Abstract
Suppression of androgen receptor (AR) activity in prostate cancer by androgen depletion or direct AR antagonist treatment, although initially effective, leads to incurable castration resistant prostate cancer (CRPC) via compensatory mechanisms including resurgence of AR and AR splice variant (ARV) signaling. Emerging evidence suggests that Sigma1 (also known as sigma-1 receptor) is a unique chaperone or scaffolding protein that contributes to cellular protein homeostasis. We reported previously that some Sigma1-selective small molecules can be used to pharmacologically modulate protein homeostasis pathways. We hypothesized that these Sigma1 mediated responses could be exploited to suppress AR protein levels and activity. Here we demonstrate that treatment with a small molecule Sigma1 inhibitor prevented 5α-dihydrotestosterone (DHT)-mediated nuclear translocation of AR and induced proteasomal degradation of AR and ARV, suppressing the transcriptional activity and protein levels of both full-length and splice-variant AR. Consistent with these data, RNAi knockdown of Sigma1 resulted in decreased AR levels and transcriptional activity. Furthermore, Sigma1 physically associated with ARV7 and ARv567es as well as full-length AR. Treatment of mice xenografted with ARV-driven CRPC tumors with a drug-like small molecule Sigma1 inhibitor significantly inhibited tumor growth associated with elimination of AR and ARV7 in responsive tumors. Together, our data show that Sigma1 modulators can be used to suppress AR/ARV-driven prostate cancer cells via regulation of pharmacologically responsive Sigma1-AR/ARV interactions both in vitro and in vivo.
Keywords: Sigma1, small molecule, androgen receptor, splice variant, ErbB receptor, chaperone, scaffold, prostate cancer, castration resistant prostate cancer (CRPC)
INTRODUCTION
Androgen receptor (AR) is the primary driver of prostate cancer progression, and at all stages of disease prostate cancer cells are dependent upon AR signaling for growth and survival (1, 2). First line therapy involves suppression of AR activity by androgen depletion or direct AR ligand treatment. However, incurable castration resistant tumors develop as a result of resurgent AR activity attributable to a series of molecular alterations including: increased expression of AR or AR co-factors, mutations that facilitate promiscuous AR activation by non-androgen steroid hormones, or constitutively active AR splice variants that are non-responsive to androgen ablation or AR ligand treatment (1–4). Furthermore, intratumoral androgen biosynthesis and compensatory activation of multiple kinase pathways have been implicated in reactivation of the AR axis in castration resistant prostate cancer (CRPC) (2, 3). The adaptive nature of the AR axis in prostate cancer cells underscores the importance of discovering and developing novel approaches to overcome the inevitable resistance to current AR-targeted therapies by targeting AR and the networks on which it depends.
In prostate cancer cells the cascade of events leading to AR transcriptional activity is initiated by binding to dihydrotestosterone (DHT), which is converted from testosterone by the enzyme 5α-reductase (2, 4). DHT binding to the carboxy-terminal androgen binding domain results in AR conformational changes, dimerization, and transport from the cytoplasm to the nucleus wherein it interacts with transcriptional co-factors and is assembled into DNA bound protein complexes that produce AR-driven transcriptional activity (2). In the absence of bound androgens newly synthesized AR is held in the cytoplasm by multi-chaperone complexes, including heat shock proteins (HSPs), which are thought to stabilize cytoplasmic AR. Protein chaperones, including heat shock protein 90 (HSP90), bind to and facilitate the processing, assembly, and transport of AR and associated proteins. Disruption of AR-chaperone associations can destabilize AR and result in subsequent degradation by the ubiquitin proteasome system (UPS) (5, 6).
Multiple AR splice variants (ARV) have been identified in CRPC tumors and cell lines (2, 4). The best characterized ARV are the ARV7 and ARv567es splice variants, both of which lack much of the carboxy-terminal region of AR, including the ligand-binding domain (LBD) (2, 4). Therefore the activities of these ARV are hormone independent, and LBD targeted therapeutic agents such as enzalutamide are ineffective against these variants (2, 4).
Sigma1 (traditionally known as the sigma-1 receptor) is a unique 26 kilodalton integral membrane protein that more recently has been described as a molecular chaperone (7–9). Sigma1 is highly expressed in various cancer cell lines and can be detected throughout the cytoplasm, primarily as an endoplasmic reticulum (ER) protein with a long cytoplasmic tail (7, 8, 10). The physiological role of Sigma1 in cancer cells is still unclear. However, emerging evidence from our laboratory and others (9, 11) suggest that Sigma1 may in fact function as a novel molecular chaperone or a scaffolding protein in cancer cells. We have found that Sigma1 is involved in aspects of cancer cell protein homeostasis including protein synthesis, folding, transport, and degradation (12, 13).
Furthermore, we have found that these Sigma1 protein homeostasis regulating functions can be pharmacologically modulated by selective small molecule compounds. Negative modulators of Sigma1 (inhibitors) disrupt protein homeostasis in cancer cells and thus suppress their growth and survival. Here we asked whether we could exploit these small molecule Sigma1 modulator responses to suppress AR levels and activity in hormone-sensitive and CRPC cells.
MATERIALS AND METHODS
Chemicals
IPAG (1-(4-Iodophenyl)-3-(2-adamantyl) guanidine) was purchased from Tocris. Bafilomycin A1, inhibitor of the vacuolar type H+-ATPase (V-ATPase), 17-AAG (17-N-allylamino-17-demethoxygeldanamycin), MG132 (Z-Leu-Leu-Leu-al), and 5α-dihydrotestosterone (DHT) were purchased from Sigma-Aldrich. Cycloheximide was purchased from Alfa Aesar.
Cell lines and transfections
The LNCaP, VCaP, 22Rv1, and PC3 prostate cancer cell lines were acquired directly from ATCC. C4-2 cells were acquired directly from MD Anderson’s Characterized Cell line Core. All of these cell lines were authenticated by short tandem repeat (STR) profiling. Cell lines were acquired within the past six years. LAPC4 cells were a kind gift from Dr. Charles Sawyers (Memorial Sloan-Kettering Cancer Center). Under standard culture conditions, cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (Corning). For DHT induction assays, cells were maintained for 3–5 days in phenol-red free IMEM supplemented with 5% charcoal stripped serum (CSS) (Corning) prior to DHT induction. For experiments performed under standard growth, steady-state conditions, cells were seeded approximately 24 hours prior to start of drug treatment in most assays.
Control-A siRNA was purchased from Santa Cruz Biotechnology. The two human Sigma1 siRNA tested here were purchased from Santa Cruz Biotechnology and Dharmacon. Human ATG5 siRNA was purchased from Cell Signaling Technologies. For all Sigma1 and ATG5 siRNA knockdown experiments, 100 nanomoles of siRNA per approximately 100,000 cells was transfected with INTERFERin transfection reagent (PolyPlus), and 48 hours later, cells were reseeded, allowed to attach and recover for 16–24 hours and transfected again.
The FLAG-AR, FLAG-ARV7, and FLAG-ARv567es plasmid constructs were gifts from Dr. Stephen Plymate (University of Washington School of Medicine, Seattle, WA) and have been described elsewhere(14). Plasmid transfections were performed with jetPRIME transfection reagent (PolyPlus) according to manufacturer’s procedures.
Colony formation, cell survival, and death assays
Trypan blue exclusion assays were used to quantify cell death. Values were generated from at least 3 independent determinations, and statistical significance was determined as described below.
For the 3-D colony formation in soft agar assay, cells in culture medium were gently mixed at a 1:1 ratio with 0.6% agar and seeded at a density of 5,000 cells per milliliter of soft agar per 35mm well and layered on top of a solid footer comprising 1% agar (BD Biosciences). The soft agar layer was allowed to solidify, cell culture medium was added to cover the soft agar layer, and 24 hours later drug treatment was started. Cells were allowed to grow for approximately 4 weeks with weekly medium changes until visible colonies formed in untreated control cells. The suspended colonies were fixed with methanol and stained with 0.01% crystal violet (Fisher). Colonies were counted automatically using the GelCount Colony Counter (Oxford Optronix).
For 2-D colony formation and crystal violet assays, 2 or 4 × 103 cells were seeded into a tissue culture-treated 12-well dish. Twenty-four hours after seeding drug treatment was applied to the wells and medium was replaced weekly until visible colonies formed in untreated control cells. Subsequently, the cells were washed with DPBS then fixed in 4% formaldehyde. The fixed cells were stained with 0.4% crystal violet (Fisher), washed with water and allowed to air-dry. Plates were photographed for analysis and colony counting. Crystal violet was extracted from cells by methanol and absorbance measurements at OD 540 nm were acquired with a VersaMax plate reader (Molecular Devices). The relative number of surviving cell colonies was quantified and normalized data presented as the ratio of 540 nm values in the experimental treatment wells to 540 nm absorbance values in the untreated control wells.
Immunoblots and antibodies
Cell lysis, protein extraction, SDS-PAGE, and immunoblotting were performed as described previously (13), with a few modifications. The Luminata Western HRP Substrate chemiluminescence kit (Millipore) was used to reveal immunoblotted proteins. The rabbit polyclonal anti-Sigma1 antibody was generated in our laboratory as described elsewhere (13). The anti-β-actin and anti-HA (Y-11) antibodies were purchased from Santa Cruz Biotechnology. The rabbit anti-FLAG, anti-GAPDH, anti-LC3B, anti-ATG5, anti-p97/VCP, anti-PSMB5, anti-polyubiquitin (clone P4D1), anti-HSP90, anti-HSP70, anti-HSP27, anti-glucocorticoid receptor (GR), anti-ErbB2/HER2, anti-ErbB3/HER3, anti-Akt antibodies, and horseradish peroxidase conjugated secondary antibodies were all purchased from Cell Signaling Technology. The anti-androgen receptor (D6F11) rabbit monoclonal antibody (Cell Signaling Technology) used here was raised against an epitope in the amino-terminus of AR. The anti-ARV7 antibody (Abcam, EPR15656) was raised against an epitope specific to the ARV7 splice variant, and the anti-ARv567es antibody (Abcam, EPR15657) was raised against an epitope specific to the ARv567es splice variant. We confirmed the specificity of the commercial ARV7 and ARv567es antibodies (Supplemental Figure 1).
Confocal microscopy
Cells were seeded onto #1.5 (0.17 mm) borosilicate glass coverslips (Electron Microscopy Sciences) coated with 0.1 mg/mL 75,000–150,000 MW poly-d-lysine substrate (Sigma-Aldrich) 36 hours prior to fixation. The cells were washed with room temperature Dulbecco’s modified phosphate buffered saline solution (DPBS) and fixed in 4% formaldehyde (Pierce) for 15 minutes. After DPBS washes, cells were subjected to antigen retrieval using a 10% (v/v) citrate-based unmasking solution (Vector) for 10 minutes at 95°C. To reduce nonspecific secondary staining, cells were incubated with Image-iT FX solution (Molecular Probes) for 30 minutes at room temperature and washed with DPBS. Cells were blocked in 5% (v/v) normal goat serum (Invitrogen), 0.3% (v/v) Triton X-100 in DPBS for 1 hour. Sigma1 immunostaining was performed with our rabbit polyclonal anti-Sigma1 antibody (described previously (13)) in 1% (w/v) bovine serum albumin (BSA), 0.3% (v/v) Triton X-100 in DPBS, overnight at 4°C. After the primary stain, cells were washed with DPBS and stained with the corresponding secondary Alexa Fluor 488 goat anti-rabbit IgG (H+L) (Molecular Probes) in 1% (w/v) BSA, 0.3% (v/v) Triton X-100 in DPBS for 2 hours at room temperature. Cells were washed with DPBS, nuclear counter-stained with 0.1 μg/mL DAPI (Pierce), then mounted onto glass slides using Prolong Gold (Molecular Probes). Coverslips were sealed using nail polish 36 hours after mounting. Images were acquired using the Olympus FV1000 inverted confocal microscope using a 60x 1.42 NA oil immersion objective at a scanning resolution of 0.051 μm/pixel. Five randomly selected microscopic fields were taken for each condition.
AR transcriptional activity assay by quantitative RT-PCR (qRT-PCR)
RNA templates for qRT-PCR consisted of total RNA isolated using the RNeasy kit (Qiagen) per manufacturer’s protocol. Taq-Man primer probe sets were purchased from Life Technologies and the genes and catalog numbers used for the qRT-PCR experiments are the following: KLK3/PSA, (Hs03063374-m1), TMPRSS2 (Hs01120965-m1), and GAPDH (Hs99999905-m1). qRT-PCR was performed using the 7900HT- Fast Real Time PCR System (Applied Biosystems) and the reactions were performed in triplicate using the Brilliant II qRT-PCR master mix one step (Agilent Technologies) following the manufacturer’s instructions. Data were normalized to GAPDH transcript levels and presented as fold increase or percent decrease of KLK3/PSA and TMPRSS2 transcripts relative to non-treated or DMSO treated controls.
AR transcriptional activity assay by AR response element dependent luciferase reporter
The Human Androgen Receptor Reporter Assay System (Indigo Biosciences, product #IB03001) was used to evaluate AR transcriptional activity. Briefly, this assay uses Chinese hamster ovary cells expressing human AR and an Androgen Responsive Element (ARE)-firefly luciferase construct. The manufacturers protocol was adhered to with the following exceptions: cells were seeded in CRM and compound-free CSM and allowed to adhere for 24 hours, at which time the media was replaced with CSM containing compounds at the listed concentrations and 400 pM of the androgen 6αFl-Testosterone (6αFlT). The CRM and CSM media are described in the manufacturer product materials. The synthetic androgen analog, 6αFIT, was used as a reference agonist for the AR system. Once the combination treatment was applied the plates were incubated for 16 hours. The plate was read with an integration time of 500 milliseconds per well and 3 consecutive whole-plate reads.
AR nuclear localization assay
LNCaP cells were transfected with a GFP-AR plasmid construct and grown in selection medium containing 1 mg/mL G418 sulfate for approximately 3 weeks. Resistant colonies were selected and pooled, and stably-transfected GFP-positive cells were isolated by cell sorting.
For the DHT induced nuclear localization assay, LNCaP(GFP-AR) cells were seeded as described above for confocal microscopy. Cells were cultured in phenol-red free IMEM containing 5% charcoal stripped serum (CSS) for 3–5 days. Subsequently, cells were pre-incubated for 30 minutes with 10 μM IPAG, then 1nM DHT was added and treatment continued for 3 hours. The cells were washed with room temperature DPBS and fixed in 4% formaldehyde (Pierce) for 15 minutes. After fixation, cells were washed with DPBS, nuclear counter-stained with 0.1 μg/mL DAPI (Pierce), then mounted onto glass slides using Prolong Gold (Molecular Probes).
Images were acquired using the Olympus FV1000 inverted confocal microscope using a 60x 1.42 NA oil immersion objective at a scanning resolution of 0.051 μm/pixel in 0.7 μm z-stack slices. Five randomly selected microscopic fields were taken for each condition with at least 10 z-stack slices per field. Quantification was performed in Fiji-ImageJ by tracing an outline of the nucleus and whole cell and measuring the intensity of GFP-AR staining, generating relative % nuclear and cytoplasmic GFP-AR levels. For each experimental condition, 3 z-stack slices of at least 5–10 cells were analyzed per field in at least 5 fields from 3 independently performed experiments.
Isopycnic density-gradient centrifugation
LNCaP cells were seeded approximately 24 hours prior to treatment with DMSO (drug vehicle) or IPAG (10μM) for 3 hours. Subsequently, the cells were harvested by scraping with a rubber policeman and washed with DPBS. The cells were pelleted by centrifugation at 200 gmax for 7 minutes at 4°C in a GH-3.8 rotor (Beckman-Coulter), washed with DPBS, and pelleted once more. The cells were resuspended in ice-cold isotonic homogenization buffer (HB; 250mM D-sucrose, 150 mM sodium chloride, 25 mM Tris-HCl pH 7.6, 1 mM EDTA, supplemented with Halt Protease and Phosphatase Inhibitors (Pierce)) with the volume normalized against the pellet size. Cells were mechanically lysed using a syringe-driven stainless-steel encased Balch ball-bearing homogenizer (Isobiotec, Heidelberg, Germany). The cells were passed though the tungsten carbide ball bore 14 times with a high tolerance clearance of 12μm. The post-nuclear supernatant was generated by separation of the homogenate using centrifugation at 1000 gmax for 15 minutes at 4°C in a F45-30-11 rotor (Eppendorf). The iodinated density gradient media OptiPrep (Axis-Shield) was used to create a discontinuous density gradient. Stock (60% w/v) Optiprep was diluted in HB to obtain a 0.9 mL bottom layer of 50% (1.272 g/mL) Optiprep, a 0.8 mL middle layer of 30% (1.175 g/mL) Optiprep, and a 2 mL top layer of 10% w/v (1.079 g/mL) Optiprep loaded into OptiSeal polypropylene tubes (Beckman-Coulter). A 1 mL aliquot of PNS was loaded on top of the OptiPrep and separated by ultracentrifugation at 48,000 gmax for 19 hours at 4°C in a TLA 100.4 fixed angle rotor (Beckman-Coulter). Fourteen 300 μL fractions were collected by tube puncture and gravity flow.
Co-immunoprecipitation assay
PC3 cells (AR-negative) were transiently transfected with either pcDNA3.1 (empty vector plasmid), Sigma1-HA, FLAG-AR, FLAG-ARV7, or FLAG-ARv567es plasmid constructs using jetPRIME Transfection Reagent (Polyplus). Forty-eight hours post-transfection, cells were harvested and lysed in HB as described above. Subsequently, lysates were solubilized in an equal volume of 2X NP-40 wash buffer (300 mM sodium chloride, 25 mM Tris pH 7.6, 10% glycerol, 2% NP-40 detergent) supplemented with both Halt Protease and Phosphatase Inhibitors (Pierce). Detergent solubilized lysates were pre-cleared on Protein G Magnetic Beads (BioRad) coupled with mouse serum (Sigma-Aldrich). The pre-cleared lysates were then layered on Protein G Magnetic Beads coupled with mouse monoclonal (H-5) FLAG antibody (Santa Cruz Biotechnology) and incubated for 1 hour at 4°C with constant rotation. The Protein G Magnetic Beads were then washed with 1X NP-40 wash buffer (300 mM sodium chloride, 25 mM Tris pH 7.6, 10% glycerol, 1% NP-40 detergent). The immunoprecipitated protein complexes were eluted by heating at 95°C in Laemmli sample loading buffer supplemented with dithiothreitol (DTT) and β-mercaptoethanol.
Xenograft and in vivo drug efficacy study
Tumors were initiated by injecting 1 × 106 22Rv1 cells in 20% Matrigel into the right flank of castrated male athymic nude mice. Tumors were measured by caliper three times a week, and their volumes were calculated using the formula (a x b2)/2, where a is the longest dimension of the tumor, and b is the width. When tumors reached ~150 mm3, mice were randomly sorted and treatment was started. CT-189 (30 mg/kg) in a solution of 1% sodium carboxymethyl cellulose, 0.5% Tween-80 and DPBS or vehicle alone was administered orally using a gavage needle that had been dipped in 50% sucrose in water. Mice were dosed every other day until vehicle tumors reached ~1,000 mm3, and were euthanized 48 hours after their final dose. Tumors were dissected and flash frozen. Statistical analysis of tumor growth was performed using analysis of variance (ANOVA) (vehicle N = 8, CT-189 N = 8). Mice were weighed on tumor measurement days. All studies involving animals were done in accordance with protocols approved by the Drexel University College of Medicine Institutional Animal Care and Use Committee.
Protein from flash frozen tumors was extracted and solubilized in a solution of RIPA, 10% glycerol, EDTA, and Halt Protease and Phosphatase Inhibitors (Pierce) using a Bead-Ruptor tissue homogenizer (Omni). CT-189 treatment associated changes in proteins of interest in tumors (i.e., pharmacodynamics marker) were detected by immunoblot and quantified by densitometry.
Statistical analysis
For statistical analysis of immunoblot densitometry, ratios of protein band density in drug vehicle versus Sigma1 inhibitor treated samples were compared to an average of the vehicle treated samples. An unpaired, two-tailed t-test was performed to evaluate the statistical significance of differences between the two groups.
Statistical significance of multiple comparisons was determined by one-way analysis of variance (ANOVA) followed by Bonferroni’s post-test using Prism software (GraphPad). This was applied to the 22Rv1 xenografted tumor growth inhibition study.
RESULTS
Sigma1 is required for prostate cancer cell growth and survival
First, we confirmed that RNAi or pharmacological inhibition of Sigma1 suppresses prostate cancer cell growth and survival. In these studies, we evaluated a panel of clinically relevant prostate cancer cell lines, including a hormone-therapy sensitive model of prostate cancer, LNCaP, which exhibits ligand-dependent AR activity in response to dihydrotestosterone (DHT). LNCaP harbors a gain-of-function AR mutation (T877A) that occurs with significant frequency in human disease (15). We also tested the CRPC model cell lines, C4-2 (16, 17), VCaP (18), and 22Rv1 (19–21). The VCaP and 22Rv1 cells express splice variant-derived forms of the AR, including the ARv567es and ARV7 splice variants that are constitutively active and refractory to AR-targeted therapeutics (19–21).
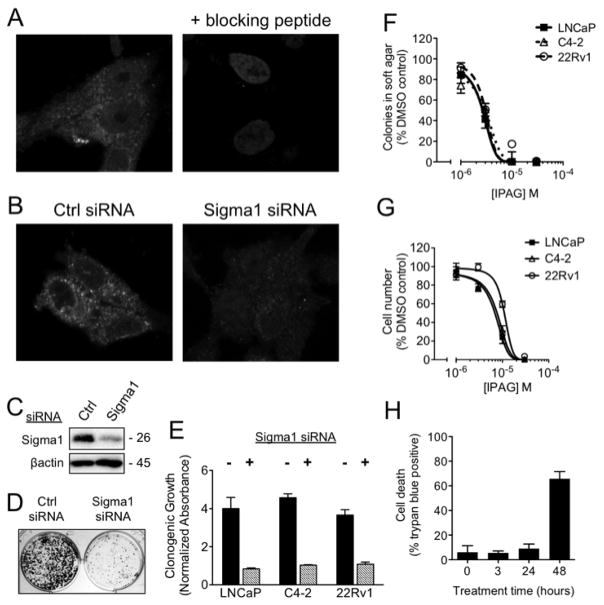
Using an anti-Sigma1 antibody generated in our laboratory (12, 13), we first confirmed Sigma1 protein expression in the cytoplasm of AR positive prostate adenocarcinoma cell lines by confocal immunofluorescence microscopy (data shown for C4-2 in Fig. 1A–B). We confirmed specificity of the Sigma1 immunostain by Sigma1 blocking peptide pre-adsorption (Fig. 1A). We then confirmed the ability of treatment with Sigma1 siRNA to significantly reduce Sigma1 protein expression (Fig. 1B–C).
Figure 1. Suppression of AR positive prostate adenocarcinoma cell growth and survival in response to siRNA knockdown and small molecule inhibition of Sigma1.
(A) Left panel: Immunofluorescence micrograph demonstrating cytoplasmic expression of Sigma1 protein in C4-2 cells. Sigma1 and DAPI staining of the nuclei. Right panel: Preadsorption of Sigma1 antibody with 1 mg/mL of an epitope blocking peptide to determine specificity of antibody staining. (B) Immunofluorescence micrograph showing Sigma1 siRNA knockdown in C4-2 cells; non-specific control (Ctrl siRNA) or Sigma1 siRNA indicated. (C) Immunoblot confirming Sigma1 knockdown in C4-2 cells. (D–E) Clonogenic growth of LNCaP, C4-2, and 22Rv1 cells transfected with Ctrl siRNA or Sigma1 siRNA. Representative image for C4-2 shown in (D) and quantification of clonogenic growth by crystal violet stain shown in (E). Data are presented as ratio of clonogenic growth determined by crystal violet absorbance compared to non-treated control (Clonogenic Growth, Normalized Absorbance). (F) Soft agar assay quantifying dose-associated suppression of 3-dimensional, clonogenic growth of LNCaP, C4-2, and 22Rv1 cell colonies in increasing concentrations of IPAG. (G) Alamar Blue assay quantifying dose-related suppression of 2-dimensional growth and survival of LNCaP, C4-2, and 22Rv1 cells in the presence of increasing concentrations of IPAG. All of the above data are presented as mean ± standard error of the mean (S.E.M.). (H) Time course of LNCaP cell death in response to 10 μM IPAG treatment as measured by trypan blue exclusion.
Next we performed siRNA-mediated knockdown experiments to determine whether Sigma1 is required for prostate cancer cell growth and survival. Clonogenic growth was suppressed in Sigma1 knockdown cell cultures, shown for C4-2 in Figure 1D and quantified for C4-2, LNCaP, and 22Rv1 using a crystal violet staining assay in Figure 1E. Consistent with the RNAi results we found that treatment with a prototypic, selective small molecule inhibitor of Sigma1, IPAG, suppressed prostate cancer cell growth and survival in a dose-responsive manner in both 3-dimensional (3-D) soft agar growth and 2-dimensional (2-D) monolayer growth assays (Fig. 1F–G, respectively). Interestingly, IPAG inhibited prostate cancer cell growth more potently in 3-D culture compared to 2-D monolayer culture (Fig. 1F–G), with an IC50 of 3 μM for all cell lines under 3-D growth conditions compared to IC50 values of 6, 7, and 10 μM for LNCaP, C4-2, and 22Rv1, respectively, under 2-D growth conditions. The difference in response may be due to the growth conditions as well as the time of treatment: the 2-D assay measures inhibition of cell survival after 72 hours of treatment with IPAG, whereas the 3-D assay measures inhibition of cell survival after approximately 3 – 4 weeks of treatment. The Sigma1 selective actions of IPAG have been confirmed elsewhere (13, 22).
Importantly, small molecule inhibition of Sigma1 produced time-dependent cell death. Significant cell death was observed between 24 and 48 hours after initiation of treatment of LNCaP cells with 10 μM IPAG (Fig. 1H). Based on this data, we performed all subsequent mechanism-focused experiments well within the 24 hour time point to minimize potential confounds associated with cell death.
Androgen receptor transcriptional activity is suppressed by RNAi mediated knockdown and small molecule inhibition of Sigma1
Subsequently, we asked whether the growth and survival suppressing effects of Sigma1 inhibitor treatment corresponded with changes to AR transcriptional activity. We used quantitative reverse transcriptase-PCR (qRT-PCR) to evaluate changes in transcript levels of the AR target genes KLK3/PSA and TMPRSS2. For these studies we evaluated the same panel of prostate cancer cell lines described above, with the addition of VCaP cells which harbor an amplification of the AR locus and the TMPRSS2-ERG fusion, which renders ERG expression under AR control (23).
We found significant reduction of KLK3/PSA and TMPRSS2 transcript levels in both LNCaP and VCaP cells following treatment with 20 μM IPAG for 16 hours under steady-state, standard culture conditions – i.e., in fetal bovine serum containing growth medium (Fig. 2A).
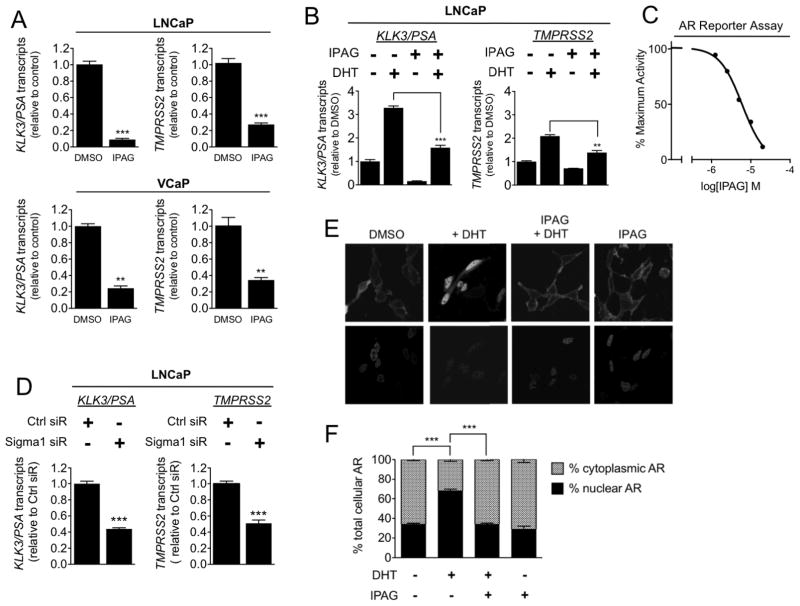
Figure 2. Small molecule Sigma1 inhibitor suppresses AR transcriptional activity and DHT-induced AR nuclear translocation.
(A) Detection of KLK3/PSA and TMPRSS2 mRNA transcripts by qRT-PCR from LNCaP and VCaP cells treated for 16 hours with 20 μM IPAG under steady-state, normal culture conditions. (B) Detection of KLK3/PSA and TMPRSS2 mRNA transcript levels by qRT-PCR from LNCaP cells grown in CSS medium for 72 hours then subjected to co-treatment of 1 nM DHT with DMSO or 10 μM IPAG for 3 hours. (C) AR transcriptional activity luciferase reporter assay. Dose-related inhibition of androgen agonist, 6α-Fl Testosterone (6α FlT) stimulated AR transcriptional activity by IPAG (2.5, 5, 10, 20, 40 μM), 6 μM IC50. (D) Detection of KLK3/PSA and TMPRSS2 mRNA transcript levels by qRT-PCR from LNCaP cells wherein Sigma1 was knocked down by siRNA. Immunoblot confirmation of Sigma1 knockdown in LNCaP shown in Fig. 4A. Data are presented as mean ± S.E.M from at least three independent experiments. (E) Confocal micrographs of LNCaP(GFP-AR) cells grown in charcoal-stripped medium for 3 days followed by 3 hour treatment with 1 nM DHT alone or in the presence of 10 μM IPAG. (F) Quantification of relative nuclear and cytoplasmic GFP-AR levels. For each condition, at least 5 cells were counted per field in at least 5 fields from 3 independently performed experiments, and data are presented as mean ± S.E.M. **P < 0.01; ***P < 0.001.
To evaluate the effects of Sigma1 inhibition specifically on androgen induced AR transcriptional activity, we maintained LNCaP cells in culture medium supplemented with charcoal stripped serum (CSS) to mimic androgen deprivation and treated these cells with DHT in the absence or presence of IPAG (Fig. 2B). Following three days of growth in CSS medium, treatment with 1nM DHT for 3 hours induced KLK3/PSA and TMPRSS2 transcript levels (Fig. 2B). Treatment with IPAG alone for 3 hours reduced KLK3/PSA and TMPRSS2 transcript levels below control levels (Fig. 2B). Co-treatment with IPAG blocked DHT induction of KLK3/PSA and TMPRSS2 transcripts, maintaining transcripts close to control levels (Fig. 2B).
To measure effects on direct AR transcriptional activity, we used a luciferase-based AR reporter assay. In this assay, 16-hour treatment with IPAG dose-responsively inhibited an AR agonist, 6αFIT, with an IC50 of 6 μM (Fig. 2C).
Consistent with small molecule inhibition of Sigma1, siRNA knockdown of Sigma1 in LNCaP cells significantly suppressed KLK3/PSA and TMPRSS2 transcript levels (Fig. 2D). Two different commercial Sigma1 siRNA produced decreases in KLK3/PSA and TMPRSS2 transcript levels (data shown for SCBT siRNA in Fig. 2D).
Altogether, these data demonstrate that both RNAi mediated knockdown and pharmacological inhibition of Sigma1 suppress AR transcriptional activity.
Treatment with a Sigma1 inhibitor prevents nuclear transport of cytoplasmic AR
As Sigma1 is present throughout the cytoplasm of these prostate cancer cells (Fig. 1), and as nascent AR is stabilized by its cognate chaperones in the cytoplasm, we asked whether the decreased transcriptional activity in response to pharmacological inhibition of Sigma1 could be explained by inhibition of DHT induced translocation of AR to the nucleus. To evaluate this effect, we generated a LNCaP cell line stably transfected with a green fluorescent protein (GFP)-tagged androgen receptor (GFP-AR) construct. Using this LNCaP(GFP-AR) cell line, we monitored and evaluated DHT-induced nuclear localization of GFP-AR in the absence and presence of IPAG (Fig. 2E–F). LNCaP(GFP-AR) cells were cultured in CSS supplemented medium for 3–5 days then treated for 3 hours with 1 nM DHT alone or in the presence of 10 μM IPAG (Fig. 2E–F). Treatment with DHT alone for 3 hours resulted in an increase from 34% ± 1% to 68% ± 2% nuclear GFP-AR (Fig. 2E–F). Co-treatment with 10 μM IPAG completely abrogated this effect, maintaining the level of nuclear GFP-AR at 33% ± 1% (Fig. 2E–F). Thus, the small molecule Sigma1 inhibitor prevented DHT-induced nuclear translocation of AR.
Sigma1 inhibitor induces ubiquitin proteasome mediated degradation of AR
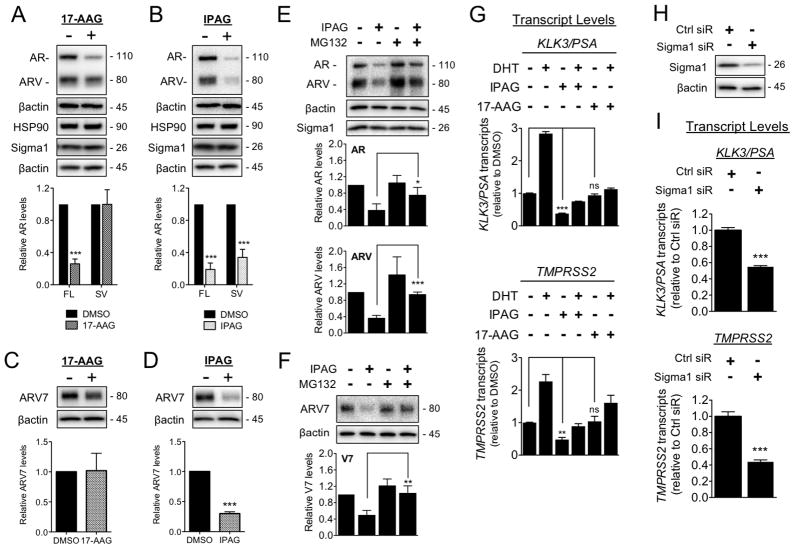
Subsequently we investigated the fate of AR following extended treatment (16 hours) with IPAG or knockdown with Sigma1 siRNA. Knockdown of Sigma1 in LNCaP cells suppressed AR protein levels, under standard culture conditions (Fig. 3A). Consistent with Sigma1 siRNA, treatment with IPAG decreases steady-state AR protein levels in all 5 cell lines examined (Fig. 3B). In LNCaP cells, treatment with IPAG resulted in a >75% decrease in AR protein levels (Fig. 3B). IPAG treatment suppressed AR to similar levels in LAPC4 (wild-type AR expressing) cells (24) and castration resistant C4-2 and 22Rv1 cells and clearly suppressed AR in VCaP cells as well (Fig. 3B). Interestingly, treatment with IPAG also eliminated splice-variant AR (ARV), including ARV7, as well as full-length AR in ARV-bearing 22Rv1 and VCaP cells under CSS medium culture conditions (Fig. 3B). Densitometry of immunoblots in Figure 3B are presented in Supplemental Figure 2.
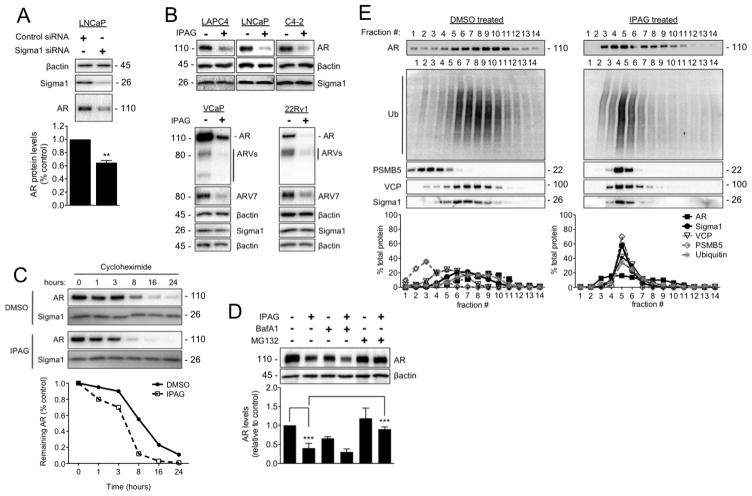
Figure 3. Ubiquitin proteasome-mediated degradation of AR in response to treatment with small molecule Sigma1 inhibitor.
(A) Immunoblot detection of Sigma1 and AR protein levels in LNCaP cells wherein Sigma1 was knocked down by siRNA. Quantification of AR protein in these cells is shown. (B) Immunoblots of AR, AR splice variants (ARV), ARV7, and Sigma1 protein levels following 16 hour treatment of the 5 indicated prostate cancer cell lines with DMSO (−) or 10 μM IPAG (+). (C) AR protein levels in LNCaP cells following treatment with cycloheximide (CHX) for indicated times. (D) Representative immunoblot and quantification of 3 independent immunoblots of AR protein levels in LNCaP cells co-treated for 16 hours with 10 μM IPAG and 1 μM MG132 or 100 nM Bafilomycin A1 (BafA1). (E) Immunoblot of 14 fractions collected following isopycnic fractionation of post-nuclear cell components from LNCaP cells treated for 3 hours with DMSO or 10 μM IPAG. Immunoblots (IB) of AR, p97/VCP (VCP), polyubiquitinated proteins (Ub), 26S proteasome subunit (PSMB5), and Sigma1. Histograms show quantification of immunoblots where the percentage of each protein per fraction is depicted relative to its total across all 14 fractions. All data are presented as mean ± S.E.M. **P < 0.01; ***P < 0.001.
We previously found that extended treatment with IPAG could suppress unfolded protein response (UPR) associated protein translation (12, 13). Therefore, we evaluated the potential and relative contribution of protein synthesis and degradation by treating with IPAG in the presence of the translation inhibitor, cycloheximide (CHX). Whereas translation arrest by CHX alone resulted in a time-dependent decrease in AR protein levels from 95% to 11% over 24 hrs, the combination of CHX and IPAG resulted in decreases that ranged from 80% to 1% of control levels (Fig. 3C). These data suggest that although translation arrest may contribute to decreased AR levels in response to IPAG treatment, protein degradation plays a significant role in the elimination of AR.
We previously found that extended treatment with IPAG could also induce autophagy (13). Therefore, we tested whether the IPAG-mediated decrease in steady-state AR protein levels was due to autophagosomal degradation by combining IPAG with bafilomycin A1 (inhibitor of autolysosomal degradation) or treating cells in which ATG5 had been knocked down with siRNA. ATG5 is a protein product of the essential autophagy gene ATG5, which is required for autophagosome formation. Inhibition of autophagosomal degradation or autophagosome formation did not block IPAG induced degradation of AR (Fig. 3D and Supplemental Figure 3, respectively).
In parallel, we examined whether the ubiquitin proteasome system (UPS) is the operative pathway in Sigma1 inhibitor-induced AR degradation. Co-treatment of LNCaP cells with the proteasome inhibitor MG132 blocked IPAG-mediated degradation of AR (Fig. 3D), providing compelling evidence in support of the hypothesis that Sigma1 inhibitor-mediated degradation of AR is UPS-mediated.
By 3 hours of treatment with IPAG steady state AR protein levels did not yet change significantly; however, DHT-induced nuclear localization was blocked (Fig. 2E–F). Subsequently, we asked whether during this time frame treatment with IPAG resulted in sequestration and targeting of cytoplasmic AR to the ubiquitin proteasome system (UPS). We used an isopycnic centrifugation technique to isolate subcellular fractions of LNCaP cells. In this assay, IPAG induces co-fractionation of AR and Sigma1 along with UPS components including PSMB5, polyubiquitinated proteins, and p97/VCP (Fig. 3E). In response to treatment with IPAG, 90% of Sigma1, 98% of PSMB5, 84% of p97/VCP, 66% of poly-ubiquitin immunoreactivity, and 46% of AR co-fractionated in fractions 4–6, from initially disparately distributed fractions (Fig. 3E). These results further support the idea that the Sigma1 inhibitor induces accumulation of AR to ubiquitin-enriched cell compartments to be targeted for proteasomal degradation. It is noteworthy that a portion of the total cellular AR within these cells shifts to Sigma1-enriched fractions, suggesting that a subpopulation of AR interacts with Sigma1 at a given time and emphasizing that IPAG-induced AR degradation is time dependent.
Selectivity of Sigma1 inhibitor induced protein elimination
Treatment with the Sigma1 inhibitor appears to be selective. Indeed, in response to IPAG treatment the levels of several proteins decrease; however, the levels of some proteins remain steady and a few increase (Fig. 4A). Because of its features, Sigma1 regulation of protein homeostasis draws comparison with HSP90. Here, we demonstrate that pharmacological modulation of Sigma1 and HSP90 involve distinct mechanisms.
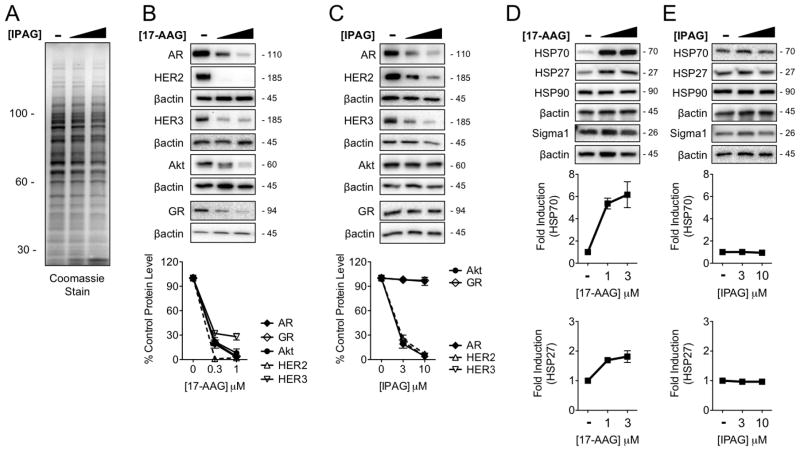
Figure 4. Selectivity of Sigma1 inhibitor induced protein degradation in comparison with HSP90 inhibitor.
(A) Coomassie stain of SDS-PAGE resolved whole cell lysates from LNCaP cells treated for 16 hours with DMSO (−) or increasing concentrations of IPAG (3, 10 μM). (B–C) Immunoblot of HSP90 client proteins from LNCaP cells treated for 16 hours with DMSO (−) or increasing concentrations of 17-AAG (0.3, 1 μM) (B) or IPAG (3, 10 μM) (C). The GR immunoblots are of whole cell lysates from VCaP cells. (D–E) Immunoblot showing induction of HSP70 and HSP27 in response to treatment with 17-AAG (1, 3 μM) (D), and absence of HSP70 and HSP27 induction in response to treatment with IPAG (3, 10 μM) (E). All data represent mean ± S.E.M. and are generated from at least 3 independent determinations.
Sigma1 modulators and HSP90 inhibitors induce the degradation of some overlapping but also distinct sets of client proteins. HSP90 chaperones a range of client proteins important in cancer cell growth and survival. We tested the effects of IPAG treatment on steady-state levels of several well-defined HSP90 client proteins that play important roles in this disease: ErbB2/HER2, ErbB3/HER3, and Akt. We found that the Sigma1 inhibitor affects some but not all of these client proteins. Treatment with IPAG elicited a similar dose-related decrease in steady-state ErbB2/HER2 and ErbB3/Her3 as the geldanamycin analog 17-AAG (Fig. 4B, C). However, in contrast to elimination of Akt produced by 17-AAG, we observed no significant change in Akt protein levels in response to IPAG treatment (Fig. 4B, C).
HSP90 is the best-characterized chaperone of glucocorticoid receptor (GR) as well as AR. In VCaP cells, treatment with 17-AAG resulted in dose-related elimination of GR (Fig. 4B). Interestingly, we observed no measurable change in GR protein levels in response to IPAG treatment (Fig. 4C). Considering the conservation and overlap in AR and GR mechanisms and support machinery, this result is somewhat unexpected. However, it further highlights the specificity and some key differences in mechanisms governing the actions of HSP90 and Sigma1.
Compensatory up-regulation of HSP70 and HSP27 are hallmarks of HSP90 inhibition (6, 25). We confirmed that treatment with 17-AAG results in salient induction of both HSP70 and HSP27 (Fig. 4D). In clear contrast, treatment with IPAG did not induce HSP70 and HSP27 (Fig. 4E).
Together these data demonstrate a degree of selectivity of Sigma1 inhibitor-mediated actions. They demonstrate that Sigma1 and HSP90 have some overlapping but also distinct sets of client proteins, and they suggest that pharmacological modulation of Sigma1 and HSP90 are governed by distinct mechanisms of action.
Sigma1 inhibitor eliminates AR and AR splice variants
Nascent cytoplasmic AR is bound and stabilized by multi-chaperone protein complexes that include HSP90. DHT binding to AR stimulates transport of this complex to the nucleus, and HSP90 is thought to play a role in the transport as well as stabilization of AR (6, 26). However, HSP90 does not bind AR splice-variants (ARVs) (14, 20, 21, 27, 28), and HSP90 inhibitors such as 17-AAG are unable to suppress ARV driven transcriptional activity (14, 29).
Consistent with previously published reports, treatment with 17-AAG resulted in decreased full-length AR levels compared to DMSO, whereas ARV levels did not significantly change compared to vehicle treated control levels (Fig. 5A). In contrast, treatment with the Sigma1 inhibitor, IPAG, resulted in a significant decrease of both full-length AR and ARV levels (Fig. 5B).
Figure 5. Sigma1 inhibitor mediated degradation of AR and ARV and suppression of AR and ARV driven transcription in comparison with HSP90 inhibitor.
(A–F) Treatment with IPAG induces proteasomal degradation of full-length and splice variant AR. (A–B) Immunoblot and densitometry of full-length AR (FL) and ARV (SV) from whole cell lysates of 22Rv1 cells treated for 16 hours with 17-AAG (3 μM) (A) or IPAG (10 μM) (B). Quantifications of immunoblots by densitometry are shown below each immunoblot panel. (C–D) Immunoblots and densitometric quantification of ARV7 in 22Rv1 cells treated for 16 hours with 17-AAG (3 μM) (C) or IPAG (10 μM) (D). (E–F) Representative immunoblots of AR and ARV protein levels (E) and ARV7 (F) in whole cell lysates from 22Rv1 cells co-treated for 8 hours with 10 μM IPAG and 1 μM MG132. Bands were quantified by densitometry and data are presented as levels of AR, ARV, or ARV7 relative to DMSO treated control. (G) Detection of KLK3/PSA and TMPRSS2 mRNA transcript levels by qRT-PCR from 22Rv1 cells grown in CSS for 5 days, then subjected to 16 hour treatment with 10 μM IPAG or 3 μM 17-AAG combined with 1 nM DHT. (H) Immunoblot confirming siRNA knockdown of Sigma1 in 22Rv1 cells from which RNA was isolated and evaluated in (I). (I) Detection of KLK3/PSA and TMPRSS2 mRNA transcript levels by qRT-PCR from CSS medium cultured 22Rv1 cells in which Sigma1 was knocked down. Bars represent mean ± S.E.M. and are generated from at least three independent determinations. *P < 0.05; **P < 0.01; ***P < 0.001.
Among the multiple ARVs that have been identified in CRPC tumors and cell lines (2, 20, 21, 27, 28) the best characterized are the ARV7 and ARv567es (2, 28). As a result of epitopes produced during splicing, these ARV contain unique amino acids that have enabled generation of antibodies specific to these variants (2, 20, 21, 27, 28). The 22Rv1 cell line expresses ARV7 (14). Using a validated antibody specifically raised against ARV7 (30, 31), we demonstrate that IPAG suppresses ARV7 compared to vehicle treated control levels in 22Rv1 cells whereas 17-AAG does not (Fig. 5C–D).
We asked whether proteasomal degradation contributes to the elimination of ARVs in response to treatment with IPAG. Co-treatment of 22Rv1 cells with IPAG and MG132 blocked the degradation of both full-length AR and ARVs (Fig. 5E), which we confirmed specifically for ARV7 as well (Fig. 5F).
Sigma1 inhibitor suppresses AR splice variant transcriptional activity
We confirmed that IPAG-mediated elimination of AR and ARV corresponds with suppression of AR and ARV transcriptional activity. When cultured in CSS medium, ARV transcriptional activity continues as evidenced by detectable KLK3/PSA and TMPRSS2 transcripts under this hormone-depleted culture condition. In this experiment, we set our baseline relative KLK3/PSA and TMPRSS2 transcript value at 1.0 (Fig. 5G). Treatment with DHT induces full-length AR transcriptional activity, increasing KLK3/PSA and TMPRSS2 transcript levels above DHT naïve cells (Fig. 5G). When treated with IPAG, KLK3/PSA and TMPRSS2 transcript levels decreased (Fig. 5G). This is consistent with the decreased ARV protein levels in response to treatment with IPAG (Fig. 3, 5). Co-treatment of DHT and with IPAG maintained KLK3/PSA and TMPRSS2 transcript levels below baseline (Fig. 5G).
In contrast to IPAG, treatment of 22Rv1 cells with 17-AAG did not decrease AR target gene transcript levels below the baseline value, with KLK3/PSA and TMPRSS2 transcript levels (Fig. 5G). Therefore, constitutive ARV transcriptional activity in 22Rv1 cells is not altered by 17-AAG. These data support previous reports demonstrating that the HSP90 inhibitor, geldanamycin, does not suppress ARV transcriptional activity (14, 29). Co-treatment with DHT and 17-AAG resulted in increased KLK3/PSA and TMPRSS2 transcript levels (Fig. 5G).
These data demonstrate that pharmacological inhibition of Sigma1 suppresses ARV as well as AR transcriptional activity. However, consistent with recent reports, HSP90 inhibitors suppress AR but not ARV transcriptional activity (Fig. 5 and (14, 29)).
Finally, we asked whether ARV transcriptional activity would be altered by Sigma1 RNAi. Consistent with small molecule inhibition of Sigma1, siRNA knockdown of Sigma1 in 22Rv1 cells grown in CSS medium (Fig. 5H) suppressed KLK3/PSA and TMPRSS2 transcript levels (Fig. 5I).
Sigma1 physically associates with AR and AR splice variants
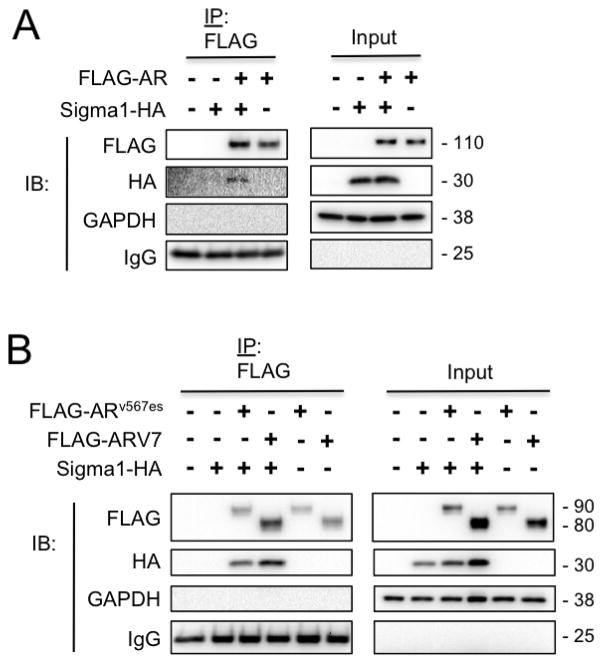
The inability of HSP90 inhibitors to degrade ARV may be attributed to the lack of physical association between the two proteins (14, 29). This is consistent with HSP90 binding to AR CTD at amino acid residues which are absent in the ARVs (14, 20, 21, 27, 28). However, as the Sigma1 inhibitor decreased the levels of both AR and ARV, we asked whether Sigma1 physically associates with ARV7 and ARv567es. To address this question, we performed co-immunoprecipitation assays using FLAG epitope tagged AR, ARV7, and ARv567es constructs (described elsewhere (14, 29)) along with an affinity tagged Sigma1 construct. The Sigma1-HA plasmid construct contains a dual carboxy-terminal affinity-tagged murine Sigma1 with tandem hemagglutinin (HA) epitope (32). The high degree of amino acid homology between murine and human Sigma1 make them functionally interchangeable (32). We transiently transfected both constructs into PC3 cells (AR-negative prostate adenocarcinoma cell line) and immunoprecipitated post-nuclear cell lysates with an anti-FLAG antibody. Whereas HSP90 binds full-length AR but not ARV7 or ARv567es (14), we demonstrate here that Sigma1 physically associates with both AR variants (Fig. 6B) as well as full-length AR (Fig. 6A). Together, our data suggest that Sigma1 may play a role in the support machinery of the AR-axis and contribute to the regulation of ARV as well as full-length AR. Our data also suggest that Sigma1 may physically associate with the N-terminal domain (NTD) of AR; however, whether this association is direct or indirect remains to be determined.
Figure 6. Sigma1 physically associates with full-length and splice variant AR.
(A) Co-immunoprecipitation of full-length AR and Sigma1. PC3 cells (AR-negative prostate adenocarcinoma) were transiently transfected with an empty vector plasmid (−), affinity tagged Sigma1 (Sigma1-HA), FLAG-AR, or both of the latter. Detergent soluble post-nuclear cell lysates were subjected to immunoprecipitation using an anti-FLAG antibody (IP: FLAG) and immunoblotted (IB) with antibodies against the indicated proteins. Input indicates post-nuclear cell lysates prior to IP. (B) Co-immunoprecipitation of ARVs and Sigma1. PC3 cells were transiently transfected with Sigma1-HA, FLAG-ARV7, and FLAG-ARv567es plasmid constructs and subsequently subjected to the same procedure as described in (A).
In vivo efficacy of CT-189, a novel Sigma1 modulator with drug-Like properties
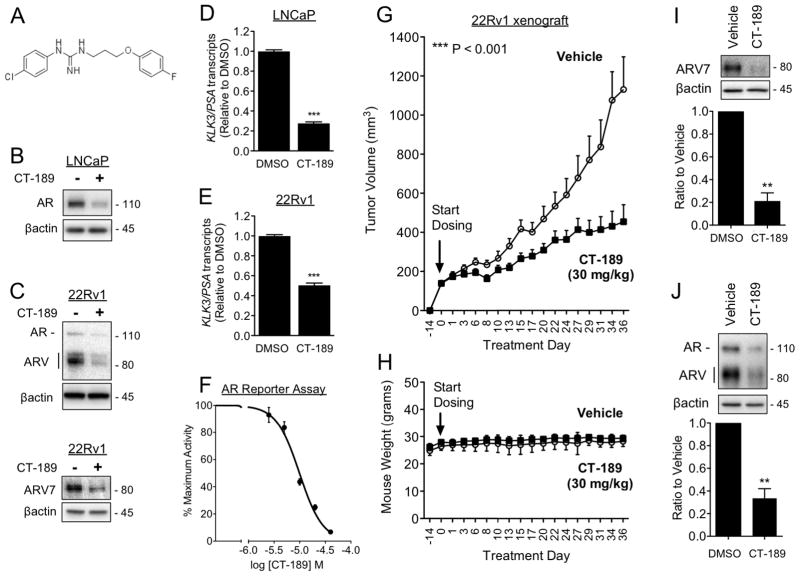
Although it is a useful chemical probe, IPAG has insufficient drug-like properties, and we were unable to use this compound in our in vivo efficacy studies. Therefore, we focused on modifications of IPAG as starting points to develop more drug-like probes to study the function of Sigma1 in vivo. We synthesized 1-(4-chlorophenyl)-3-(3-(4-fluorophenoxy)propyl)guanidine, which we named CT-189 (Fig. 7A and Supplemental Table). CT-189 binds Sigma1 with a Ki = 38 nM using a [3H](+)-pentazocine binding competition assay described elsewhere(33), maintains AR and ARV eliminating properties (Fig. 7B–C), and AR and ARV transcriptional activity suppressing activities (Fig. 7D–F).
Figure 7. In vivo efficacy: Small molecule Sigma1 inhibitor suppresses growth of ARV driven CRPC tumor xenografted mice.
(A) Chemical structure of 1-(4-chlorophenyl)-3-(3-(4-fluorophenoxy)propyl)guanidine (CT-189). (B) Immunoblots of AR in LNCaP cells treated for 16 hours with CT-189 (20 μM). (C) Immunoblots of AR, ARV, and ARV7 from 22Rv1 cells, grown in CSS medium, treated for 16 hours with CT-189 (20 μM). (D–E) Detection of KLK3/PSA mRNA transcript levels by qRT-PCR from LNCaP cells (D) and 22Rv1 cells grown in CSS medium for 5 days (E), then subjected to 16 hour treatment with 20 μM CT-189. (F) Luciferase reporter assay showing dose-responsive inhibition of AR transcriptional activity by CT-189 (1.25, 2.5, 5, 10, 20, 40 μM). (G) In vivo tumor growth inhibition by CT-189 in castrated mice xenografted with 22Rv1 cells. Treatment with CT-189 was started 14 days post-implantation of 22Rv1 cells, when tumors reached ~150 mm3. Either drug vehicle or CT-189 (30 mg/kg) was administered by oral gavage every other day (q.a.d., quaque altera die). N ≥ 8 for each treatment arm. One-way analysis of variance (ANOVA) was used to determine statistical significance, ***P < 0.001. (H) Mouse weight (grams) during treatment course. No significant weight loss compared to vehicle was observed in CT-189 (30 mg/kg) treated mice. (I–J) Tumors were measured and harvested within 48 hours after final drug administration, at treatment day 36. Tumor proteins were extracted and immunoblotted to evaluate changes in ARV7 (I) and AR (J) protein levels. Note that in panel J only AR was quantified. Bars represent mean ± S.E.M. and are generated from at least three tumors for each treatment condition analyzed in triplicate. **P < 0.01; ***P < 0.001.
CT-189 has drug-like properties. It is well tolerated by mice at efficacious doses. Importantly, mice treated with CT-189 did not lose weight (Fig. 7H), and we observed no behavioral abnormalities compared to vehicle treated mice.
In vivo, CT-189 inhibited the growth of xenografted 22Rv1 tumors by 60% following ~36 days of treatment (Fig. 7G). A dose of 30 mg/kg CT-189 was administered by oral gavage every other day during this period. We performed biochemical analysis of the harvested tumors by immunoblot to confirm that AR and ARV7 elimination was associated with tumor growth inhibition. Compared to treatment with drug vehicle, tumors from CT-189 treated mice had decreased levels of ARV7 (79% ± 7% decrease) as well as AR (67% ± 8% decrease) protein levels (Fig. 7I and J, respectively).
DISCUSSION
Sigma1 and the AR axis
The dual goals of this study were to better understand the role of Sigma1 with regard to AR processing and function and to determine whether modulation of its activity may have therapeutic potential for the treatment of castration resistant prostate cancer. The concept of the role of Sigma1 is rapidly evolving, and emerging evidence suggests that either Sigma1 may function as an allosteric co-factor protein associated with bona fide receptor systems or as a novel chaperone (34) or scaffolding protein. We demonstrate that treatment with a prototypic Sigma1 modulator, IPAG, results in cytoplasmic sequestration of AR and proteasome mediated degradation. Consistent with these data, RNAi knockdown of Sigma1 results in decreased AR levels and transcriptional activity. Furthermore, we demonstrate that a drug-like analog of IPAG, which we named CT-189, maintains Sigma1 mediated AR/ARV eliminating activities, suppresses AR transcriptional activity, and importantly, significantly inhibits ARV driven tumor growth in a 22Rv1 xenograft model. To our knowledge this work represents the first demonstration of a direct interaction between Sigma1 and the AR axis, and it is the first evidence of in vivo efficacy of Sigma1 modulator compounds through this mechanism. An indirect connection between the AR axis and Sigma1 can be inferred from parallel lines of research showing that the endogenous steroid hormone dehydroepiandrosterone (DHEA) has affinity for both AR and Sigma1 (34–37). DHEA has been shown to upregulate AR levels and activity in the central nervous system (38), and it has been proposed to act as a Sigma1 agonist or activator (34). In the context of prostate cancer, circulating adrenal androgens, DHEA and DHEA sulfate (DHEA-S), are taken up by prostate tumors and converted into testosterone and other steroids that bind and activate AR (39). Elevated intratumoral androgens are characteristic of castration resistant tumors (39, 40). DHEA and DHEA-S contribute to the persistent synthesis of intratumoral androgens following androgen deprivation therapy which drives CRPC (39, 40). It is tempting to speculate that Sigma1 interaction with DHEA may also play a role in prostate tumor growth. Our data, along with these indirect lines of evidence, suggest a role for Sigma1 in feedback mechanisms that regulate AR-associated networks.
Sigma1 as a novel ligand-operated scaffolding protein that engages in selective protein associations
Sigma1 is expressed throughout the cytoplasm of prostate cancer cells and has been described as a primarily endoplasmic reticulum (ER) protein. Based on the recently resolved crystal structure (8), Sigma1 is an ER bound integral membrane protein with a relatively short ER luminal component and a relatively long cytoplasmic tail. The ER extends throughout the cytoplasmic space of the cell and associates with essentially all organelles (41). Therefore, it is not surprising that Sigma1 can physically associate with cytoplasmic and cytosolic proteins as well as ER luminal proteins. Here we have found that Sigma1 physically and functionally interacts with AR.
HSP90 is the most extensively characterized AR chaperone. Along with co-chaperones, HSP90 binds and stabilizes nascent AR in the cytoplasm, and upon DHT binding to AR, AR is thought to dissociate from this chaperone complex and subsequently undergo transport to the nucleus (reviewed in reference 6). In our pull-down experiments we show that AR and Sigma1 physically associate (Fig. 6). Together, our experiments demonstrate that although Sigma1 and HSP90 may share some common associated proteins and may engage in multi-protein complexes, they do not necessarily interact directly and they have distinct mechanisms of action. Three lines of evidence exemplify that pharmacological modulation of Sigma1 and HSP90 are driven by distinct mechanisms of action.
First, Sigma1 modulators such as IPAG and CT-189 eliminate both AR and ARV in 22Rv1 cells whereas the HSP90 inhibitor 17-AAG eliminates only the full-length AR (Fig. 5, 7, and (14)). Among the multiple ARVs that have been identified in CRPC tumors and cell lines (2, 20, 21, 27, 28) the best characterized are the ARV7 and ARv567es splice variants (2, 28). Recent reports demonstrate that HSP90 inhibitors suppress AR but not ARV transcriptional activity(14, 29). Whereas HSP90 binds full-length AR but not ARV7 or ARv567es (14), Sigma1 physically associates with both AR variants as well as full-length AR (Fig. 6). Furthermore, full-length AR and ARV are eliminated by IPAG but not 17-AAG (14, 29). This is consistent with HSP90 binding to AR CTD at amino acid residues that are absent in the AR variants (14, 20, 21, 27, 28). Our results with 22Rv1 cells suggest that Sigma1 binds to AR, either directly or indirectly, however, by a mechanism distinct from HSP90.
Second, our discovery that IPAG destabilizes and eliminates some, but not all, HSP90 client proteins provides further evidence that Sigma1 and HSP90 have distinct physiological roles and mechanisms of action. Notably, glucocorticoid receptor (GR) protein levels do not change in response to treatment with IPAG under conditions wherein AR is nearly eliminated (Fig. 4B, C). This was a surprising finding considering the degree of overlap between AR and GR mechanisms. It highlights the selectivity of Sigma1 modulator actions and further distinguishes Sigma1 and HSP90 mechanisms of action. Treatment with IPAG also eliminates ErbB2/HER2 and ErbB3/HER3, whereas the level of other HSP90 clients such as Akt is not altered (Fig. 4B, C).
The third finding providing evidence that Sigma1 and HSP90 function through distinct mechanisms is the lack of HSP70 and HSP27 induction in response to treatment with Sigma1 modulators (Fig. 4E). Inhibition of HSP90 with small molecule inhibitors results in heat shock factor 1 (HSF1)-dependent transcriptional up-regulation of HSP70 and HSP27 (6, 25). This cytoprotective, compensatory induction of HSP70 and HSP27 is a pharmacodynamic marker of HSP90 inhibition. This response is absent in Sigma1 modulation and thus further distinguishes HSP90 and Sigma1 mediated effects (Fig. 4D, E).
Our data suggest that Sigma1 engages in a number of multi-protein complexes, some of which include AR. Whether Sigma1 directly or indirectly binds AR remains to be determined. Furthermore, it remains to be determined whether Sigma1 modulators directly alter protein-protein associations or the intracellular transport and localization of Sigma1-associated complexes. Evidence of physical association with AR along with convergence of Sigma1, UPS components, and a subpopulation of AR to the same subcellular fractions suggest that both contribute to the pharmacological mechanism of Sigma1 modulator actions.
Implications for discovery of novel therapeutic agents and strategies to treat advanced prostate cancer
Resistance to first- and second-generation AR–targeting agents is invariably associated with reactivation of the AR axis, either via induction of intratumoral steroidogenesis or increased expression of AR and truncated AR splice variants (2). This is further complicated by compensatory up-regulation or feedback regulation of associated pathways, including ErbB receptor and PI3K (phosphatidyl inositol-3-kinase) activation in PTEN (phosphatase and tensin homolog) deficient prostate cancers (42). These examples demonstrate the importance of discovering and developing novel approaches to co-targeting the AR axis and the networks on which it depends (2).
It is noteworthy that Sigma1 inhibition suppresses growth of PTEN mutant LNCaP and C4-2 and PTEN null PC3 cells (Fig. 1 and (12)). These data suggest that Sigma1 inhibitors engage mechanisms downstream of PTEN or mechanisms that cooperate with but are distinct from canonical PI3K/Akt and ERK growth and survival signaling pathways. PTEN deficiency, by mutation or loss of PTEN, has a significant impact on prostate cancer progression. Indeed, over 50% of advanced prostate cancers are PTEN deficient (42–44). Suppression of growth signaling in PTEN deficient prostate cancers (12, 13) as well as suppression of pathways that compensate for AR-targeted inhibition demonstrate that Sigma1 modulators may be used more broadly than just to target the AR axis.
Although Sigma1 modulators alter multiple processes and systems in cancer cells, our data provide evidence that Sigma1 is a specific but multi-functional target. Our comparison of IPAG and 17-AAG provides evidence in support of this notion. Additionally, in LNCaP and C4-2 cells, the steady-state protein levels of other nuclear hormone receptors including retinoic acid receptors and retinoid X receptors were not affected by treatment with IPAG (Supplemental Figure 4). We are just beginning to learn how selective Sigma1 modulator compounds can be used to regulate specific but multiple cellular processes. Here we demonstrate that Sigma1-mediated protein homeostasis can be pharmacologically regulated to inhibit AR transport and stability. The three CRPC lines (C4-2, VCaP, and 22Rv1) evaluated here were all responsive to small molecule Sigma1 inhibition. The Sigma1 inhibitor also reduced ErbB2/HER2 and ErbB3/HER3 protein levels (Fig. 4C), thus abrogating compensatory up-regulation of ErbB2/HER2 and ErbB3/HER3 in response to AR axis targeted treatments (2, 42, 45). The ability to pharmacologically modulate multi-functional targets such as Sigma1 is advantageous in cancer as it imposes a barrier to compensatory response mechanisms to targeted therapies without the broad and often toxic effects of chemotherapy.
Here, we demonstrate in vivo efficacy with a drug-like Sigma1 compound, CT-189, in an ARV driven CRPC tumor xenograft model (Fig. 7 and Supplemental Table). Importantly, this Sigma1 inhibitor produced no detectable side-effects at efficacious doses - no weight loss and no behavioral abnormalities were observed under these study conditions. Altogether, our data provide evidence in support of the potential value of this therapeutic approach and Sigma1 as a drug target in the treatment of AR driven cancers.
Supplementary Material
Acknowledgments
We thank Drs. Paul McGonigle and Paul Campbell for critical reading of the manuscript. FJK was supported by an American Cancer Society Institutional Research Grant, Drexel University Clinical and Translational Research Institute Grant, Sidney Kimmel Cancer Center Pilot Study Award, and a Coulter-Drexel Translational Research Partnership Program Award. KK was supported by NIH grants CA099996, CA159945, CA176401, P30CA056036-14.
Footnotes
Disclosure of Potential Conflicts of Interest: F.J.K. has ownership interest (including patents) in Context Therapeutics.
References
- 1.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009 Aug 1;15(15):4792–8. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mostaghel EA, Plymate SR, Montgomery B. Molecular pathways: targeting resistance in the androgen receptor for therapeutic benefit. Clin Cancer Res. 2014 Feb 15;20(4):791–8. doi: 10.1158/1078-0432.CCR-12-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knudsen KE, Penning TM. Partners in crime: deregulation of AR activity and androgen synthesis in prostate cancer. Trends Endocrinol Metab. 2010 May;21(5):315–24. doi: 10.1016/j.tem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferraldeschi R, Welti J, Luo J, Attard G, de Bono JS. Targeting the androgen receptor pathway in castration-resistant prostate cancer: progresses and prospects. Oncogene. 2015 Apr 2;34(14):1745–57. doi: 10.1038/onc.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solit DB, Zheng FF, Drobnjak M, Munster PN, Higgins B, Verbel D, et al. 17-Allylamino-17-demethoxygeldanamycin induces the degradation of androgen receptor and HER-2/neu and inhibits the growth of prostate cancer xenografts. Clin Cancer Res. 2002 May;8(5):986–93. [PubMed] [Google Scholar]
- 6.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nature reviews Cancer. 2010 Aug;10(8):537–49. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanner M, Moebius FF, Flandorfer A, Knaus HG, Striessnig J, Kempner E, et al. Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proc Natl Acad Sci U S A. 1996 Jul 23;93(15):8072–7. doi: 10.1073/pnas.93.15.8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt HR, Zheng S, Gurpinar E, Koehl A, Manglik A, Kruse AC. Crystal structure of the human sigma receptor. Nature. 2016 Apr 4; doi: 10.1038/nature17391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007 Nov 2;131(3):596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 10.Aydar E, Onganer P, Perrett R, Djamgoz MB, Palmer CP. The expression and functional characterization of sigma (sigma) 1 receptors in breast cancer cell lines. Cancer Lett. 2006 Oct 28;242(2):245–57. doi: 10.1016/j.canlet.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Crottes D, Martial S, Rapetti-Mauss R, Pisani DF, Loriol C, Pellissier B, et al. Sig1R protein regulates hERG channel expression through a post-translational mechanism in leukemic cells. J Biol Chem. 2011 Aug 12;286(32):27947–58. doi: 10.1074/jbc.M111.226738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim FJ, Schrock JM, Spino CM, Marino JC, Pasternak GW. Inhibition of tumor cell growth by Sigma1 ligand mediated translational repression. Biochem Biophys Res Commun. 2012 Sep 21;426(2):177–82. doi: 10.1016/j.bbrc.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrock JM, Spino CM, Longen CG, Stabler SM, Marino JC, Pasternak GW, et al. Sequential cytoprotective responses to Sigma1 ligand-induced endoplasmic reticulum stress. Mol Pharmacol. 2013 Nov;84(5):751–62. doi: 10.1124/mol.113.087809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillis JL, Selth LA, Centenera MM, Townley SL, Sun S, Plymate SR, et al. Constitutively-active androgen receptor variants function independently of the HSP90 chaperone but do not confer resistance to HSP90 inhibitors. Oncotarget. 2013 May;4(5):691–704. doi: 10.18632/oncotarget.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun C, Shi Y, Xu LL, Nageswararao C, Davis LD, Segawa T, et al. Androgen receptor mutation (T877A) promotes prostate cancer cell growth and cell survival. Oncogene. 2006 Jun 29;25(28):3905–13. doi: 10.1038/sj.onc.1209424. [DOI] [PubMed] [Google Scholar]
- 16.Thalmann GN, Anezinis PE, Chang SM, Zhau HE, Kim EE, Hopwood VL, et al. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994 May 15;54(10):2577–81. [PubMed] [Google Scholar]
- 17.Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int J Cancer. 1994 May 1;57(3):406–12. doi: 10.1002/ijc.2910570319. [DOI] [PubMed] [Google Scholar]
- 18.Korenchuk S, Lehr JE, LMC, Lee YG, Whitney S, Vessella R, et al. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001 Mar-Apr;15(2):163–8. [PubMed] [Google Scholar]
- 19.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008 Jul 1;68(13):5469–77. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009 Mar 15;69(6):2305–13. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009 Jan 1;69(1):16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spruce BA, Campbell LA, McTavish N, Cooper MA, Appleyard MV, O’Neill M, et al. Small molecule antagonists of the sigma-1 receptor cause selective release of the death program in tumor and self-reliant cells and inhibit tumor growth in vitro and in vivo. Cancer Res. 2004 Jul 15;64(14):4875–86. doi: 10.1158/0008-5472.CAN-03-3180. [DOI] [PubMed] [Google Scholar]
- 23.Mertz KD, Setlur SR, Dhanasekaran SM, Demichelis F, Perner S, Tomlins S, et al. Molecular characterization of TMPRSS2-ERG gene fusion in the NCI-H660 prostate cancer cell line: a new perspective for an old model. Neoplasia. 2007 Mar;9(3):200–6. doi: 10.1593/neo.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craft N, Shostak Y, Carey M, Sawyers CL. A mechanism for hormone-independent prostate cancer through modulation of androgen receptor signaling by the HER-2/neu tyrosine kinase. Nat Med. 1999 Mar;5(3):280–5. doi: 10.1038/6495. [DOI] [PubMed] [Google Scholar]
- 25.Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012 Jan 1;18(1):64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georget V, Terouanne B, Nicolas JC, Sultan C. Mechanism of antiandrogen action: key role of hsp90 in conformational change and transcriptional activity of the androgen receptor. Biochemistry. 2002 Oct 1;41(39):11824–31. doi: 10.1021/bi0259150. [DOI] [PubMed] [Google Scholar]
- 27.Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010 Aug;120(8):2715–30. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, Van der Steen T, Tindall DJ. Are androgen receptor variants a substitute for the full-length receptor? Nat Rev Urol. 2015 Mar;12(3):137–44. doi: 10.1038/nrurol.2015.13. [DOI] [PubMed] [Google Scholar]
- 29.Shafi AA, Cox MB, Weigel NL. Androgen receptor splice variants are resistant to inhibitors of Hsp90 and FKBP52, which alter androgen receptor activity and expression. Steroids. 2013 Jun;78(6):548–54. doi: 10.1016/j.steroids.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hickey TE, Irvine CM, Dvinge H, Tarulli GA, Hanson AR, Ryan NK, et al. Expression of androgen receptor splice variants in clinical breast cancers. Oncotarget. 2015 Dec 29;6(42):44728–44. doi: 10.18632/oncotarget.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA, et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016 Jun 4; doi: 10.1001/jamaoncol.2016.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan YX, Mei J, Xu J, Wan BL, Zuckerman A, Pasternak GW. Cloning and characterization of a mouse sigma1 receptor. J Neurochem. 1998 Jun;70(6):2279–85. doi: 10.1046/j.1471-4159.1998.70062279.x. [DOI] [PubMed] [Google Scholar]
- 33.Ryan-Moro J, Chien CC, Standifer KM, Pasternak GW. Sigma binding in a human neuroblastoma cell line. Neurochem Res. 1996 Nov;21(11):1309–14. doi: 10.1007/BF02532372. [DOI] [PubMed] [Google Scholar]
- 34.Maurice T, Su TP. The pharmacology of sigma-1 receptors. Pharmacol Ther. 2009 Nov;124(2):195–206. doi: 10.1016/j.pharmthera.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mo Q, Lu SF, Simon NG. Dehydroepiandrosterone and its metabolites: differential effects on androgen receptor trafficking and transcriptional activity. The Journal of steroid biochemistry and molecular biology. 2006 Apr;99(1):50–8. doi: 10.1016/j.jsbmb.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 36.Arnold JT. DHEA metabolism in prostate: For better or worse? Mol Cell Endocrinol. 2009 Mar 25;301(1–2):83–8. doi: 10.1016/j.mce.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbs TT, Farb DH. Dueling enigmas: neurosteroids and sigma receptors in the limelight. Science’s STKE: signal transduction knowledge environment. 2000 Nov 28;2000(60):pe1. doi: 10.1126/stke.2000.60.pe1. [DOI] [PubMed] [Google Scholar]
- 38.Lu SF, Mo Q, Hu S, Garippa C, Simon NG. Dehydroepiandrosterone upregulates neural androgen receptor level and transcriptional activity. Journal of neurobiology. 2003 Nov;57(2):163–71. doi: 10.1002/neu.10260. [DOI] [PubMed] [Google Scholar]
- 39.Mostaghel EA. Steroid hormone synthetic pathways in prostate cancer. Translational andrology and urology. 2013 Sep;2(3):212–27. doi: 10.3978/j.issn.2223-4683.2013.09.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montgomery RB, Mostaghel EA, Vessella R, Hess DL, Kalhorn TF, Higano CS, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008 Jun 1;68(11):4447–54. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voeltz GK, Rolls MM, Rapoport TA. Structural organization of the endoplasmic reticulum. EMBO reports. 2002 Oct;3(10):944–50. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011 May 17;19(5):575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997 Mar 28;275(5308):1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 44.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011 Jun 14;19(6):792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao S, Ye H, Gerrin S, Wang H, Sharma A, Chen S, et al. ErbB2 Signaling Increases Androgen Receptor Expression in Abiraterone-Resistant Prostate Cancer. Clin Cancer Res. 2016 Mar 2; doi: 10.1158/1078-0432.CCR-15-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.