Abstract
The human microbiome is an intriguing potentially modifiable risk factor in our arsenal against Mycobacterium tuberculosis, the leading infectious disease killer globally. Previous studies have shown associations between the human microbiome and pulmonary disease states; however, etiological links between the microbiome and tuberculosis (TB) infection or disease remain unclear. Immunomodulatory roles of the microbiome may prove to be a critical asset in the host response against TB, including in preventing TB infection, reducing progression from latency, mitigating disease severity, and lowering the incidence of drug resistance and coinfections. This review examined the associations between TB and the gut and lung microbiome. Eight studies were identified through a PubMed database search, including one animal study (N = 1), case report (N = 1), and case–control studies (N = 6). TB infection and disease were associated with reduced gastrointestinal microbial diversity in a murine model and human case report. Sputum microbial diversity differed by TB status in case–control studies, although some reported heterogeneous findings. Current evidence suggests that the gut and lung microbiome are associated with TB infection and disease. However, as studies are limited, etiological and longitudinal research is needed to determine clinical relevance.
Introduction
Mycobacterium tuberculosis caused 1.5 million deaths in 2014,1 and one-third of the global population has latent tuberculosis (TB) infection.2 The scourge of TB infection and disease has been documented since early human history, and highlights the substantial remaining challenges of TB control and eradication efforts. In addition to current strategies, successfully addressing TB infection and disease may require targeting other modifiable risk factors, including host microbiota.
The number of host cells comprising the human body is vastly outnumbered by the number of symbiotic microorganisms,3 and therefore interactions between host cells and microbes occur constantly. The microbiome refers to “the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space.”4 Previous studies have highlighted potential roles of the gastrointestinal and lower respiratory tract's microbiome in the immunological response against TB infection and disease. Generally, bidirectional linkages between the gut microbiome and immune system have been well documented.5,6 Gut microbiota have been associated with a number of disease states, including asthma, autoimmune disorders, cardiovascular diseases, and nonalcoholic fatty liver disease.5–7
This review assessed the association between TB infection and disease and the gut and lung microbiome. The microbiome may have important implications for addressing major challenges to effective TB control, including through immunomodulation, to reduce and address TB transmission, progression from latency, disease severity, and drug resistance.
TB pathology and immune response.
Mycobacterium tuberculosis bacilli are transmitted from patients with active TB through airborne droplets.2 In lungs, the innate host response includes the detection (by Toll-like receptors) and elimination (via alveolar macrophages and dendritic cells inducing antimicrobial peptides [cathelicidin] and autophagy) of M. tuberculosis.8 Gamma-interferon and tumor necrosis factor-alpha activate the antimycobacterial capacity of macrophages, through producing nitric oxide, reactive oxygen, and nitrogen intermediates.8 Immune cells (macrophages, fibroblasts, T and B cells) accumulate to form granulomas, surrounding M. tuberculosis and restricting growth.8 Despite these host defenses, survival mechanisms of M. tuberculosis (including inhibiting phagolysosome fusion) allow some bacteria to persist within granulomas.8
In terms of adaptive immunity, cell-mediated responses by cluster of differentiation (CD)4+ and CD8+ T cells are critical to successfully address M. tuberculosis.8 Primary effector functions of CD4+ and CD8+ T cells include producing gamma-interferon and cytokines to activate macrophages, and lysing infected macrophages.8 The T helper (Th1) cytokine expression pattern is important to eliminate M. tuberculosis9; conversely, Th2 and regulatory T (Treg) cell responses support M. tuberculosis survival.8,10 However, an enhanced Th1 response simultaneously causes inflammation and host tissue damage,11 which suggests a balanced Th1/Th2 immune response is ideal for patient health outcomes. In summary, the dynamic interplay between host immune response and M. tuberculosis survival mechanisms (such as T cell homeostasis) is modulated through numerous factors, potentially including the microbiome.
The gut and lung microbiome.
Gut bacteria play important roles in nutrient metabolism, intestinal homeostasis (through preventing overgrowth of intestinal pathogens), and immunity.5,12,13 Several studies have examined the associations between altered gut microbiota and clinical pathologies including inflammatory bowel disease, diabetes, and obesity.5,6,14
Historically, the lower respiratory tract has been considered sterile in healthy individuals, but recent culture-independent studies have shown evidence to the contrary.15,16 Given the limited available studies focusing on the lung microbiome, one key research gap is determining whether the observed microbiota are simply a continuation of the upper respiratory tract or separate. Preliminary findings corroborate that microbial populations present in the lung are distinct from the upper respiratory tract,17 and show intra-lung heterogeneity.16
Immunomodulation of microbiota.
Murine studies involving gut microbiota have provided evidence of the modulation of microbiota by the immune system. In one study, transgenic mice with expression of human defensin 5, antimicrobial polypeptides secreted by Paneth cells in the small intestine, received a virulent Salmonella typhimurium challenge.18 The transgenic mice had lower bacterial burden in their terminal ilea, compared with wild-type controls.18 In addition, mice lacking specific proteins involved in innate immunity (Toll-like receptor 5, nucleotide-binding oligomerization domain–like receptor family pyrin domain-containing 6 inflammasome) had intestinal dysbiosis and associated pathologies, including colitis or insulin sensitivity.19,20 Subsequently, the transfer of altered microbiota to wild-type mice was observed to lead to disease.20 Based on preliminary data, hypotheses include that immune responses (including against M. tuberculosis) facilitate cross talk between microbial populations (e.g., between the lung and gut microbiome); and dysbiotic microbiota may adversely influence some clinical pathologies.
Microbial diversity is hypothesized to affect the growth of particular opportunistic pathogens, due to resource competition. Krishna and others (2016) observed that opportunistic pathogens (Rothia mucilaginosa) were associated with increased complexity and diversity of sputum microbiota among patients with active TB.21 Iwai and others (2014) reported that lower bacterial burden was associated with increased community richness (taxa per sample) and phylogenetic diversity among Ugandans with human immunodeficiency virus and acute pneumonia.22
Influence of the microbiome on immunity.
Conversely, specific commensal gut microorganism species have been observed to modulate the immune system.23–26 Gut bacteria species (Bacteroides fragilis species, Clostridium genus) were associated with altered Treg cell counts, function, and development.23,24 Previous studies have reported the influence of bacteria on respiratory health, including 1) airway inflammation and 2) lung damage (in the context of influenza and probiotic supplementation). In germ-free mice, increased airway inflammation was rescued by the administration of certain bacterial species (Faecalibacterium, Lachnospira, Rothia, and Veillonella).26 Similarly, greater abundance of bacteria species (Veillonella, Prevotella) in the supraglottic-characteristic taxa were associated with higher indicators of airway inflammation.27
Moreover, two studies showed links between gut bacteria and lung damage. Disrupted gut microbiota (decreased Bifidobacterium and Lactobacillus) was associated with altered immune response to influenza A infection and increased lung damage in a murine model.28 Separately, probiotic supplementation of Bifidobacterium was associated with improved Toll-like receptor 7 response and reduced lung damage.28 One hypothesis is that probiotics are protective against pneumococcal disease through modulating the upper respiratory tract microbiome.29
In summary, given these dynamic bidirectional interactions between the microbiome and host immunity, commensal bacteria could have a role in the immunological response to pulmonary TB infection and disease. Specific commensal microorganisms may facilitate the cell-mediated immune response to TB infection and disease, and modulate inflammation and lung damage through functional changes of the metagenome and metabolite production.12,13
TB and the Microbiome
In this review, eight studies examined the association between pulmonary TB infection and disease and the microbiome, including an animal study,30 a case report,31 and 6 case–control studies (Table 1).21,32–36 Key study findings are highlighted in the following two sections (regarding TB and the gut or lung microbiome, respectively), and study methodology is compared in the third section.
Table 1.
Studies assessing the association between TB and the lung and gut microbiome*
| Study design | Sample size | Location | Exposure | Outcome | Key findings | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Assessment | Biological specimen | Microbial assessment method(s) | Hypervariable regions† | ||||||
| Murine model | 5 female BALB/c mice | – | Maryland | TB infection (aerosolized Mycobacterium tuberculosis CDC1551 strain) | Bacterial diversity‡ | Stool | 16S rRNA sequencing | V1–V2 | Pre- vs. postinfection | • Majority of differential OTUs: Firmicutes phyla | 30 |
| • Alpha diversity (Shannon diversity index): postinfection decrease (in all mice) | • Relative species abundance of Lachnospiraceae and Ruminococcaceae and Bacteriodales order greater preinfection and among controls | ||||||||||
| • Beta diversity (PCoA; UniFrac): P < 0.001 | |||||||||||
| • 88 OTUs more abundant preinfection: q < 0.01 | |||||||||||
| 5 female BALB/c mice | 5 mice (1:1 age-matched) | TB infection (aerosolized M. tuberculosis H37Rv strain) | TB vs. control | ||||||||
| • Differential clustering (PCoA; UniFrac distances): P < 0.001 | |||||||||||
| • 73 different OTUs: q < 0.01 | |||||||||||
| Case study | Patient with multidrug-resistant TB | – | Marseille, France | – | Microbial diversity | Stool | 16S and 18S rRNA sequencing; culture | V6 | • Depleted gut microbiota: 39 bacterial species, 18 phylotypes, 19 OTUs | 31 | |
| • Most phyla (from pyrosequencing) confirmed with culture (Firmicutes, Actinobacteria, Proteobacteria), except Fusobacteria species (only culture) and Cyanobacteria/Chloroplast (only pyrosequencing) | |||||||||||
| • New species detected by culture (Candidatus “Paenibacillus antibioticophila”) and 18S rRNA sequencing (3 fungal species) | |||||||||||
| Case control | 31 patients with pulmonary TB | 24 controls | Shanghai, China | TB status | Bacterial diversity | Sputum (cases); saliva/pharyngeal secretions (controls) | 16S rRNA sequencing | V3 | TB vs. control | 32 | |
| • Greater phyla and genera richness among patients with TB (24 phyla; 564 genera) vs. controls (17 phyla; 235 genera) | |||||||||||
| • Most prevalent phyla: Firmicutes, Proteobacteria, Bacteriodetes, Crenarchaeota, and Actinobacteria | |||||||||||
| • Lower Bacteriodetes and higher Actinobacteria prevalence | |||||||||||
| • Certain genera (Stenotrophomonas, Cupriavidus, Pseudomonas, Thermus) and phyla (Aquificae, Planctomycetes) only in TB samples | |||||||||||
| • Differential clustering (PCoA; UniFrac distances) | |||||||||||
| 32 patients with unilateral pulmonary TB | 24 healthy controls | Jiangsu, China | TB status | Bacterial diversity | Bronchoalveolar lavage (cases); saliva, pharyngeal secretions (controls) | 16S rRNA sequencing | V3 | TB vs. control | 36 | ||
| • Clustering (PCoA; UniFrac distances): distinct | |||||||||||
| • Genus: most abundant genus differed | |||||||||||
| TB (uninfected vs. infected lungs) | |||||||||||
| • Clustering (PCoA; UniFrac distances): similar | |||||||||||
| • Genus: Mycobacteria and Porphyromonas abundances greater inside TB lesions | |||||||||||
| • Shannon diversity index: higher in bronchoalveolar lavage fluid from lungs with lesions, compared with controls | |||||||||||
| 22 patients with TB | 14 controls with TB-like coughing | Hong Kong SAR, China | TB status | Bacterial diversity | Sputum | 16S rRNA sequencing | V1–V2 | TB vs. control | 33 | ||
| • Phyla: lower Firmicutes, and higher Proteobacteria and Bacteriodetes abundance | |||||||||||
| • Genra: lower Streptococcus, and higher Neisseria and Prevotella | |||||||||||
| • OTUs: 8 more prevalent, 2 less prevalent | |||||||||||
| • No differences in clustering (PCoA; UniFrac distances) | |||||||||||
| Overall | |||||||||||
| • 98% of identified sequences accounted for 5 phyla (Proteobacteria, Firmicutes, Bacteriodetes, Fusobacteria, Actinobacteria) | |||||||||||
| • Most prevalent bacterial genera: Neisseria, Prevotella, Streptococcus | |||||||||||
| Group 1: Newly diagnosed pulmonary TB (N = 25) | Group 4: Healthy controls (N = 20) | Shanghai, China | TB status | Bacterial diversity | Sputum (cases); throat swabs (controls) | 16S rRNA sequencing | V1–V2 | • Clustering (PCoA; UniFrac distances): differed between Groups 1 and 3 vs. 4; similar between Groups 1 and 3 | 34 | ||
| Group 2: Recurrent TB (N = 30) | • Relative species abundance (phyla, genera): heterogeneous between groups | ||||||||||
| Group 3: Treatment failure (N = 20) | • Some genera (Bergeyella, Sharpea) only in Groups 1–3 | ||||||||||
| 6 patients with TB | 6 controls without TB | Medellìn, Colombia | TB status | Bacterial and fungi diversity | Sputum, oropharyngeal, nasal samples | 16S rRNA sequencing (bacteria; fungi [fungal nuclear ribosomal internal transcribe spacer 1 regions]) | V1–V2 | Bacteria: TB vs. control | 35 | ||
| • Shannon diversity index: bacterial diversity (in nasal samples) less diverse | |||||||||||
| • Clustering (PCoA; UniFrac distances): sputum and oropharyngeal samples clustered together, distinct from nasal samples | |||||||||||
| • Oropharyngeal: Streptococcaceae more abundant | |||||||||||
| Bacteria: overall | |||||||||||
| • Phyla: relative abundance differed across sample type (bacterial diversity higher in sputum and lower in nasal samples) | |||||||||||
| Fungi: TB vs. control | |||||||||||
| • Genus: Cryptococcus (in oropharyngeal samples) significantly lower | |||||||||||
| Fungi: overall | |||||||||||
| • Shannon diversity index: fungal diversity greatest in nasal samples | |||||||||||
| • Phyla: 90% sequences in Ascomycota and Basidiomycota; Ascomycota significantly differed between study participants | |||||||||||
| 25 patients with pulmonary TB | 16 controls | Madhya Pradesh, India | TB status | Bacterial diversity | Sputum | 16S rRNA sequencing | V6–V7 | TB vs. control | 21 | ||
| • Clustering (PCoA): distinct | |||||||||||
| • Phyla: higher Firmicutes and Actinobacteria, lower Proteobacteria and Fusobacteria | |||||||||||
| • Most prevalent genera (cases: Streptococcus, Neisseria, Veillonella; controls: Gammaproteobacteria, Streptococcus, Neisseria, Haemophilus) | |||||||||||
| • Species: Veillonella dispar and Rothia mucilaginosa greater | |||||||||||
| • Shannon diversity index: lower for TB (3.88; 602 OTUs) than controls (4.13; 490 OTUs) | |||||||||||
HIV = human immunodeficiency virus; OTU = operational taxonomic units; PCoA = principal coordinate analysis; rRNA = ribosomal RNA; SAR = Special Administrative Region; TB = tuberculosis.
PubMed search strategy was based on the following terms: “((microbiota[mesh] OR microbiota[tw] OR microbiome*[tw] OR virome*[tw] OR gut bacteria[tw]) AND (tuberculosis[mesh] OR tuberculosis[tw] OR TB[tw])),” and restricted by publication date (before June 6, 2016). Exclusion criteria included 1) not primary data source (including reviews, editorials); 2) no whole microbiome sequencing techniques; and 3) not among patients or animals with TB or exposure to TB (including assessing vaccine response among healthy volunteers, microbiome among patients with HIV, or reproductive tract inflammation). Among the 44 studies initially identified, eight studies were within the scope of this review and included.
Assessed by 16S rRNA sequencing.
First experiment: baseline (preinfection) to 179 days (postinfection). Second experiment: 0–46 days.
TB and the gut microbiome.
Low bacterial species richness and abundance were found in gastrointestinal tract samples from mice and a patient with active TB (Table 1).30,31 Five female BALB/c mice were infected with aerosolized M. tuberculosis (CDC1551 strain).30 Pre- and postinfection samples differed, in terms of bacterial abundance (among 88 operational taxonomic units [OTUs]; q < 0.01) and composition (beta-diversity indices of weighted and unweighted UniFrac distances by Principal Coordinate Analysis [PCoA]; both P ≤ 0.005).30 In a second experiment, mice were infected with a different strain of M. tuberculosis (H37Rv; N = 5), and compared with 1:1 age-matched controls.30 Fecal samples from a single time point postinfection similarly showed differential clustering and bacterial abundance among 73 OTUs; q < 0.01), compared with uninfected samples.30 In the case report, Dubourg and others (2013) obtained stool samples from a patient with multidrug-resistant active TB who previously received multiple oral antibiotic regimens.31 Gut microbiota were severely depleted (39 bacterial species, 18 phylotypes, and 19 OTUs).31
Other studies have corroborated the potential role of intestinal microbiota in TB. Perry and others (2010) reported that individuals with latent TB and Helicobacter pylori had more interferon gamma and Th1-like cytokines, compared with those without H. pylori.37 Actinobacteria in infant stool samples was associated with increased T cell responses to vaccination, including Bacille Calmette–Guérin vaccination for TB.25 Furthermore, cynomolgus macaques with H. pylori infection were less likely to develop active TB if inoculated with M. tuberculosis.37 Another murine study demonstrated how specific enteric bacteria (Helicobacter hepaticus) modulated the immune system to alter susceptibility to M. tuberculosis and vaccine response in a mouse model.38 One hypothesis was that activation of the innate immune system facilitates an enhanced response to other pathogens, such as M. tuberculosis.
Several recent studies have provided evidence of the immunomodulatory mechanism of the gut microbiome, which produces short-chain fatty acid (SCFA) metabolites that may affect the host response against M. tuberculosis.39 Intestinal microbiota produce SCFAs through the fermentation of resistant starches and dietary fiber.40 SCFAs have roles in host metabolism (as substrate for de novo lipid and glucose synthesis) and immunomodulation (through downregulating pro-inflammatory cytokines and Treg cells).41 In a study involving M. tuberculosis stimulation of human peripheral blood mononuclear cells, physiological concentrations of the SCFA butyrate significantly decreased pro-inflammatory cytokine production.39 Given the importance of the cell-mediated response (including Treg cells42) against TB infection, SCFA production is one hypothesized mechanism of the link between the microbiome and TB infection and disease.
Overall, the gut microbiome of TB samples had lower bacterial abundance and composition, relative to controls. However, given the limited data and heterogeneous study designs, preliminary evidence suggests the need for further studies to confirm etiology through mechanistic and clinical studies. Potential next steps include improving our understanding of the role of metabolites in mediating the association between the gut microbiome and TB disease and infection.
TB and the lung microbiome.
Several case–control studies in this review showed that bacterial diversity (richness, abundance, OTU clustering, Shannon index) in the respiratory tract differs between individuals with active TB, compared with controls (Table 1).21,32–34,36 Based on PCoA, four studies found distinct OTU clustering of sputum samples from study participants with TB disease, compared with controls.21,32,34,36 However, one study observed no differences in OTU clustering.33 All case–control studies identified differences in bacterial richness or relative abundance, based on taxonomic categorizations (phyla, genus, species), in sputum from individuals with and without active TB. However, the specific differential taxonomic groups were inconsistent and not replicated across studies, which may reflect the relatively small and diverse sample populations.
In five case–control studies, sputum was considered an indicator for the microbiome of the lung and lower respiratory tract among study participants with active TB. Given that sputum is likely to be contaminated by the upper respiratory tract during expectoration, the continued use of sputum reflects the challenge of directly obtaining samples from the lung.
Sample collection methods for referent groups varied, including samples from deep coughing of healthy individuals,32,36 throat swabs,34 bronchoalveolar lavages,36 and sputum from individuals with TB-like coughing.33 Botero and others (2014) reported that the microbial compositions of sputum and oropharyngeal samples were similar, which supports the use of throat swabs from controls.35 Limitations include deep cough, throat swab, and sputum samples may represent the upper respiratory tract (instead of the lung and lower respiratory tract microbial composition); TB-like coughing could be caused by other diseases that affect the microbiome; and the invasiveness of bronchoalveolar lavages. Thus, interpretations of study findings need to account for potential sample contamination and the appropriate selection of controls.
As a brief summary, several studies showed distinct bacterial richness or relative abundance of the lung microbiome among patients with active TB, compared with controls. Further studies are needed, particularly in light of differences in study designs and discrepant observations.
Comparison of study methods.
Strengths of studies in this review included the consistent use of next-generation platforms for 16S ribosomal RNA (rRNA) sequencing to assess the bacterial microbiome in all eight studies. Seven studies used 454 instruments (GS FLX, GS FLX-Titanium [Roche Diagnostics, Base, Switzerland]), and one study used an Ion Torrent PGM (Thermo Fisher Scientific, San Jose, CA). Two studies additionally evaluated fungal species,31,35 and one study compared 16S rRNA results with culture and 18S rRNA sequencing.31 In all studies, sequence analyses were conducted through software (including Quantitative Insights Into Microbial Ecology,43 Mothur44), and OTUs were identified through aligning sequences with reference databases (Ribosomal Database Project,45 Greengenes46), to eliminate chimeras. Some studies considered differences (including sociodemographic characteristics) of sample populations, which are supported by previous literature. In addition, the eight studies included study participants from several geographic locations, which allowed for comparison and confirmation of similar findings across diverse populations.
Available studies had several limitations, in addition to aforementioned challenges. Compared with Sanger sequencing (such as with ABI 3730 Genetic Analyzer [Applied Biosystems, Foster City, CA]), 454 sequencing has been reported to have higher error rates due to greater insertion and deletion rates.47 Importantly, methodological heterogeneity in data analyses rendered comparisons across studies difficult; these included differences in hypervariable regions (V1–V3, V6, V7), diversity indices (including alpha- [Shannon index, Chao1] and beta-diversity [unweighted and weighted UniFrac, Jaccard]), and additional analyses (PCoA, hierarchical clustering heatmap).
Separately in the case–control studies, the sample sizes (12–95) and geographic locations (Asia [China, Hong Kong, India], South America [Colombia]) ranged widely. Therefore, results were difficult to generalize to other countries. In addition, numerous other potential confounding factors (antibiotic use [including anti-TB treatment], comorbidities, environmental factors, diet) were either unaccounted for or considered differently across study populations. Standard anti-TB medications are a combination of antibiotics, which are expected to alter the microbiome, and therefore need to be considered while interpreting study results. Four of the six case–control studies excluded participants with recent antibiotic use (1 or 3 months before baseline).32,33,35,36 However, two studies did not report the exact length of time that participants did not receive antibiotics before sample collection,21 or prior antibiotic use.34 In the case report, the patient was receiving anti-TB treatment of multidrug-resistant TB at the time of sample collection.31
Discussion
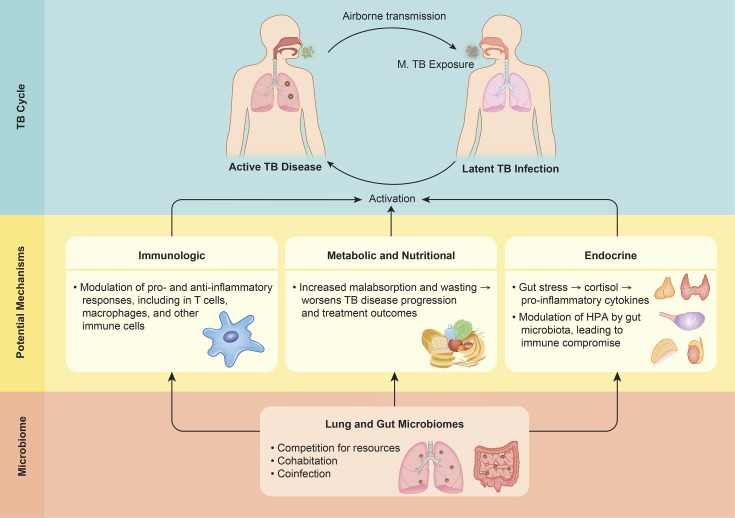
In this review, there is limited evidence regarding the key question of the bidirectional associations between TB infection and disease and the lung and gut microbiome. Potential mechanistic pathways need to be considered through epidemiological and mechanistic studies, including immunological, nutritional, metabolic, and endocrine factors (Figure 1 ). Furthermore, there are a number of other research gaps and related questions that have not been directly addressed by studies to date. One of these overarching knowledge gaps involves interindividual variability, which is a key focus area of the Human Microbiome Project.48 What is the extent that a core set of human microbiome genes or species is shared between individuals?48 In addition, what are the temporal changes of lung and gut microbiota (including throughout TB disease progression and treatment)? Separately, to what extent does cross talk occur between the lung and gut microbiome, particularly in response to TB infection and disease-related perturbances? Followingly, do these interactions between the respiratory and gastrointestinal microbiome modulate immune responses to TB infection and disease?
Figure 1.
Potential mechanistic pathways between the microbiome and tuberculosis (TB).
Conclusion
Based on preliminary evidence from studies in this review, the lung and gut microbiome were associated with TB infection and disease. The microbiome is a potential modifiable risk factor for TB infection and disease; however, the number of available studies is limited. Most studies have focused on characterizing the microbial profile among individuals with and without active TB disease. Future studies are necessary to further elucidate etiology, key mechanisms, and potential clinical significance. Specifically, it is important to assess 1) the sputum and gut microbiome as risk factors for TB infection and disease susceptibility, disease progression, and treatment outcomes and 2) the effects of TB infection and disease on the sputum and gut microbiome, which can subsequently impact health via alteration of immune responses.
ACKNOWLEDGMENTS
We thank Michael S. Glickman for his comments and suggestions, and TNQ (tnq.co.in) for their assistance with the graphic design of Figure 1.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) or the National Institutes of Health.
Footnotes
Financial support: Research reported in this publication was supported by the Human Ecology Alumni Association of Cornell University (for Madeleine R. Wood) and the National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases; T32-DK007158 award; for Elaine A. Yu).
Disclosure: Madeleine R. Wood and Elaine A. Yu have no conflicts of interest. Saurabh Mehta is an unpaid board member of and has an equity interest in a diagnostic start-up focused on developing assays for low-cost and point-of-care measurement of certain nutrients from a drop of blood using results from his research as a faculty member at Cornell University.
Authors' addresses: Madeleine R. Wood and Elaine A. Yu, Division of Nutritional Sciences, College of Human Ecology, Cornell University, Ithaca, NY, E-mails: mrw245@cornell.edu and eay27@cornell.edu. Saurabh Mehta, Division of Nutritional Sciences, College of Human Ecology, Cornell University, Ithaca, NY, and Institute for Nutritional Sciences, Global Health, and Technology, Cornell University, Ithaca, NY, E-mail: smehta@cornell.edu.
References
- 1.World Health Organization . Global Tuberculosis Report 2015. Geneva, Switzerland: World Health Organization; 2015. [Google Scholar]
- 2.World Health Organization . Tuberculosis Fact Sheet. Geneva, Switzerland: World Health Organization; 2015. [Google Scholar]
- 3.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 4.Lederberg J, McCray AT. ‘Ome sweet ’omics: a genealogical treasury of words. Scientist. 2001;15:8. [Google Scholar]
- 5.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 6.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 9.Mosmann TR, Sad S. The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol Today. 1996;17:138–146. doi: 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya D, Dwivedi VP, Maiga M, Van Kaer L, Bishai WR, Das G. Small molecule-directed immunotherapy against recurrent infection by Mycobacterium tuberculosis. J Biol Chem. 2014;289:16508–16515. doi: 10.1074/jbc.M114.558098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 12.Minarrieta L, Ghorbani P, Sparwasser T, Berod L. Metabolites: deciphering the molecular language between DCs and their environment. Semin Immunopathol. 2016;39:177–198. doi: 10.1007/s00281-016-0609-6. [DOI] [PubMed] [Google Scholar]
- 13.Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol. 2016;35:8–15. doi: 10.1016/j.mib.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 15.Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, Kaess H, Deterding RR, Accurso FJ, Pace NR. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA. 2007;104:20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L, Jablonski K, Kleerup E, Lynch SV, Sodergren E, Twigg H, Young VB, Bassis CM, Venkataraman A, Schmidt TM, Weinstock GM. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 19.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishna P, Jain A, Bisen PS. Microbiome diversity in the sputum of patients with pulmonary tuberculosis. Eur J Clin Microbiol Infect Dis. 2016;7:1205–1210. doi: 10.1007/s10096-016-2654-4. [DOI] [PubMed] [Google Scholar]
- 22.Iwai S, Huang D, Fong S, Jarlsberg LG, Worodria W, Yoo S, Cattamanchi A, Davis JL, Kaswabuli S, Segal M, Huang L, Lynch SV. The lung microbiome of Ugandan HIV-infected pneumonia patients is compositionally and functionally distinct from that of San Franciscan patients. PLoS One. 2014;9:e95726. doi: 10.1371/journal.pone.0095726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134:e362–e372. doi: 10.1542/peds.2013-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, Investigators CS, Mohn WW, Turvey SE, Brett Finlay B. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 27.Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Chen H, Berger KI, Goldring RM, Rom WN, Blaser MJ, Weiden MD. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:1–12. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S, Jiang ZY, Sun YF, Yu B, Chen J, Dai CQ, Wu XL, Tang XL, Chen XY. Microbiota regulates the TLR7 signaling pathway against respiratory tract influenza A virus infection. Curr Microbiol. 2013;67:414–422. doi: 10.1007/s00284-013-0380-z. [DOI] [PubMed] [Google Scholar]
- 29.Licciardi PV, Toh ZQ, Dunne E, Wong S-S, Mulholland EK, Tang M, Robins-Browne RM, Satzke C. Protecting against pneumococcal disease: critical interactions between probiotics and the airway microbiome. PLoS Pathog. 2012;8:e1002652. doi: 10.1371/journal.ppat.1002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winglee K, Eloe-Fadrosh E, Gupta S, Guo H, Fraser C, Bishai W. Aerosol Mycobacterium tuberculosis infection causes rapid loss of diversity in gut microbiota. PLoS One. 2014;9:e97048. doi: 10.1371/journal.pone.0097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubourg G, Lagier JC, Armougom F, Robert C, Hamad I, Brouqui P, Raoult D. The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur J Clin Microbiol Infect Dis. 2013;32:637–645. doi: 10.1007/s10096-012-1787-3. [DOI] [PubMed] [Google Scholar]
- 32.Cui Z, Zhou Y, Li H, Zhang Y, Zhang S, Tang S, Guo X. Complex sputum microbial composition in patients with pulmonary tuberculosis. BMC Microbiol. 2012;12:276. doi: 10.1186/1471-2180-12-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung MK, Lam WY, Fung WY, Law PT, Au CH, Nong W, Kam KM, Kwan HS, Tsui SK. Sputum microbiota in tuberculosis as revealed by 16S rRNA pyrosequencing. PLoS One. 2013;8:e54574. doi: 10.1371/journal.pone.0054574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Liu W, He L, Huang F, Chen J, Cui P, Shen Y, Zhao J, Wang W, Zhang Y, Zhu M, Zhang W, Zhang Y. Sputum microbiota associated with new, recurrent and treatment failure tuberculosis. PLoS One. 2013;8:e83445. doi: 10.1371/journal.pone.0083445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Botero LE, Delgado-Serrano L, Cepeda ML, Bustos JR, Anzola JM, Del Portillo P, Robledo J, Zambrano MM. Respiratory tract clinical sample selection for microbiota analysis in patients with pulmonary tuberculosis. Microbiome. 2014;2:29. doi: 10.1186/2049-2618-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y, Lin F, Cui Z, Zhang X, Hu C, Shen T, Chen C, Zhang X, Guo X. Correlation between either Cupriavidus or Porphyromonas and primary pulmonary tuberculosis found by analysing the microbiota in patients' bronchoalveolar lavage fluid. PLoS One. 2015;10:e0124194. doi: 10.1371/journal.pone.0124194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry S, de Jong BC, Solnick JV, de la Luz Sanchez M, Yang S, Lin PL, Hansen LM, Talat N, Hill PC, Hussain R, Adegbola RA, Flynn J, Canfield D, Parsonnet J. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS One. 2010;5:e8804. doi: 10.1371/journal.pone.0008804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold IC, Hutchings C, Kondova I, Hey A, Powrie F, Beverley P, Tchilian E. Helicobacter hepaticus infection in BALB/c mice abolishes subunit-vaccine-induced protection against M. tuberculosis. Vaccine. 2015;33:1808–1814. doi: 10.1016/j.vaccine.2015.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lachmandas E, van den Heuvel CN, Damen MS, Cleophas MC, Netea MG, van Crevel R. Diabetes mellitus and increased tuberculosis susceptibility: the role of short-chain fatty acids. J Diabetes Res. 2016;2016:6014631. doi: 10.1155/2016/6014631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bird AR, Brown IL, Topping DL. Starches, resistant starches, the gut microflora and human health. Curr Issues Intest Microbiol. 2000;1:25–37. [PubMed] [Google Scholar]
- 41.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shafiani S, Tucker-Heard G, Kariyone A, Takatsu K, Urdahl KB. Pathogen-specific regulatory T cells delay the arrival of effector T cells in the lung during early tuberculosis. J Exp Med. 2010;207:1409–1420. doi: 10.1084/jem.20091885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Challenger GG, Van Horn DJ, Weber CF. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Suske CR, Tiedje JM. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jumpstart Consortium Human Microbiome Project Data Generation Working G Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One. 2012;7:e39315. doi: 10.1371/journal.pone.0039315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The Human Microbiome Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]