression in lung homogenates of immunized mice at 48 or 72 hours after intranasal inoculation with A/Shanghai/2/2013(H7N9) influenza virus and sequential group B streptococcus serotype II exposure (n = 5-6 in each group). All data presented as mean ± SEM. We used the 100 pg of total RNA for reverse transcription. Real-time polymerase chain reaction was performed using SybrGreen as a fluorophore. Mouse glyceraldehyde-3-phosphate dehydrogenase gene expression was used for normalization, and lungs homogenates of untreated mice were used as a control. P values provided compared with PBS group. GBSV indicates group B streptococcus vaccine; IL-6, interleukin 6; LAIV, live attenuated influenza vaccine; PBS, phosphate-buffered saline; SEM, standard error of the mean.
Discussion
The most severe influenza diseases are often accompanied by bacterial complications, which are the main cause of death during influenza infection. One of the new approaches to microbial vaccines development is the use of recombinant polypeptides corresponding to conserved portions of surface proteins of bacterial human pathogens.
The effectiveness of excess serum HI titers against H7N9 influenza virus compared with those against homologous H7N3 after LAIV or LAIV + GBSV in boosting immunization has confirmed a number of previously published data about cross-reaction of existing H7N3 cold-adapted vaccines against divergent H7N9 strains possessing pandemic potential,17–19 despite the fact that the HI has been considered a strain-specific reaction. This may be attributed to the changes in viral receptor-binding characteristics.20 The study of a number of vaccines against influenza H7 subtype, both inactivated and live, has shown that a double vaccination is required to achieve an adequate antibody response.21–23 Recent studies have shown that after vaccination with inactivated vaccines in patients primed with LAIV, specific antibodies to the globular part of H7 increased dramatically and were characterized by more pronounced affinity and avidity, thereby evidencing the importance of LAIV development for pre-pandemic vaccination.24
Evaluation of serum IgG antibodies against individual GBSV peptides confirmed the earlier findings of the greatest immunogenicity of P6 after intranasal administration.13 The most poorly immunogenic when administered intranasally was Stv peptide mass 130 kDa consisting of 2 parts, the immunogenicity of which after subcutaneous vaccination has been shown previously.14 Perhaps the lack of an immune response to the polypeptides caused by their structure led to rapid destruction by the proteolytic enzymes, which are in abundance in mucous secretion.
The subclasses of serum IgG induced after immunization with viral or bacterial vaccines may indicate the relative contribution of Th2-type versus Th1-type cytokines.25 Previously it was reported that after viral infections, the major antibody isotype in the sera of various strains of mice was presented with IgG2a26 stimulated during Th1-type immune responses.27 Huber et al28 have shown that stimulation of IgG2a antibodies after vaccination with influenza HA-expressing vectors provided effective protection against lethal challenge with H3N2 influenza virus based on PR8/34 in the absence of strong neutralizing antibody titers. At the same time, the authors argue the importance of both IgG1 and IgG2a expression in vaccination against influenza. In our study, the outbred mice vaccinated using H7N3 LAIV displayed a significant increase in both IgG isotypes against H7N9 influenza with a greater increase in titers of IgG1, although against homologous H7N3 equal IgG1/IgG2 immune responses were obtained. Notably, we had previously detected delayed clearance of the GBS after infection with homologous wild-type virus A/mallard/Netherlands/12/00(H7N3) at an early period of re-infection,15 whereas after H7N9 virus reinfection such a delay was not observed. These differences can be partially explained by the fact that IgG2a isotype is the most effective activating Fc receptor–mediated effector functions, which include the stimulation of antibody-dependent cell-mediated cytotoxicity.29,30
After viral and bacterial challenge, the results of quantitative viral and bacteriological studies were consistent with the clinical and morphological study. Postchallenge dynamics of the weight loss have shown that LAIV or LAIV + GBSV-vaccinated mice did not lose weight which was associated with no infectious virus isolation from the lungs in these vaccine groups. This may indicate the predominant viral lesions of the lung, detected at a morphological examination of mice vaccinated using GBSV only or nonvaccinated. Indeed, in the absence of vaccination, viral and bacterial infections lead to the development of alveolar edema, severe damage to the blood vessel endothelium, severe exudative process with massive damage to the alveolar epithelium, and the evidence of viral damage associated with the highest isolation of the virus from the lungs. However, in the group of GBSV-vaccinated mice, the challenge virus also was isolated from the lungs in high titers, although the damage was limited by focal pneumonia wherein the GBS burden was 25 times lower compared with nonvaccinated animals. These data may indicate that GBS, though left from the lungs in 2 days after the secondary bacterial infection, was still able to complicate the course of infection in nonvaccinated mice, whereas specific GBS antibodies due to GBSV immunization prevented this.
The enhanced lung protection after challenge in the LAIV + GBSV group, compared with mice from other vaccine groups, was associated with higher expression of IFN-γ, which may indicate induction T-cell immunity although IFN-γ is often used to monitor CD8 + cell-mediated immunity. Data on the increased IFN-gamma expression levels after the viral and bacterial challenge of the LAIV + GBSV immunized mice were also confirmed after A/PR8 and GBS challenge of mice vaccinated with LAIV + GBSV (data not shown). In addition, in this study, we used the infectious virus based on A/PR8/34 background with the A/Shanghai/2/2013(H7N9) influenza virus surface antigens. Previously, Sun and Metzger31 reported that pulmonary IFN-γ produced during T-cell responses to A/PR8/34 influenza infection in mice inhibited initial S pneumoniae clearance from the lung by alveolar macrophages due to suppression of phagocytosis and leads to enhanced susceptibility to secondary pneumococcal infection, which was prevented by IFN-γ neutralization after influenza infection. In our study, the IFN-γ expression in control mice after viral and bacterial challenges was 1.3 times higher compared with intact animals, although after the LAIV + GBSV vaccination this increase was generally more pronounced.
Although the mouse influenza pneumonia model is often considered unsuitable for extrapolating the effects of IFNs in humans due to physiological differences in IFN responsiveness between species,32 the level of interferon in the lungs is associated with the reproduction of infectious influenza virus in both humans and mice. Moreover, in both humans and mice, the same receptor complex is used for both IFN-1α and IFN-1β.33 Despite previously reporting no major contribution for IFN-1α/IFN-1β pathways in recovery from influenza virus infection in mice, the type 1 IFNs can play roles in determining the rate of virus replication in the initial stages of infection and in shaping the initial inflammatory immune response.32 Mok et al34 have shown that day 3 p.i., in mice infected with A/Shanghai/2/2013(H7N9) virus, lung viral titers were positively correlated with elevated levels of proinflammatory cytokines TNF-α, IFN-1α, IFN-γ–induced protein 10 (IP-10), although there was no significant correlation between virus replication and cytokine levels at day 5 p.i. Yoo et al35 demonstrated that IFN-1β treatment elicited enhanced Th1 effector and cytolytic T-cell responses in the lungs of mice infected with A/WSN/33(H1N1) influenza virus. Associated with the polarization toward a type 1 immune response, IFN-1β treatment of mice resulted in accelerated viral clearance and diminished pulmonary eosinophilia in infected lung tissues. Chan et al36 reported a significant reduction in IFN-1 responses and an associated lymphocyte activation following a homotypic virus challenge due to vaccine-induced antibody responses. Although interferon signaling may be beneficial in response to a virus, it is still not clear how it affects the development of secondary bacterial pneumonia. Shahangian et al37 considered that elevated type 1 interferon after primary influenza infection may play a role in increasing the lung’s susceptibility to secondary pneumococcal pneumonia. In this regard, we tried to follow the lung IFN-1β mRNA expression during the onset of double viral and bacterial infections influenced by LAIV and GBSV or merged vaccination. In this study, at 72 hours post primary virus infection, we detected 1.5 to 3.4 increase of IFN-1β expression in all groups of mice compared with the control intact animals with the noticeably lower elevation after LAIV + GBSV vaccination compared with nonvaccinated mice and the LAIV- or GBSV-vaccinated mice.
Smith et al38 found that a range of proinflammatory cytokines was elevated in mice sequentially infected with influenza and S pneumoniae. The production of IL-6 and TNF-α has been shown to enhance vascular permeability and may enable the extrapulmonary spread of bacteria.39 In our work, we have revealed notable differences in IL-6 expression after viral and GBS challenge between nonvaccinated animals and any of the vaccine groups 48 hours p.i. with the least significant decrease being in the LAIV + GBSV group (P = .25). The TNF-α mRNA expression was heightened after vaccination with LAIV only or GBSV only.
Taken together, these data suggest that intranasal immunization using LAIV and GBSV may modulate innate immunity pathways, thus reducing the primary viral and secondary bacterial infection.
It is not uncommon to believe that even influenza viruses producing minimal epithelial cell damage still enhance subsequent bacterial infection in mice.40,41 Influenza neuraminidase and upregulation of platelet-activating factor receptor expression during murine viral infection may increase bacterial adherence.42 In our study, the use of H7N3 LAIV had a positive effect on the course of influenza and GBS infections. All methods of vaccination improved the course of the disease after viral challenge and bacterial invasion. Vaccination using LAIV decreased primary viral outcomes, and this effect can aid in the prevention of secondary bacterial infections
In conclusion, the combined vaccination with LAIV and GBS polypeptides provided advantageous protection against infections with A/Shanghai/2/2013(H7N9) CDC-RG influenza virus followed by serotype II GBS infection and improved bacterial clearance from the lungs of mice compared with LAIV or GBSV, whereas mock-vaccinated animals displayed severe morbidity, including significant weight loss and pathomorphological manifestations of severe viral and bacterial lung injury. Use of mucosal routes for vaccine administration ensured the formation of systemic and local immunity is one of the possible strategies to improve the vaccine’s immunogenicity.43 In this regard, the bacterial peptides could be optimized for mucosal delivery by insertion into virus vector constructions to better mimic the natural route of virus infection and potentially elicit mucosal immune responses.
Footnotes
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2535 words, excluding any confidential comments to the academic editor.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the budget of Federal State Budgetary Scientific Institution “Institute of Experimental Medicine”, Saint Petersburg, Russia and the subsidy funds from the Federal budget under the agreement No. 14.613.21.0023.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions
YAD and GFL conceived and designed the experiments. TAK, TAS, KBG, and GOL analyzed the data. YAD and GFL wrote the first draft of the manuscript. TAK and ANS contributed to the writing of the manuscript. YAD, GFL, TAK, and ANS jointly developed the structure and arguments for the paper. ANS and LGR made critical revisions and approved the final version. All authors reviewed and approved the final manuscript.
REFERENCES
- 1.World Health Organization http://www.who.int/csr/don/17-august-2016-ah7n9-china/en/
- 2.Wu Z, Sha J, Yu Z, et al. Epidemiological and virological differences in human clustered and sporadic infections with avian influenza A H7N9. Int J Infect Dis. 2016;49:9–17. doi: 10.1016/j.ijid.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 3.Zeng H, Belser JA, Goldsmith CS, et al. A(H7N9) virus results in early induction of proinflammatory cytokine responses in both human lung epithelial and endothelial cells and shows increased human adaptation compared with avian H5N1 virus. J Virol. 2015;89:4655–4667. doi: 10.1128/JVI.03095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajagopal L. Understanding the regulation of Group B streptococcal virulence factors. Future Microbiol. 2009;4:201–221. doi: 10.2217/17460913.4.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention Bacterial coinfections in lung tissue specimens from fatal cases of 2009 pandemic influenza A (H1N1)—United States. Morb Mortal Wkly Rep. 2009;58:1071–1074. [PubMed] [Google Scholar]
- 6.Ustinovitch I, Vlasov G, Totolyan A, Suvorov A. Cloning and expression of gene fragment IgA-binding protein of group B streptococci. Folia Microbiol. 1999;44:726–728. doi: 10.1007/BF02825670. [DOI] [PubMed] [Google Scholar]
- 7.Kling DE, Gravekamp C, Madoff LC, Michel JL. Characterization of two distinct opsonic and protective epitopes within the alpha C protein of the group B Streptococcus. Infect Immun. 1997;65:1462–1467. doi: 10.1128/iai.65.4.1462-1467.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganeshkumar N, Arora N, Kolenbrander PE. Saliva-binding protein (SsaB) from Streptococcus sanguis 12 is a lipoprotein. J Bacteriol. 1993;175:572–574. doi: 10.1128/jb.175.2.572-574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vorobieva EI, Meringova LF, Leontieva GF, Grabovskaya KB, Suvorov AN. Analysis of recombinant group B streptococcal protein ScaAB and evaluation of its immunogenicity. Folia Microbiol. 2005;50:172–176. doi: 10.1007/BF02931468. [DOI] [PubMed] [Google Scholar]
- 10.José RJ, Periselneris JN, Brown JS. Community-acquired pneumonia. Curr Opin Pulm Med. 2015;21:212–218. doi: 10.1097/MCP.0000000000000150. [DOI] [PubMed] [Google Scholar]
- 11.Berry AM, Paton JC. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect Immun. 1996;64:5255–5262. doi: 10.1128/iai.64.12.5255-5262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleary PP, Handley J, Suvorov AN, Podbielski A, Ferrieri P. Similarity between the group B and A streptococcal C5a peptidase genes. Infect Immun. 1992;60:4239–4244. doi: 10.1128/iai.60.10.4239-4244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desheva YA, Leontieva GF, Kramskaya TA, et al. Evaluation in mouse model of combined virus-bacterial vaccine based on attenuated influenza A(H7N3) virus and the group B streptococcus recombinant polypeptides. Open Microbiol J. 2016;10:168. doi: 10.2174/1874285801610010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suvorov A, Ustinovitch I, Meringova L, Grabovskaya K. Construction of recombinant polypeptides based on beta antigen C (Bac) protein & their usage for protection against group B streptococcal infection. Indian J Med Res. 2004;119:228. [PubMed] [Google Scholar]
- 15.Rowe T, Abernathy RA, Hu-Primmer J, et al. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J Clin Microbiol. 1999;37:937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X, Tumpey TM, Morken T, Zaki SR, Cox NJ, Katz JM. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J Virol. 1999;73:5903–5911. doi: 10.1128/jvi.73.7.5903-5911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Q, Chen Z, Cheng X, Xu L, Jin H. Evaluation of live attenuated H7N3 and H7N7 vaccine viruses for their receptor binding preferences, immunogenicity in ferrets and cross reactivity to the novel H7N9 virus. PLoS ONE. 2013;8:e76884. doi: 10.1371/journal.pone.0076884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph T, McAuliffe J, Lu B, et al. A live attenuated cold-adapted influenza A H7N3 virus vaccine provides protection against homologous and heterologous H7 viruses in mice and ferrets. Virology. 2008;378:123–132. doi: 10.1016/j.virol.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krammer F, Albrecht RA, Tan GS, et al. Divergent H7 immunogens offer protection from H7N9 virus challenge. J Virol. 2014;88:3976–3985. doi: 10.1128/JVI.03095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gambaryan AS, Matrosovich TY, Philipp J, et al. Receptor-binding profiles of H7 subtype influenza viruses in different host species. J Virol. 2012;86:4370–4379. doi: 10.1128/JVI.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fries LF, Smith GE, Glenn GM. A recombinant viruslike particle influenza A (H7N9) vaccine. N Engl J Med. 2013;369:2564–2566. doi: 10.1056/NEJMc1313186. [DOI] [PubMed] [Google Scholar]
- 22.Mulligan MJ, Bernstein DI, Winokur P, et al. Serological responses to an avian influenza A/H7N9 vaccine mixed at the point-of-use with MF59 adjuvant: a randomized clinical trial. JAMA. 2014;312:1409–1419. doi: 10.1001/jama.2014.12854. [DOI] [PubMed] [Google Scholar]
- 23.Shcherbik S, Pearce N, Balish A, et al. Generation and characterization of live attenuated influenza A (H7N9) candidate vaccine virus based on russian donor of attenuation. PLoS ONE. 2015;10:e0138951. doi: 10.1371/journal.pone.0138951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halliley JL, Khurana S, Krammer F, et al. High-affinity H7 head and stalk domain–specific antibody responses to an inactivated influenza H7N7 vaccine after priming with live attenuated influenza vaccine. J Infect Dis. 2015;212:1270–1278. doi: 10.1093/infdis/jiv210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura SI, Kurata T. Defense mechanisms against influenza virus infection in the respiratory tract mucosa. Jpn J Infect Dis. 2004;57:236–247. [PubMed] [Google Scholar]
- 26.Coutelier JP, Van der Logt JT, Heessen FW, Warnier G, Van Snick J. IgG2a restriction of murine antibodies elicited by viral infections. J Exp Med. 1987;165:64–69. doi: 10.1084/jem.165.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snapper CM, Paul WE. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236:944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- 28.Huber VC, McKeon RM, Brackin MN, et al. Distinct contributions of vaccine-induced immunoglobulin G1 (IgG1) and IgG2a antibodies to protective immunity against influenza. Clin Vaccine Immunol. 2006;13:981–990. doi: 10.1128/CVI.00156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuberger MS, Rajewsky K. Activation of mouse complement by monoclonal mouse antibodies. Eur J Immunol. 1981;11:1012–1016. doi: 10.1002/eji.1830111212. [DOI] [PubMed] [Google Scholar]
- 30.Kipps TJ, Parham P, Punt J, Herzenberg LA. Importance of immunoglobulin isotype in human antibody-dependent, cell-mediated cytotoxicity directed by murine monoclonal antibodies. J Exp Med. 1985;161:1–17. doi: 10.1084/jem.161.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun K, Metzger DW. Inhibition of pulmonary antibacterial defense by interferongamma during recovery from influenza infection. Nat Med. 2008;14:558–564. doi: 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- 32.Price GE, Gaszewska-Mastarlarz A, Moskophidis D. The role of alpha/beta and gamma interferons in development of immunity to influenza A virus in mice. J Virol. 2000;74:3996–4003. doi: 10.1128/jvi.74.9.3996-4003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Díaz MO. The human type I interferon gene cluster. Semin Virol. 1995;6:143–149. [Google Scholar]
- 34.Mok CKP, Lee HHY, Chan MCW, et al. Pathogenicity of the novel A/H7N9 influenza virus in mice. MBio. 2013;4:e00362–e00413. doi: 10.1128/mBio.00362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoo JK, Baker DP, Fish EN. Interferon-β modulates type 1 immunity during influenza virus infection. Antiviral Res. 2010;88:64–71. doi: 10.1016/j.antiviral.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Chan J, Babb R, David SC, McColl SR, Alsharifi M. Vaccine-induced antibody responses prevent the induction of interferon type I responses upon a homotypic live virus challenge. Scand J Immunol. 2016;83:165–173. doi: 10.1111/sji.12410. [DOI] [PubMed] [Google Scholar]
- 37.Shahangian A, Chow EK, Tian X, et al. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest. 2009;119:1910–1920. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith MW, Schmidt JE, Rehg JE, Orihuela CJ, McCullers JA. Induction of pro- and anti-inflammatory molecules in a mouse model of pneumococcal pneumonia after influenza. Comp Med. 2007;57:82–89. [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Le TQ, Kurihara N, et al. Influenza Virus—cytokine-protease cycle in the pathogenesis of vascular hyperpermeability in severe influenza. J Infect Dis. 2010;202:991–1001. doi: 10.1086/656044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong HH, Chen Y, James M, Van Deusen J, Welling DB, DeMaria TF. Expression of cytokine and chemokine genes by human middle ear epithelial cells induced by formalin-killed Haemophilus influenzae or its lipooligosaccharide htrB and rfaD mutants. Infect Immun. 2001;69:3678–3684. doi: 10.1128/IAI.69.6.3678-3684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alymova IV, Portner A, Takimoto T, Boyd KL, Babu YS, McCullers JA. The novel parainfluenza virus hemagglutinin-neuraminidase inhibitor BCX 2798 prevents lethal synergism between a paramyxovirus and Streptococcus pneumoniae. Antimicrob Agents Chemother. 2005;49:398–405. doi: 10.1128/AAC.49.1.398-405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCullers JA, Bartmess KC. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J Infect Dis. 2003;187:1000–1009. doi: 10.1086/368163. [DOI] [PubMed] [Google Scholar]
- 43.Song L, Xiong D, Hu M, Kang X, Pan Z, Jiao X. Immunopotentiation of different adjuvants on humoral and cellular immune responses induced by HA1-2 subunit vaccines of H7N9 influenza in mice. PLoS ONE. 2016;11:e.0150678. doi: 10.1371/journal.pone.0150678. [DOI] [PMC free article] [PubMed] [Google Scholar]