Abstract
Crimean–Congo hemorrhagic fever is a tick-borne disease caused by the arbovirus Crimean–Congo hemorrhagic fever virus (CCHFV, family Bunyaviridae, genus Nairovirus). CCHFV can cause a severe hemorrhagic fever with high-case fatality rates in humans. CCHFV has a wide geographic range and has been described in around 30 countries in the Middle East, Asia, Europe, and Africa including Mali and neighboring countries. To date, little is known about the prevalence rates of CCHFV in Mali. Here, using banked bovine serum samples from across the country, we describe the results of a seroepidemiological study for CCHFV aimed at identifying regions of circulation in Mali. In total, 1,074 serum samples were tested by a modified in-house CCHFV-IgG-enzyme-linked immunosorbent assay (ELISA) with confirmatory testing by commercial ELISA and immunofluorescence assay. Overall, 66% of samples tested were positive for CCHFV-specific IgG antibodies. Regional seroprevalence rates ranged from 15% to 95% and seemed to correlate with cattle density. Our results demonstrate that CCHFV prevalence is high in many regions in Mali and suggest that CCHFV surveillance should be established.
Introduction
The genus Nairovirus (family Bunyaviridae) contains seven serogroups consisting of 34 predominantly tick-borne viruses of which Crimean–Congo hemorrhagic fever virus (CCHFV) is the most prevalent.1,2 In humans, CCHFV causes an acute infection referred to as Crimean–Congo hemorrhagic fever, a disease which is often associated with varying degrees of bleeding such as petechiae or ecchymosis as well as other clinical manifestations including encephalitis and multi-organ failure. Disease onset typically follows an incubation period of up to 7 days and is characterized by nondescript indicators including a rapid onset of high-grade fever, fatigue, cephalalgia, dizziness, photophobia, and myalgia, often with nausea, vomiting, and diarrhea.3–7 Death occurs in 10–50% of cases.8,9 This variation might be due to a certain diversity of individual and general awareness, effectiveness of the public health system, and the circulating virus strain in the different regions.10
CCHFV has a wide geographic range and has been described in approximately 30 countries in the Middle East, Asia, Europe, and Africa including Mali and neighboring countries.3,8,11–13 The vector and natural reservoir of CCHFV are the wide spread Hyalomma subspecies (ssp.) ticks.7 The predominant vector of CCHFV varies geographically and includes Hyalomma anatolicum spp. (especially Hyalomma anatolicumanatolicum) throughout Eurasia and the northern half of Africa; Hyalomma marginatum subspecies (Hyalomma marginatum marginatum, Hyalomma marginatum rufipes, Hyalomma marginatumturanicum, and Hyalomma marginatumisaaci) also in Eurasia and Africa; and Hyalomma truncatum, Hyalomma impeltatum, and Hyalomma impressum primarily within Africa.12,14–19 A wide range of wild and domesticated agricultural animals, such as hares, hedgehogs, cattle, sheep, and goats, can serve as transient reservoirs of CCHFV and play an important role in the natural life cycle of the virus.19–22 Humans are most commonly infected via direct tick bites; however, infections following exposure to tissues, blood, or body fluids of infected animals or human patients are also well described.23 Infected animals do not show clinical signs, but a viremia of up to 2 weeks and seroconversion can be detected.18
CCHFV is prevalent in west Africa with documented human fatal cases occurring in Senegal and in Mauritania.11,24 In recent years, the virus appears to be spreading to new areas in many countries with the report of first human cases of the disease.24–27 To date, CCHFV has been understudied in Mali. In 2005, Traoré and others reported a CCHFV seroprevalence rate of 4.5% in inhabitants of Baguineda town in the Koulikoro Region of Mali.28 In 2014, CCHFV was detected by polymerase chain reaction (PCR) in ticks collected in Kati Daral cattle market in the same region of Mali. Genetic analyses demonstrated the amplified sequences were highly related to a previously identified strain in Mauritania and confirmed the presence of CCHFV positive vectors in Mali.29
Given the prominent involvement of ruminants in the CCHFV life cycle, these animals provide a convenient sentinel group of species that is often used to gauge CCHFV epizootic activity in specific areas and therewith to define areas of possible risk.30–32 Seroepidemiological studies in livestock are important as they can determine the prevalence of CCHFV circulation in a region and help to define potential risk areas. Unfortunately, few serological assays for CCHFV that are compatible with cattle or other livestock have been published so far, which hinders these types of epidemiological studies.10 Here, we use an indirect in-house CCHFV-IgG-enzyme-linked immunosorbent assay (ELISA) and two commercially available but species (bovine)–adapted ELISA and immunofluorescence assay kits for a retrospective CCHFV seroprevalence study testing bovine samples collected from across Mali. Our results indicate that CCHFV prevalence is high in many regions in Mali and suggest that CCHFV surveillance should be established to monitor the presence and distribution of this virus.
Materials and Methods
Study site.
Mali is a landlocked country in west Africa with its capital Bamako. Geographically, it is the eighth largest country in Africa covering more than 1,240,000 km2. Mali has eight administrative regions in addition to the capital city, the district of Bamako.
Serum samples.
For the seroepidemiological study, 1,075 bovine serum samples from the Central Veterinary Laboratory bovine serum bank were retrospectively tested for the presence of IgG antibodies reactive to CCHFV antigens. The samples originated from different sites across Mali including Bamako (N = 20), Gao (N = 20), Kayes (N = 20), Kidal (N = 20), Koulikoro (N = 63), Mopti (N = 575), Segou (N = 252), Sikasso (N = 85), and Tombouctou (N = 20). The samples were originally collected between 2005 and 2014 as a part of Malian national transboundary animal diseases surveillance program.
For the validation of the in-house CCHFV-IgG-ELISA, 303 serum samples from cattle in Germany were used as a negative reference panel, as Germany is outside of the CCHFV endemic zone. The positive reference serum panel was composed of serum samples from cattle from different European and African countries: 19 serum samples collected in Albania, 22 collected in Macedonia, 62 collected in Turkey, and 50 collected in Mauretania. Sera were included in the positive reference serum panel if they were positive in the adapted commercial CCHFV-IgG-ELISA (Vector-Best, Novosibirsk, Russia) and in the adapted commercial CCHFV-IgG-IFA (Euroimmun, Lübeck, Germany)18 (Table 1).
Table 1.
Validation results of the indirect in-house CCHFV-IgG-ELISA
| Positive reference sera | Negative reference sera | |
|---|---|---|
| Positive | 146 | 2 |
| Negative | 2 | 294 |
| Inconclusive | 5 | 7 |
CCHFV = Crimean–Congo hemorrhagic fever virus; ELISA = enzyme-linked immunosorbent assay.
In-house CCHFV-IgG-ELISA.
The indirect in-house CCHFV-IgG-ELISA used in this study is a modification of a previously described protocol.18 Briefly, 96-well Greiner F immunoplates (Greiner Bio-One, Kremsmünster, Austria) were coated with 0.2 μg/well recombinant nucleocapsid protein of the CCHFV strain KosovoHoti (accession no. DQ133507) diluted in 100 μL coating buffer (PBS, 0.5% BSA, pH 9) and incubated overnight at 4°C. The sera were diluted 1/40 in dilution buffer no. 11 (100 μL/well; IDVet, Grabels, France) and incubated for 1 hour at 37°C. Hundred micro liters/well goat antibovine IgG-horseradish peroxidase conjugate (SouthernBiotech, Birmingham, AL) were used for detection of bound anti-CCHFV antibodies (1/1,000 in conjugate dilution buffer no. 3, IDVet, 2 hours at 37°C). Color change was induced by 3,3′,5,5′-Tetramethylbenzidine reaction and stopped after 10 minutes with H2SO4 (each 100 μL/well). All washing steps were performed with phosphate-buffered saline–Tween 20 0.1% (three times). Optical density values were measured with the Tecan infinite F200 PRO reader (Tecan group, Männedorf, Switzerland) at 420 nm wavelength. The additionally detected reference wavelength was automatically subtracted by the corresponding computer software Tecani-control (Tecan group). The final result was obtained by building a ratio between the OD value of the tested sample with the OD value of the positive control (Result [%] = OD sample/OD positive control × 100).
Commercial ELISA and IFA.
The CCHFV-IgG-ELISA from Vector Best and the CCHFV-IgG-IFA from Euroimmun are commercially available kits for the detection of CCHFV-specific antibodies in human serum samples. The commercial ELISA from VectorBest uses an inactivated virus from an Uzbek strain (UZ10145) as antigen, whereas the commercial IFA from Euroimmun is based on nucleocapsid and glycoprotein precursor proteins (IbAr10200) transfected cells. The performance of both serological assays was evaluated with human sera from three different continents by Vanhomwegen and others; the detected specificity was excellent (100%), but both assays showed a lack of sensitivity (ELISA: 80.4%; IFA: 86.1%).13 The commercial assays were also previously adapted and validated for testing west African cattle sera (ELISA: 98% sensitivity and 100% specificity; IFA: 95% sensitivity and 98% specificity).18,33
Seroepidemiological investigation.
One thousand seventy-five samples from Mali were tested with the in-house CCHFV-IgG-ELISA according to a flowchart for seroepidemiological studies.34 Briefly, all samples showing positive or inconclusive results in the first in-house CCHFV-IgG-ELISA series were tested a second and, if necessary, a third time for confirmation; all samples that tested negative in the first CCHFV-IgG-ELISA series were eliminated from further analysis. All the sera were also tested once with the commercial CCHFV-IgG-ELISA. Finally, inconclusive ELISA results were clarified by IFA. Samples were considered to be positive if they showed positive results in both ELISAs or if they were positive in one ELISA and inconclusive in the other one.
Results and Discussion
Using the reference serum panels, a negative cutoff of 16% and a positive cutoff of 19% were defined for the newly modified in-house ELISA. A receiver operating characteristic (ROC)–based analysis was used to determine the cutoffs of the indirect in-house CCHFV-IgG-ELISA. The ROC analysis was performed in R targeting a maximal diagnostic specificity and sensitivity with a confidence level of 95%. A cutoff of 17% was determined. The diagnostic sensitivity and specificity were 99% and 97%, respectively. However, the known antibody positive and antibody negative samples showed a considerable overlap. The 17% cutoff did not classify these samples accurately. Therefore, we used two cutoffs as suggested in the Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2016.35 A negative cutoff (16%) and a positive cutoff (19%), framing the overlapping area, were set. The values between the cutoffs were classified inconclusive. These samples were retested in the indirect in-house CCHFV-IgG-ELISA. All in-house CCHFV-IgG-ELISA samples with a final inconclusive result could be classified by the commercial CCHFV-IgG-ELISA. Samples with a final result lower than 16% were considered to be “negative,” samples with a final result more than 19% were considered to be “positive,” and all samples in between were termed “inconclusive.” The diagnostic sensitivity and the diagnostic specificity were 99%.
ELISA platforms are considered the frontline assay for seroepidemiological studies on CCHFV prevalence in animals.36 Although we acknowledge the possibility of the detection of cross-reactive antibodies generated against a closely related virus, in the current study we used a tripartite diagnostic approach consisting of the commercial CCHFV-IgG-ELISA, the in-house CCHFV-IgG-ELISA, and the commercial IFA to determine the final result and minimize the possibility of false positives. After both ELISAs, 964 samples showed a definite result. IFA was necessary for determining the final result of the remaining 111 serum samples, which had divergent results in the ELISAs. This detection difference could be caused by the different antigens used in the ELISAs and may suggest different cross-reactive potentials of the Malian strain of CCHFV with the Kosovo and the Uzbek antigens.
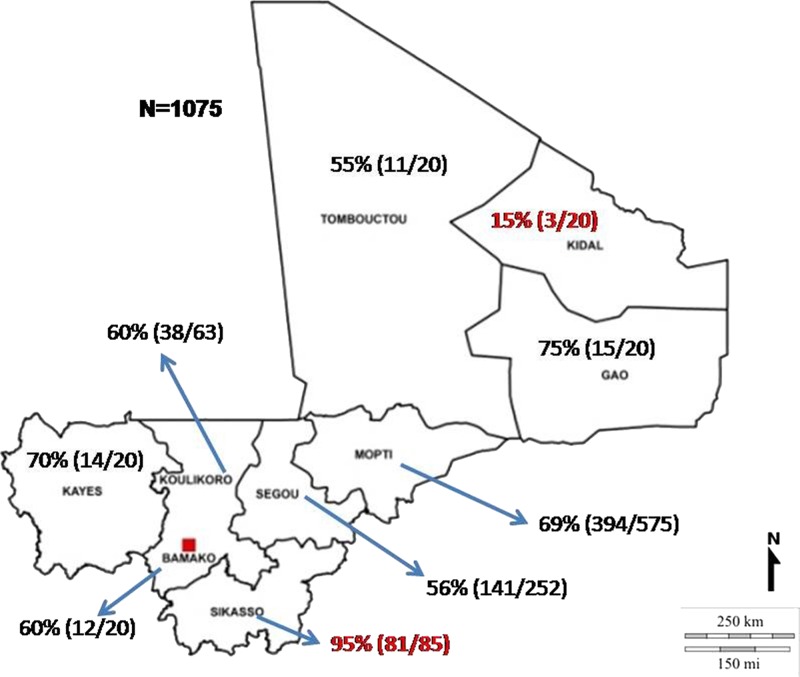
In the here presented seroepidemiological study, the overall prevalence of CCHFV-specific IgG antibodies in the cattle samples was 66% (706/1,075; 95% confidence interval [CI]: 63–69%) in Mali. The highest percentage was detected in the predominantly agricultural region of Sikasso with 95% (81/85; CI: 88–99%) and the lowest percentage was detected in northern Saharan region of Kidal with 15% (3/20; CI: 3–38%) (Figure 1 ). With the exception of Gao (75% prevalence; CI: 51–91%) with 2–4 bovines/km2, the seroprevalence was proportional to the cattle density. The highest prevalence rates in Sikasso (95% prevalence) and Mopti (69% prevalence; CI: 65–72%) correlated with greater than 9 bovines/km2; whereas the lowest seroprevalence rates in Kidal (15%; CI: 3–38%) and Tombouctou (40% prevalence; CI: 19–64%) correlated with a density of 1–4 bovines/km2.37 Kidal shows a very low prevalence even if the low cattle density is taken into account, though it is possible that this is a sampling artefact as just 20 sera were tested from this region.
Figure 1.
Geographical distribution of the Crimean–Congo hemorrhagic fever virus seroprevalence in Mali.
Our results suggest that the majority of cattle tested positive and consequently have been exposed to CCHFV in the past. The presence of CCHFV has been reported in a previous study in Mali in which Zivcec and others29 collected 80 unfed Hyalomma spp. ticks from 20 cattle at the Daral livestock market at Kati, Koulikoro, Mali. They performed reverse transcription-PCR using RNA from tick homogenates and found a viral sequence highly related (98% identity) to that of a strain from Mauritania. Livestock movement across long and porous borders between Mali and its neighboring countries is a real concern and challenge. High numbers of cattle cross borders to Côte d'Ivoire and Burkina Faso for pasture reasons; others are brought to Mauritania and Senegal for sale. The prevalence in Mauritania (67%) is very similar to the one detected in this study for Mali (66%).33 This indicates that the livestock trade plays an important role in the movement of CCHFV between these countries. Since this study is retrospective, acute infections cannot be investigated. Our study is the first to document serological evidence of the circulation of CCHFV in Mali. One implication is that the disease is spreading to new areas and countries as suggested by the recent human cases in Spain.38 It could also indicate that CCHFV was underreported so far and is actually circulating in the tick and mammal population for some time. More knowledge can be gained if studies focusing on CCHFV and its vectors are extended.
An estimated 80% of Mali's economy is attributed to agricultural practices, primarily raising herds of cattle. The high prevalence of CCHFV-specific antibodies documented here in bovines combined with the past finding of CCHFV-positive ticks collected directly from cattle at a live animal market suggests the risk of human infections with CCHFV in Mali is considerable.28,29 Recently documented CCHFV-specific IgG and IgM antibodies in suspected yellow fever cases from Mali support this statement.39 Although further studies are required to determine the prevalence of infection in humans, the results of this work strongly suggests public health measures aimed at reducing the risk of contracting CCHFV from livestock and the tick vector should be established in Mali.
ACKNOWLEDGMENTS
We thank Dick Sakai for logistical support. Also, we would like to acknowledge Pavlo Maksimov for statistical analysis of the CCHFV-IgG-ELISA.
Footnotes
Financial support: This work was supported by the Institute of Novel and Emerging Infectious Diseases, Friedrich-Loeffler-Institut, and the International Centers for Excellence in Research, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Authors' addresses: Ousmane Maiga, Entomology Unit, International Center of Excellence in Research, University of Science, Techniques and Technologies of Bamako, Bamako, Mali, E-mail: ousmanem@icermali.org. Miriam Andrada Sas, Marc Mertens, and Martin H. Groschup, Institute for Novel and Emerging Infectious Diseases, Friedrich-Loeffler-Institut, Isle of Riems–Greifswald, Germany, E-mails: miriam.sas@fli.bund.de, marc.mertens@fli.bund.de, and martin.groschup@fli.bund.de. Kyle Rosenke and Heinz Feldmann, Laboratory of Virology, National Institute of Allergy and Infectious Diseases, Hamitlon, MT, E-mails: rosenkek@niaid.nih.gov and feldmannh@niaid.nih.gov. Badian Kamissoko, Abdallah Traore, and Mamadou Niang, Central Veterinary Laboratory, Bamako, Mali, E-mails: kbadian2001@yahoo.fr, abdalltraor@yahoo.fr, and mniangm@yahoo.fr. Nafomon Sogoba and Hamidou M. Maiga, Faculty of Medicine and Dentistry, University of Sciences, Techniques and Technologies of Bamako, Bamako, Mali, E-mails: nafomon@icermali.org and maiga_m_hamidou@yahoo.fr. Modibo Sangare, Entomology Unit, International Center for Excellence in Research Mali, Bamako, Mali, E-mail: mouadib@gwmail.gwu.edu. Tom G. Schwan, National Institute of Allergy and Infectious Diseases, Hamilton, MT, E-mail: tschwan@niaid.nih.gov. Sekou F. Traore, Filariasis Research and Training Unit, International Center of Excellence in Research, Bamako, Mali, E-mail: cheick@icermali.org. David Safronetz, Division of Zoonotic diseases and Special Pathogens, National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, Manitoba, Canada, E-mail: david.safronetz@phac-aspc.gc.ca.
References
- 1.Flusin O, Vigne S, Peyrefitte CN, Bouloy M, Crance JM, Iseni F. Inhibition of Hazara nairovirus replication by small interfering RNAs and their combination with ribavirin. Virol J. 2011;8:249. doi: 10.1186/1743-422X-8-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honig JE, Osborne JC, Nichol ST. Crimean-Congo hemorrhagic fever virus genome L RNA segment and encoded protein. Virology. 2004;321:29–35. doi: 10.1016/j.virol.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 3.Ergonul O. Crimean-Congo haemorrhagic fever. Lancet Infect Dis. 2006;6:203–214. doi: 10.1016/S1473-3099(06)70435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornet JP, Kittayapong P, Gonzalez JP. Risk of arbovirus transmission by ticks in Thailand [in French] Med Trop (Mars) 2004;64:43–49. [PubMed] [Google Scholar]
- 5.Molinas A, Mirazimi A, Holm A, Loitto VM, Magnusson KE, Vikström E. Protective role of host aquaporin 6 against Hazara virus, a model for Crimean-Congo hemorrhagic fever virus infection. FEMS Microbiol Lett. 2016;363:fnw058. doi: 10.1093/femsle/fnw058. [DOI] [PubMed] [Google Scholar]
- 6.Ergonul O. Crimean-Congo hemorrhagic fever virus: new outbreaks, new discoveries. Curr Opin Virol. 2012;2:215–220. doi: 10.1016/j.coviro.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Whitehouse CA. Crimean-Congo hemorrhagic fever. Antiviral Res. 2004;64:145–160. doi: 10.1016/j.antiviral.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Atkinson B, Chamberlain J, Logue CH, Cook N, Bruce C, Dowall SD, Hewson R. Development of a real-time RT-PCR assay for the detection of Crimean-Congo hemorrhagic fever virus. Vector Borne Zoonotic Dis. 2012;12:786–793. doi: 10.1089/vbz.2011.0770. [DOI] [PubMed] [Google Scholar]
- 9.Wolfel R, Paweska JT, Petersen N, Grobbelaar AA, Leman PA, Hewson R, Georges-Courbot MC, Papa A, Heiser V, Panning M, Günther S, Drosten C. Low-density macroarray for rapid detection and identification of Crimean-Congo hemorrhagic fever virus. J Clin Microbiol. 2009;47:1025–1030. doi: 10.1128/JCM.01920-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mertens M, Schmidt K, Ozkul A, Groschup MH. The impact of Crimean-Congo hemorrhagic fever virus on public health. Antiviral Res. 2013;98:248–260. doi: 10.1016/j.antiviral.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Nabeth P, Cheikh DO, Lo B, Faye O, Vall IO, Niang M, Wague B, Diop D, Diallo M, Diallo B, Diop OM, Simon F. Crimean-Congo hemorrhagic fever, Mauritania. Emerg Infect Dis. 2004;10:2143–2149. doi: 10.3201/eid1012.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinikar S, Ghiasi SM, Hewson R, Moradi M, Haeri A. Crimean-Congo hemorrhagic fever in Iran and neighboring countries. J Clin Virol. 2010;47:110–114. doi: 10.1016/j.jcv.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Vanhomwegen J, Alves MJ, Zupanc TA, Bino S, Chinikar S, Karlberg H, Korukluoğlu G, Korva M, Mardani M, Mirazimi A, Mousavi M, Papa A, Saksida A, Sharifi-Mood B, Sidira P, Tsergouli K, Wölfel R, Zeller H, Dubois P. Diagnostic assays for Crimean-Congo hemorrhagic fever. Emerg Infect Dis. 2012;18:1958–1965. doi: 10.3201/eid1812.120710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahzounieh M, Dincer E, Faraji A, Akin H, Akkutay AZ, Ozkul A. Relationship between Crimean-Congo hemorrhagic fever virus strains circulating in Iran and Turkey: possibilities for transborder transmission. Vector Borne Zoonotic Dis. 2012;12:782–785. doi: 10.1089/vbz.2011.0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camicas JL, Cornet JP, Gonzalez JP, Wilson ML, Adam F, Zeller HG. Crimean-Congo hemorrhagic fever in Senegal. Latest data on the ecology of the CCHF virus [in French] Bull Soc Pathol Exot. 1994;87:11–16. [PubMed] [Google Scholar]
- 16.Tishkova FH, Belobrova EA, Valikhodzhaeva M, Atkinson B, Hewson R, Mullojonova M. Crimean-Congo hemorrhagic fever in Tajikistan. Vector Borne Zoonotic Dis. 2012;12:722–726. doi: 10.1089/vbz.2011.0769. [DOI] [PubMed] [Google Scholar]
- 17.Zeller HG, Cornet JP, Camicas JL. Experimental transmission of Crimean-Congo hemorrhagic fever virus by west African wild ground-feeding birds to Hyalommamarginatumrufipes ticks. Am J Trop Med Hyg. 1994;50:676–681. doi: 10.4269/ajtmh.1994.50.676. [DOI] [PubMed] [Google Scholar]
- 18.Mertens M, Vatansever Z, Mrenoshki S, Krstevski K, Stefanovska J, Djadjovski I, Cvetkovikj I, Farkas R, Schuster I, Donnet F, Comtet L, Tordo N, Ben Mechlia M, Balkema-Buschmann A, Mitrov D, Groschup MH. Circulation of Crimean-Congo hemorrhagic fever virus in the former Yugoslav Republic of Macedonia revealed by screening of cattle sera using a novel enzyme-linked immunosorbent assay. PLoS Negl Trop Dis. 2015;9:e0003519. doi: 10.1371/journal.pntd.0003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoogstraal H. The epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa. J Med Entomol. 1979;15:307–417. doi: 10.1093/jmedent/15.4.307. [DOI] [PubMed] [Google Scholar]
- 20.Camicas JL, Deubel V, Heme G, Robin Y. Ecological and nosological study of tick-borne arboviruses in Senegal. II. Experimental study of the pathogenicity of the Bhanja virus in small domestic ruminants [in French] Rev Elev Med Vet Pays Trop. 1981;34:257–261. [PubMed] [Google Scholar]
- 21.Appannanavar SB, Mishra B. An update on Crimean Congo hemorrhagic fever. J Glob Infect Dis. 2011;3:285–292. doi: 10.4103/0974-777X.83537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmeti S, Raka L. Crimean-Congo haemorrhagic fever in Kosova: a fatal case report. Virol J. 2006;3:85. doi: 10.1186/1743-422X-3-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas S, Thomson G, Dowall S, Bruce C, Cook N, Easterbrook L, O'Donoghue L, Summers S, Ajazaj L, Hewson R, Brooks T, Ahmeti S. Review of Crimean Congo hemorrhagic fever infection in Kosova in 2008 and 2009: prolonged viremias and virus detected in urine by PCR. Vector Borne Zoonotic Dis. 2012;12:800–804. doi: 10.1089/vbz.2011.0776. [DOI] [PubMed] [Google Scholar]
- 24.Tall A, Sall AA, Faye O, Diatta B, Sylla R, Faye J, Faye PC, Faye O, Ly AB, Sarr FD, Diab H, Diallo M. Two cases of Crimean-Congo haemorrhagic fever (CCHF) in two tourists in Senegal in 2004 [in French] Bull Soc Pathol Exot. 2009;102:159–161. [PubMed] [Google Scholar]
- 25.Dunster L, Dunster M, Ofula V, Beti D, Kazooba-Voskamp F, Burt F, Swanepoel R, DeCock KM. First documentation of human Crimean-Congo hemorrhagic fever, Kenya. Emerg Infect Dis. 2002;8:1005–1006. doi: 10.3201/eid0809.010510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceianu CS, Panculescu-Gatej RI, Coudrier D, Bouloy M. First serologic evidence for the circulation of Crimean-Congo hemorrhagic fever virus in Romania. Vector Borne Zoonotic Dis. 2012;12:718–721. doi: 10.1089/vbz.2011.0768. [DOI] [PubMed] [Google Scholar]
- 27.Rada AG. First outbreak of Crimean-Congo haemorrhagic fever in western Europe kills one man in Spain. BMJ. 2016;354:i4891. doi: 10.1136/bmj.i4891. [DOI] [PubMed] [Google Scholar]
- 28.Traoré AK, Doa S, Jouanelle JC, Bougoudogo F, Toure YT, Maiga KL. A propos des premieres observations serologiques de la fievrehemorragique de Crimee Congo au Mali. Mali Med. 2005;20:52–53. [PubMed] [Google Scholar]
- 29.Zivcec M, Maïga O, Kelly A, Feldmann F, Sogoba N, Schwan TG, Feldmann H, Safronetz D. Unique strain of Crimean-Congo hemorrhagicfever virus, Mali. Emerg Infect Dis. 2014;20:911–913. doi: 10.3201/eid2005.131641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton KC, Wasfy M, Samaha H, Abdel-Rahman B, Safwat S, Abdel Fadeel M, Mohareb E, Dueger E. Serosurvey for zoonotic viral and bacterial pathogens among slaughtered livestock in Egypt. Vector Borne Zoonotic Dis. 2014;14:633–639. doi: 10.1089/vbz.2013.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adam IA, Mahmoud MA, Aradaib IE. A seroepidemiological survey of Crimean Congo hemorrhagic fever among cattle in North Kordufan State, Sudan. Virol J. 2013;10:178. doi: 10.1186/1743-422X-10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohamed M, Said AR, Murad A, Graham R. A serological survey of Crimean-Congo haemorrhagic fever in animals in the Sharkia Governorate of Egypt. Vet Ital. 2008;44:513–517. [PubMed] [Google Scholar]
- 33.Sas MA, Mertens M, Isselmou E, Reimer N, Mamy BOEL, Doumbia B, Groschup MH. Crimean-Congo hemorrhagic fever virus specific antibody detection in cattle in Mauritania. Vector Borne Zoonotic Dis. 2017 doi: 10.1089/vbz.2016.2084. (In press) [DOI] [PubMed] [Google Scholar]
- 34.Mertens M, Wölfel R, Ullrich K, Yoshimatsu K, Blumhardt J, Römer I, Esser J, Schmidt-Chanasit J, Groschup MH, Dobler G, Essbauer SS, Ulrich RG. Seroepidemiological study in a Puumala virus outbreak area in South-East Germany. Med Microbiol Immunol (Berl) 2009;198:83–91. doi: 10.1007/s00430-009-0106-9. [DOI] [PubMed] [Google Scholar]
- 35.OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2016. Chapter 2.1.5 Crimean-Congo Hemorrhagic Fever. 2016. http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.01.05_CCHF.pdf/ Available at. Accessed March 1, 2017.
- 36.Spengler JR, Bergeron E, Rollin PE. Seroepidemiological studies of Crimean-Congo hemorrhagic fever virus in domestic and wild animals. PLoS Negl Trop Dis. 2016;10:e0004210. doi: 10.1371/journal.pntd.0004210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.FAO . Note Technique: Analyse des incitations par les prix pour les bovins au Mali 2005–2012. Rome, Italy: 2014. FAO. [Google Scholar]
- 38.European Centre for Disease Prevention and Control . Crimean–Congo Haemorrhagic Fever in Spain: 8 September 2016. Stockholm, Sweden: ECDC; 2016. [Google Scholar]
- 39.Safronetz D, Sacko M, Sogoba N, Rosenke K, Martellaro C, Traoré S, Cissé I, Maiga O, Boisen M, Nelson D, Oottamasathien D, Millett M, Garry RF, Branco LM, Doumbia S, Feldmann H, Traoré MS. Vectorborne infections, Mali. Emerg Infect Dis. 2016;22:340–342. doi: 10.3201/eid2202.150688. [DOI] [PMC free article] [PubMed] [Google Scholar]