Abstract
Despite a wealth of genomic information, a comprehensive alternative splicing (AS) analysis of pancreatic ductal adenocarcinoma (PDAC) has not been performed yet. In the present study, we assessed whole exome-based transcriptome and AS profiles of 43 pancreas tissues using Affymetrix exon array. The AS analysis of PDAC indicated on average two AS probe-sets (ranging from 1–28) in 1,354 significantly identified protein-coding genes, with skipped exon and alternative first exon being the most frequently utilised. In addition to overrepresented extracellular matrix (ECM)-receptor interaction and focal adhesion that were also seen in transcriptome differential expression (DE) analysis, Fc gamma receptor-mediated phagocytosis and axon guidance AS genes were also highly represented. Of note, the highest numbers of AS probe-sets were found in collagen genes, which encode the characteristically abundant stroma seen in PDAC. We also describe a set of 37 ‘hypersensitive’ genes which were frequently targeted by somatic mutations, copy number alterations, DE and AS, indicating their propensity for multidimensional regulation. We provide the most comprehensive overview of the AS landscape in PDAC with underlying changes in the spliceosomal machinery. We also collate a set of AS and DE genes encoding cell surface proteins, which present promising diagnostic and therapeutic targets in PDAC.
Introduction
Alternative splicing (AS) is one of the key regulatory events leading to transcriptome and proteome diversity. From bacteria and archaea, AS has, through increased prevalence, contributed to driving speciation and shaped the evolution of all multicellular organisms, including our own species1. Multiple distinct transcript variants are encoded by the majority, if not all human multi-exon genes, and more than half of all AS events show tissue specificity, thus highlighting a critical role of AS in creating phenotypic complexity2–4.
The functional consequences of most AS events are still unknown, with some resulting in non-functional products, some in the splice variant product(s) that assume completely different function from the wild-type protein, and in some cases the proteins even acquire antagonistic functions (for review see ref. 5). It is not surprising then that cancer has harnessed this important regulatory mechanism, which is implicated in all the ‘hallmark’ pathways of cancer6, and can hence be considered an additional cancer hallmark itself7. Some of the spliced variants in pre-malignant lesions have also been found in advanced cancers, suggesting their potential as drivers of cancer development and progression8.
Distinct patterns of cancer-specific splicing when compared to normal tissue counterparts have been reported for a variety of tumours9, 10, some of which could play a role as promising diagnostic, prognostic and/or therapeutic targets5. In pancreatic ductal adenocarcinoma (PDAC), in contrast to a wealth of other genomic data, except for a cell line study11, no AS data on a whole genome scale have yet been reported. A detailed understanding of the alternative splicing landscape in PDAC is therefore warranted; with this in mind, we have undertaken an in-depth investigation of AS events combined with whole transcriptome analysis, and explored the alterations in the splicing machinery itself in normal and pancreatic cancer specimens.
Results
Exon-based whole transcriptome profiling in PDAC
We used Affymetrix exon array to generate whole exome-based profiles for 28 PDAC (Table 1) and six normal pancreas bulk tissues. Although widely used as a normal comparator, we are aware that this can be a potential source of bias when comparing non-ductal acinar tissue versus ductal-differentiated carcinoma.
Table 1.
Demographic and clinical information of the analysed PDAC samples.
| PDAC case number | Gender | Age | Grade | pT | pN | pM | Analysis |
|---|---|---|---|---|---|---|---|
| 13 | F | 73 | G2 | T3 | N1 | M0 | EA |
| 14 | M | 64 | G2 | T3 | N1 | M0 | EA |
| 16 | F | 50 | G2 | T3 | N1 | M0 | EA/qPCR |
| 17 | F | 59 | G3 | T3 | N1 | M0 | EA/qPCR |
| 19 | M | 62 | G2 | T3 | N1 | M0 | EA/qPCR |
| 20* | M | 63 | G3 | T3 | N1 | M0 | EA |
| 21 | F | 77 | G2 | T3 | N1 | M0 | EA |
| 22 | M | 80 | G2 | T3 | N1 | M0 | EA/qPCR |
| 23 | M | 73 | G2 | T3 | N1 | M0 | EA/qPCR |
| 24 | F | 59 | G3 | T3 | N1 | M0 | EA/qPCR |
| 25 | F | 71 | G2 | T3 | N1 | M0 | EA |
| 27 | F | 42 | G2 | T2 | N1 | M0 | EA/qPCR |
| 32 | M | 67 | G2 | T3 | N1 | M0 | EA/qPCR |
| 35 | F | 68 | G2 | T4 | N0 | M0 | EA |
| 46 | M | 68 | G2 | T1 | N0 | M1 | EA/qPCR |
| 54 | F | 41 | G3 | T2 | N0 | M0 | EA/qPCR |
| 55 | M | 55 | G2 | T3 | N1 | M0 | EA |
| 60 | F | 65 | G3 | T3 | N1 | M0 | EA/qPCR |
| 63 | M | 67 | G3 | T4 | N1 | M0 | EA/qPCR |
| 64 | M | 59 | G3 | T3 | N1 | M0 | EA |
| 80 | M | 74 | G2 | T3 | N1 | M0 | EA |
| 100 | F | 73 | G2 | T3 | N0 | M0 | EA/qPCR |
| 102 | F | 73 | G2 | T3 | N1 | M0 | EA |
| 108 | M | 59 | G3 | T4 | N1 | M0 | EA |
| 150 | M | 53 | G3 | T3 | N1 | M0 | EA |
| 163 | M | 64 | G2 | T3 | N1 | M0 | EA |
| 169 | M | 66 | G2 | T4 | N1 | M0 | EA |
| 173 | M | 60 | G3 | T3 | N1 | M0 | EA |
| 89 | M | 81 | G2 | T4 | N1 | M0 | qPCR |
| 71 | F | 67 | G3 | T3 | N1 | M0 | qPCR |
| 73 | F | 74 | G2 | T3 | N1 | M0 | qPCR |
| 75 | F | 32 | G2 | T3 | N0 | M0 | qPCR |
| 95 | M | 68 | G3 | T3 | N1 | M0 | qPCR |
| 152 | M | 62 | G3 | T3 | N0 | M0 | qPCR |
| 166 | F | 65 | G2 | T3 | N1 | M0 | qPCR |
| 11 | F | 59 | G3 | T3 | N1 | M0 | qPCR |
| 15 | F | 67 | G3 | T3 | N1 | M0 | qPCR |
*This sample was derived from the patient that received neoadjuvant therapy (Gemcitabine + Oxaliplatin; all the remaining samples were obtained from naive patients. EA = exon array; qPCR = quantitative polymerase chain reaction; pTNM (pathology staging: Tumour, lymph Nodes, Metastasis).
Firstly, we analysed the expression profiles for 225,925 probe-sets across all exons and 17,528 transcript clusters after intensity filtering for all samples (see Methods). Unsupervised hierarchical clustering demonstrated a clear separation of the control and PDAC sample groups based on both probe-set and transcript level expression (Supplementary Fig. S1). In total, 1,887 transcript clusters were differentially expressed (DE) in PDAC compared to normal pancreas, of which 1071 (56.8%) were upregulated and 816 (43.2%) downregulated. Among these, 1,600 corresponded to protein-coding genes, with 917 (57.3%) upregulated and 683 (42.7%) downregulated transcript clusters in PDAC (Fig. 1A, Supplementary Table S1A). The KEGG pathway enrichment test using DAVID12 suggested that the identified overexpressed genes in PDAC were highly enriched for extracellular matrix (ECM)-receptor interaction (adj. p = 1.22e-05) and focal adhesion (adj. p = 2.37e-05), while for downregulated genes in PDAC, glycine, serine and threonine metabolism (adj. p = 9.40e-05) and maturity onset diabetes of the young (adj. p = 8.75e-05) were highly overrepresented (Fig. 1B, Supplementary Table S1B). The top deregulated canonical pathways based on Ingenuity pathways analysis (IPA) included small GTPases (CDC42, Rho), integrin and actin cytoskeleton signalling, again pointing to ECM-interaction, adhesion and cytoskeletal regulation. A full list of the pathways with the corresponding key genes is provided in Supplementary Table S1C. Analysis of the obtained differential expression profiles using the Pancreatic Expression Database13 revealed that out of 917 upregulated and 683 downregulated protein-coding genes in PDAC, 657 (71%) and 219 (32%) were previously reported displaying the same directions of changes, respectively (Supplementary Table S1D ). Among the newly discovered deregulated genes (701/1600; 44%), IPA revealed ephrin receptor and G-protein regulatory Gαi signalling as top deregulated canonical pathways (Supplementary Table S1E). Therefore, our exon-based expression analysis, with over four-fold increase in probe density in comparison to the ‘classical’ 3′-based arrays, provided a far more accurate and comprehensive portrait of pancreatic cancer transcriptome.
Figure 1.
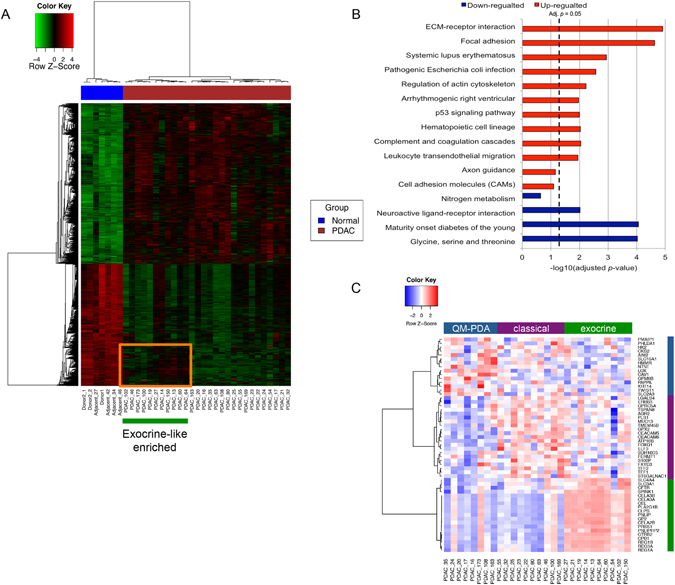
Differentially expressed protein-coding genes in PDAC. (A) Heatmap of the 1,887 differentially expressed transcript clusters in PDAC. The heatmap represents relative over- (red) and under-expressed (green) genes. The covariates at the top represent tumour (red) and normal (blue) samples. A subgroup of 11 PDAC samples is indicated by the orange box, determined by the exocrine-like gene signature by Collisson et al. with higher expression in a cluster of 255 transcripts than other PDAC samples. (B) KEGG pathway enrichment test for significantly up- and down-regulated genes in PDAC. The BH adjusted p-values were used (x-axis). (C) Heatmap and sample clustering based on the Collisson signature dividing samples into three molecular subtypes.
Interestingly, the transcriptomic profile of the 28 analysed PDAC samples also indicated the existence of two PDAC subgroups (Fig. 1A). This has prompted us to interpret our analysis in light of the previously reported PDAC subclassification by Collisson et al.14; the three PDAC groups: exocrine-like (n = 10), classical (n = 10) and QM-PDA (n = 8) are shown on Fig. 1C. The 11 closely grouped PDAC samples based on all 1,887 DE transcript clusters as indicated by the orange block in Fig. 1A were highly enriched for exocrine-like samples (n = 8, 73%). This group showed a higher level of expression of 255 transcripts compared to the rest of the PDAC samples, including pancreatic lipase-related protein PNLIPRP2 and chymotrypsin-like elastase family, member 2B (CELA2B) genes, classical acinar-originating genes (Supplementary Table S1F). KEGG pathway analysis further showed, in contrast to the other, ‘non-exocrine’ PDAC subgroup, that this cluster of genes was also highly enriched for glycine, serine and threonine metabolism, glutathione metabolism and selenoamino acid metabolism as well as Maturity onset diabetes of the young (Supplementary Table S1F), pointing to potential underlying metabolic and functional differences between the two subgroups. However, no difference in survival between exocrine-like enriched and ‘non-exocrine’ patients’ groups was seen (log rank P = 0.822). Furthermore, no significant difference in survival was present between any of the three Collisson subgroups shown in Fig. 1C (log rank P = 0.277).
After integration of our data with the data from Zhang et al. (n = 70 in the merged dataset)15 (Supplementary Fig. S2), no significant association of any of the three PDAC subgroups with survival was identified. This is in agreement with the study by Moffitt et al.16, where the sample grouping was determined by the histology (as similarly confirmed recently by Bailey et al.17) with the presence of the stable exocrine-like signature in PDAC reflecting the remnant exocrine-like compartment in the samples. Therefore, our results are not inconsistent with the cited studies16, 17, and have to be interpreted in light of the interrogated material, which in large part ( > 60%) comprised the malignant cell component.
Alternative splicing landscape in pancreatic cancer
After transcriptome analysis, we have undertaken a detailed AS analysis: in total, 2,816 differentially spliced probe-sets, representing 1,354 protein-coding genes, were identified (Supplementary Table S2A). This corresponded to on average 2.08 AS probe-sets per gene, ranging from 1–28. Of note, around half (1,424/2816; 50.5%) of these splicing events have not been reported previously in the Ensembl alternative splicing event set. The major AS event types found are listed in Table 2. Cassette exon (skipped exon, 14.3%) and alternative first exon (14.0%) were the most frequent, followed by intron retention (8.4%). Hierarchical clustering based on the FIRMA scores clearly separated the PDAC and control groups (Fig. 2A), suggesting a strong PDAC-specificity of the obtained splicing pattern. Interestingly, two subgroups of PDAC specimens were clearly evident. Since AS is shown to be associated with subtypes of breast cancer18, we explored this further, but subgrouping did not appear to correspond either to the one generated by DE signatures (Fig. 1A), nor any of the three Collisson sub-groups, and no difference in survival between the two groups was seen (log rank P = 0.433). We also did not find any association between tumour cellularity and tumour sample grouping based on DE or AS signatures. In addition, no association was observed between the tumour grade and these groups. However, it still remains to be explored if any other clinical correlates would emerge from these two AS subgroups in other studies.
Table 2.
Alternative splicing event types in PDAC.
| Splicing type | Number | Percentage (%) |
|---|---|---|
| Alternative 5′ sites (A5SS) | 47 | 1.53 |
| Alternative 3′ sites (A3SS) | 43 | 1.67 |
| Alternative first exon (AFE) | 394 | 13.99 |
| Alternative last exon (ALE) | 33 | 1.17 |
| Cassette exon (Skipped exon, CE) | 402 | 14.28 |
| Constitutive exon (CNE) | 208 | 7.39 |
| Intron retention (IR) | 237 | 8.42 |
| Intron isoform (II) | 20 | 0.71 |
| Mutually exclusive exons (MXE) | 9 | 0.32 |
CE: an exon may be spliced out of the primary transcript or retained; CNE: constitutively spliced exons. All events were based on the nomenclature described in Wang et al.4.
Figure 2.
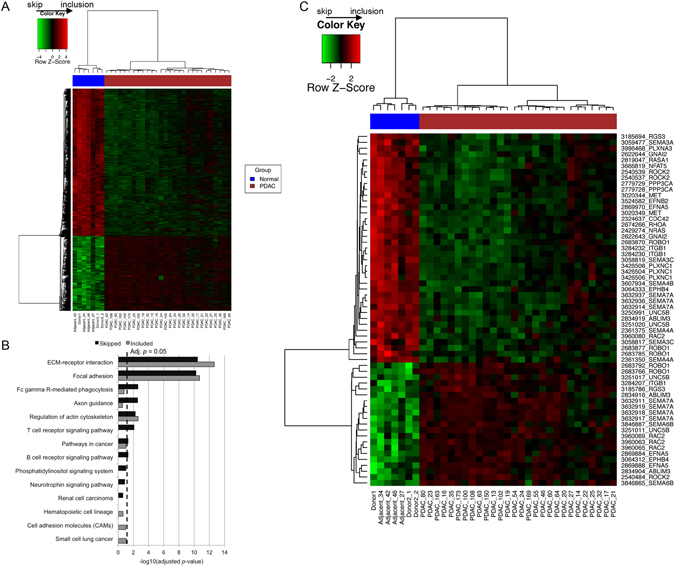
Alternative splicing in PDAC. (A) Heatmap of the 2,816 alternatively spliced probe-sets based on the FIRMA scores, with the relative inclusion shown in red and skipping shown in green. (B) KEGG pathway enrichment test for genes with included and skipped events. (C) Heatmap of 60 alternatively spliced probe-sets, representing 26 genes involved in axon guidance (KEGG). The row names are shown in the format of “probe-set” then followed by “gene symbol”.
Among differentially spliced probe-sets, 949 (33.7%) representing 555 genes were included in PDAC samples, while the remaining 66% (1,867 probe-sets/1,155 genes) were skipped in PDAC compared to normal samples (Supplementary Table S2A). Among 1,354 protein-coding genes with AS signatures, around half (638, 47%) had more than one alternatively spliced probe-set, while genes from the collagen family had the highest number of AS probe-sets, including 28 for COL5A1, 19 for COL11A1, 17 for COL5A2, 12 for COL16A1 and 11 for COL1A1 and COL17A1 (Supplementary Table S2B). For example, AS events of COL1A1 indicated by the 11 significant probe-sets in PDAC included skipped first exon (n = 1 probe-set) and highly included last exon/3′ UTR (n = 4), as well as cassette exons in the gene body (Supplementary Fig. S3A). These are significant findings, considering the abundance and increasingly recognised role of the stromal environment in PDAC.
In general, there was a weak positive correlation between the number of AS probe-sets and the number of total core probe-sets (Spearman’s rank correlation r = 0.26) and the number of exons (Spearman’s correlation r = 0.16), as noted previously for AS genes3, suggesting that selection pressure may play a role in limiting the splicing complexity in exceptionally large genes in PDAC as well.
The top ten most differentially spliced probe-sets (both skipped and included) are shown in Table 3.
Table 3.
Top differentially spliced protein-coding genes between PDAC and normal pancreas.
| Probe-set_ID | Transcript cluster_ID | Symbol | Definition | logFC (FIRMA) | Event | adj. P-Value |
|---|---|---|---|---|---|---|
| 3824875 | 3824874 | IFI30 | Interferon, gamma-inducible protein 30 | −11.93 | skipped | 6.42E-09 |
| 3861578 | 3861557 | LGALS4 | Lectin, galactoside-binding, soluble, 4 | −10.80 | skipped | 2.66E-08 |
| 2829950 | 2829947 | TGFBI | Transforming growth factor, beta-induced, 68kDa | −10.23 | skipped | 4.89E-09 |
| 2880347 | 2880292 | DPYSL3 | Dihydropyrimidinase-like 3 | −9.75 | skipped | 5.24E-14 |
| 3632030 | 3631964 | PKM2 | Pyruvate kinase, muscle | −9.71 | skipped | 9.00E-11 |
| 3119348 | 3119339 | LY6E | Lymphocyte antigen 6 complex, locus E | -9.57 | skipped | 4.96E-11 |
| 3405602 | 3405587 | GPRC5A | G protein-coupled receptor, family C, group 5, member A | −9.54 | skipped | 2.14E-15 |
| 2598263 | 2598261 | FN1 | Fibronectin 1 | −9.38 | skipped | 5.71E-08 |
| 3264344 | 3264326 | ACSL5 | Acyl-CoA synthetase long-chain family member 5 | −9.28 | skipped | 4.21E-15 |
| 3058819 | 3058759 | SEMA3C | Semaphorin-3C | −9.22 | skipped | 3.30E-11 |
| 3762200 | 3762198 | COL1A1 | Collagen, type I, alpha 1 | 13.64 | included | 3.86E-14 |
| 3013163 | 3013054 | COL1A2 | Collagen, type I, alpha 2 | 9.50 | included | 4.50E-11 |
| 3013162 | 3013054 | COL1A2 | Collagen, type I, alpha 2 | 8.44 | included | 7.09E-11 |
| 3762202 | 3762198 | COL1A1 | Collagen, type I, alpha 1 | 6.87 | included | 3.35E-13 |
| 3762203 | 3762198 | COL1A1 | Collagen, type I, alpha 1 | 6.35 | included | 4.02E-08 |
| 2535909 | 2535859 | CAPN10 | Calpain 10 | 6.30 | included | 7.07E-09 |
| 3351230 | 3351200 | TMPRSS4 | Transmembrane protease, serine 4 | 5.89 | included | 1.38E-09 |
| 3193625 | 3193482 | COL5A1 | Collagen, type V, alpha 1 | 5.81 | included | 9.30E-12 |
| 2405001 | 2404999 | MARCKSL1 | MARCKS-like 1 | 5.74 | included | 1.10E-11 |
| 2535912 | 2535859 | CAPN10 | Calpain 10 | 5.62 | included | 8.34E-07 |
To understand the putative roles of AS in PDAC, we performed detailed pathway analyses using both DAVID KEGG pathway and IPA enrichment tests. Interestingly, the KEGG pathway enrichment test suggested that AS genes were, similarly to DE genes, highly enriched for ECM-receptor interaction and focal adhesion (Fig. 2B; Supplementary Table S2C), highlighting again the importance of cancer cell-stroma interaction as a key biological feature of pancreatic cancer. In addition, Fc gamma R-mediated phagocytosis (adj. p = 2.49e-03) and axon guidance (adj. p = 2.81e-03) were also significantly overrepresented for genes with skipped probe-sets. Within the axon guidance pathway, the importance of which was recently flagged19, 26 genes (60 probe-sets) displayed significant AS signatures (Fig. 2C). While 10 of these genes (ABLIM3, EFNA5, EPHB4, ITGB1, RAC2, RGS3, ROBO1, ROCK2, SEMA7A and UNC5B) had both included and skipped probe-sets in PDAC, the majority (65%) of 60 AS probe-sets were skipped in PDAC (Fig. 2C; Supplementary Table S2D). For ITGB1, events of alternative (skipped) first exon, included and skipped exons and potential included last exon were identified. It is likely the short isoform was the preferred transcript in PDAC (Supplementary Fig. S3B ). The top AS signalling pathways in the Ingenuity pathway enrichment analysis also included integrin and axonal guidance signalling; a full list of the IPA pathways and key genes is provided in Supplementary Table S2E.
Among the 1,354 genes with AS signatures, 369 (27.3%) were genes coding for surface proteins (Supplementary Table S2F ); not surprisingly, these genes appeared to be highly enriched for cell adhesion (adj. p = 1.10e-22), cell surface receptor-linked signal transduction (adj. p = 1.60e-09), ECM-receptor interaction (adj. p = 3.7E-09), positive regulation of cell activation and proliferation (adj. p = 6.71e-07 and 9.42e-07, respectively), and the integrin-mediated signalling pathway (adj. p = 3.10e-06), comprising a number of integrins (ITGA11, ITGA3, ITGA4, ITGA5, ITGAV, ITGB1, ITGB2, ITGB4 and ITGB7 (Supplementary Table S2G).
Real-time PCR validation of alternative splicing
Real-time PCR was performed using 20 PDAC and six normal pancreas tissues for eight splicing events from randomly selected non-differentially expressed genes (fold change between 0.9–1.1): C1QTNF5, SLC17A9, NR1I2, C6orf106, ABI3, OTUD5, MDFIC and DCAKD (Fig. 3A ). As presented in Fig. 3B, the PCR results validated the exon array data with significantly higher levels for the included exons (C1QTNF5, SLC17A9, NR1I2, ABI3, OTUD5, MDFIC) and significantly lower levels for the two skipped exons (C6orf106 and DCAKD) in PDAC samples compared to normal pancreas. Thus, these data validated the accuracy of FIRMA prediction for exon skip/inclusion.
Figure 3.
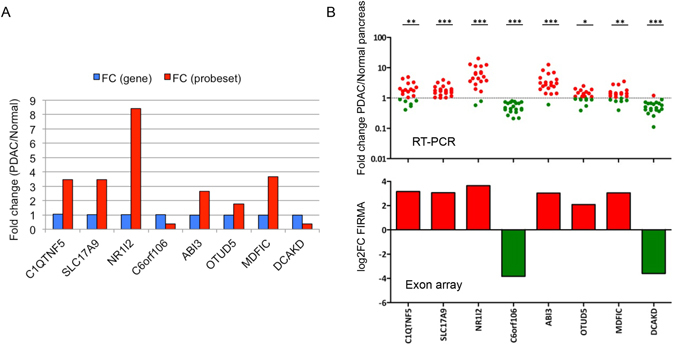
Real-time PCR validation of exon array data. (A) Selection of 8 splicing probe-sets from non-differentially expressed genes. For these 8 genes/probe-sets, there were no differences in the level of gene expression, but significant differences in the level of probe-set expression. (B) RT-PCR results of the 8 probe-sets. RT-PCR results are represented as scatter dot plots of expression fold change in PDAC compared to normal pancreas, with the significance level shown at the top (* < 0.05, ** < 0.01, *** < 0.001). Corresponding exon array results are reported in histograms of log2 FIRMA fold change (PDAC/normal) values. The two sets of results corresponded well with each other very well.
Identification of cell surface DE and AS regulated genes as candidate biomarkers and therapeutic targets
We next established which genes were commonly regulated by both differential expression (DE) and AS, by comparing the 1,354 AS to the 1,600 DE protein-coding genes (Fig. 4, Supplementary Table S3A). 360 genes showed both DE and AS signatures in PDAC, accounting for 22.5% and 26.6% of total DE and AS genes, respectively (Fig. 4A). Upon assessing the pathways that these 360 genes modulate, we found a significant enrichment for ECM-receptor interaction (adj. p = 9.78e-11) and focal adhesion (adj. p = 9.05e-06), while immunity-related pathways (particularly T cell receptor signalling (adj. p = 9.29e-04), Fc gamma receptor-mediated phagocytosis (adj. p = 1.12e-02), chemokine signalling, as well as neurotrophin signalling (which supports survival, development and function of neurons) (all with adj. p < 0.05) were predominantly affected by AS (Fig. 4B, Supplementary Table S3B). Actin cytoskeleton and axon guidance genes appeared to be regulated by both DE and AS, as well as unique AS events. This was also largely supported by IPA (Supplementary Table S3C).
Figure 4.
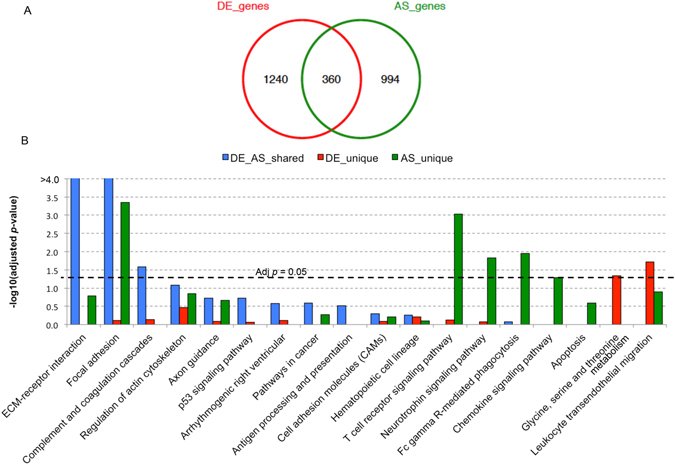
Comparison between DE and AS genes. (A) The overlapping patterns between differentially expressed (DE) genes and alternatively spliced (AS) genes. (B) The KEGG pathway enrichment for unique DE genes (red), unique AS genes (green) and genes with both DE and AS signatures (blue). The –log10 (adjusted p-values) is shown at the y-axis.
AS and DE events that affect cell surface proteins might represent an exciting opportunity for their use as biomarkers for detection and/or as therapeutic targets. A total of 135 cell surface protein-coding genes that are regulated at both gene expression and alternative splicing level are listed in Supplementary Table S3D.
The splicing pattern of two genes encoding the cell surface proteins, ROBO1 (an axon guidance gene) and LRP8 (Low Density Lipoprotein Receptor-Related Protein 8, which plays a critical role in the migration of neurons during development), were assessed by real-time PCR (Fig. 5). ROBO1 and LRP8 displayed multiple splicing events according to FIRMA prediction (Supplementary Fig. S4) and were upregulated in PDAC compared to normal tissues, with a fold change of 2.03 for ROBO1 and 1.68 for LRP8. Real-time PCR validated the exon array results for the selected splicing events in both ROBO1 (Fig. 5A) and LRP8 (Fig. 5B). Of note, while a lower expression of probe-sets 2683871 and 2413242 was seen by RT-PCR, the values did not reach statistical significance. For ROBO1, a higher skipping rate for probe-sets/exons at the 5′ end (represented by probe-set 2683870 and 2683871), but a higher inclusion rate for probe-sets/exons at the 3′ end (e.g., 2683766) and for some cassette exons in the middle (e.g., 2683792) were seen in PDAC compared to normal (Fig. 5A). Similarly for LRP8, probe-sets/exons at the 5′ end (e.g., probe-set 2413284) and cassette exons close to the 5′ side (represented by 2413242) appeared to be skipped, but exons in the middle (e.g., 2413224 and 2413229) and those close to the 3′ end tended to be included in PDAC compared to normal (Fig. 5B ). Thus, it is likely that different promoters were utilised and shorter transcripts become the primary choice in PDAC.
Figure 5.
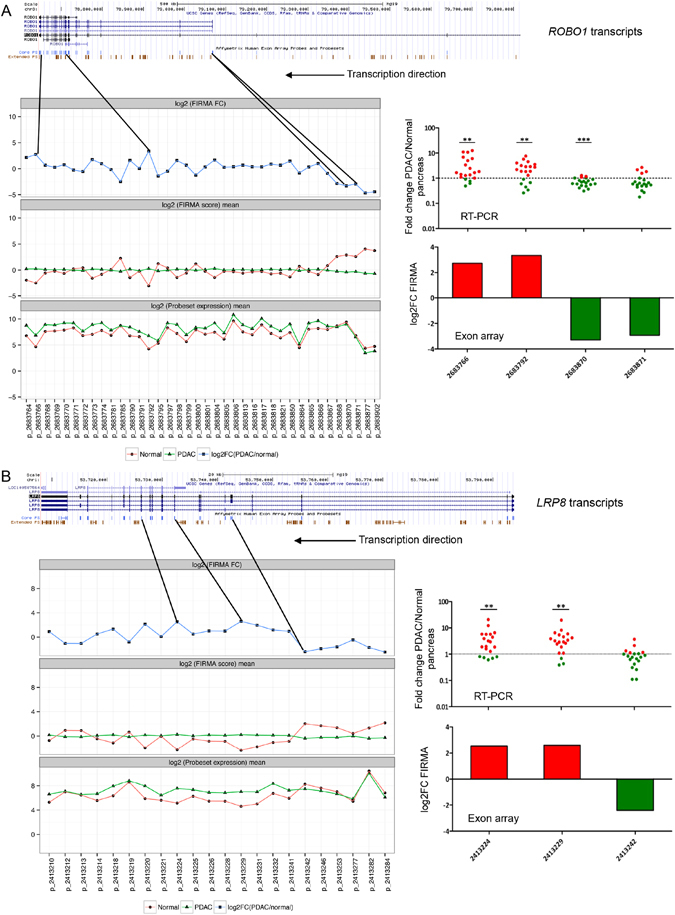
Analysis of multiple splicing events for ROBO1 and LRP8, and validation with RT-PCR. (A) Alternative splicing for ROBO1. (B) Alternative splicing for LRP8. The UCSC transcripts are shown at the top. For all quantified core probe-sets, means for probe-set expression and FIRMA values (log2 scaled), and the log2 FIRMA fold changes (PDAC/normal) were shown underneath UCSC transcripts. The probe-sets chosen for RT-PCR validation were shown with the black solid lines, with their positions within the transcripts indicated. RT-PCR results for the chosen probe-sets are represented as scatter dot plots of expression fold change in PDAC compared to normal pancreas, with the significance level shown at the top (* < 0.05, ** < 0.01, *** < 0.001). Corresponding exon array results are also reported in histograms of log2 FIRMA fold change (PDAC/normal) values. Both ROBO1 and LRP8 are coding for cell-surface proteins, and both were transcribed in the reverse strand.
In addition to surface proteins affected by DE and AS being candidate drug targets, the discovery of PDAC-associated splice variants might also represent a novel diagnostic opportunity. Using mass spectrometry, Menon et al. have detected a number of protein products of gene splice variants in the plasma of a KrasG12D/Ink4a/Arf mouse model of PDAC20. When these splice isoforms were compared to our AS signatures, 32 common AS genes were found; interestingly, these genes were highly enriched for focal adhesion (adj. p = 1.24e-03) and ECM-receptor interaction (adj. p = 2.49e-03), including TNC, several collagen genes, ZYX, MYLK and FN1. These highly conserved AS variant proteins represent an intriguing lead for further validation of their diagnostic potential.
Hypersensitive genes
We next tested the behaviour of putative driver genes recently reported by Bailey et al.17 regarding differential expression and alternative splicing events. Fifty-six mutated genes were reported using the IntoGen software (Q < 0.1, Supplementary Table S4A from ref. 17); of these, four genes showed significant DE signatures between PDAC and normal in our dataset (based on the double threshold of adjusted p-value < 1e-04 and absolute FC > 2), including three upregulated: KRAS, TGFBR1 and CALD1, and one downregulated (FBLN2) gene in PDAC. An additional eight DE genes could be identified (using just adjusted p-value < 1e-04), including four upregulated (MACF1, TNIK, TP53BP2 and PBRM1), and four downregulated genes: ACVR1B, RBM10, BCORL1 and TTC18 in PDAC compared to normal (Supplementary Table S4B).
When the AS patterns were investigated for these 56 genes, seven genes displayed AS signatures. Among them, TGFBR2, KDM6A, CALD1 and MYCBP2 all had highly skipped probe-sets, while TGFBR1, MACF1 and ITPR3 possessed both highly skipped and included probe-sets in PDAC compared to normal (Supplementary Table S4C ). Thus, a set of three significantly mutated genes that were significantly mutated and also showed both DE and AS was derived, including TGFBR1, CALD1 and MACF1 (Supplementary Fig. S5). Interestingly, both alternative first and last exons were significant events for TGFBR1, implying the differential selection of regulatory elements in PDAC development for this gene (Supplementary Fig. S5C ).
We further expanded our analysis to genes that were significantly targeted by deletions and amplifications, revealed by the copy number GISTIC analysis17. A total of 17 out of 21 genes (81.0%) in significantly deleted regions that had strong DE signatures were downregulated in PDAC based on our double threshold, demonstrating good concordance between gene expression and copy number changes, including ARHGDIG, RPL3L, SNORA64, RAB26, GOLGA8A/B and MAPK8IP2 (Supplementary Table S4D ). The 121 genes within the significantly amplified regions in PDAC were also significantly differentially expressed, with 74 (61.2%) upregulated genes, including ITGA2, VCAN, THBS2, IL7R, LOX and EDIL3 (Supplementary Table S4E ).
We then inspected the AS patterns of genes within deleted and amplified regions, and found that 12 deleted and 95 amplified genes also displayed strong AS (Supplementary Table S4F and G ). Of these, 34 genes additionally showed DE signatures, consisting of ABCA3, a deleted and downregulated gene in PDAC, and 33 amplified and upregulated genes (including ADAM19, CAST, CD180, CSTB, DOCK2, F2R, MX1, PCDH7, RUNX1, TGFBI and VCAN) are listed in Supplementary Table S4H.
The inclusion of the three significantly mutated genes (TGFBR1, CALD1 and MACF1) to the latter 34 genes resulted in 37 genes that we refer to as a set of ‘hypersensitive’ genes, as they are frequently targeted by somatic mutations, copy number aberrations, differential expression and alternative splicing, indicating their increased intrinsic susceptibility to multiple genomic aberrations in PDAC.
Spliceosomal and splicing regulator genes in PDAC
Finally, in order to understand the behaviour of the spliceosomal machinery in PDAC, we have assessed the DE and AS of genes involved in splicing. Among the 140 spliceosomal genes reported by Zhou et al.21, the expression data for 119 (85%) were investigated in our study (Supplementary Table S5A). Of these, 28 (23.5%) genes were significantly differentially expressed (using an adjusted p-value threshold of 1e-04) in PDAC, with 22 upregulated genes, the top two being PRPF40A and SNRNP27 (Supplementary Table S5A). Interestingly, unsupervised hierarchical clustering of the 119 spliceosomal genes showed clear separation between normal and PDAC samples as illustrated in Fig. 6A. To detect a potential influence of age on PDAC clustering, we investigated the association between PDAC sample clustering of spliceosomal genes and patient age, but found no significant difference in age between the two groups of patients (Wilcoxon test, p = 0.90). This was also the case when we explored the association between age and patients’ clustering based on significant AS probe-sets (Fig. 2A), Wilcoxon test, p = 0.20.
Figure 6.
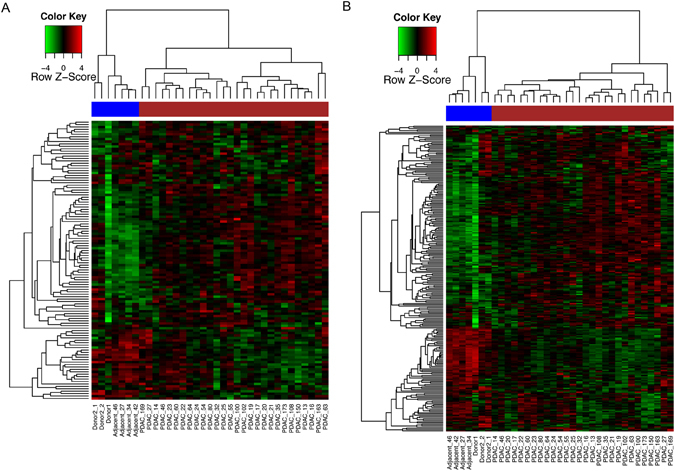
Heatmap showing the expression profiles of 119 (A) and 238 (B) spliceosomal genes, with the upregulated genes shown in red and downregulated in green. PDAC samples were indicated with the brown bar, while normal samples were indicated with the blue bar.
Previously, Carrigan et al. analysed AS in PDAC cell lines and demonstrated that 28 of 92 (~30%) spliceosomal genes had significantly decreased expression in pancreatic cancer compared with normal pancreas11; in our study, expression profiles for 18 of these 28 genes were identified and only three of them (SART1, CIRBP and HTATSF1) were significantly downregulated in PDAC (adjusted p < 0.05), while nine (DNAJC8, DHX9, DHX8, SLU7, RSRC2, MBNL2, NCBP2, PRPF4B and MBNL3) were significantly upregulated (adjusted p < 0.05) (Supplementary Fig. S6 ). This difference is likely due to the different analytical platforms and sample types utilised in the two studies. When the selection of spliceosomal genes was expanded to all 238 splicing regulators based on the Ensembl annotation (Supplementary Table S5B), 73/238 (30.7%) of these genes were found to be differentially expressed (adjusted p-value < 1e-04), with 39 upregulated and 34 downregualted genes in PDAC relative to normal. The top upregulated genes (FC > 2) included PRPF40A, SNRNP27, LGALS3, and ADAR, and the top downregulated genes (FC < 0.5) included APOBEC2, CELF3, ADARB2 and CSDC2 (Supplementary Table S5B). Unsupervised clustering based on all 238 spliceosomal genes was again able to differentiate PDAC from the healthy pancreas (Fig. 6B ).
Interestingly, 12 spliceosomal genes: AQR, C19orf43, DHX16, PPM1G, PUF60, SNRPD3, SRSF1, SRSF4, TCERG1, U2AF2, USP39 and WTAP also showed the evidence of AS - all had skipped exon signatures in PDAC (Supplementary Table S2). Thus, genes in spliceosomal machinery are themselves regulated by both DE and to a lesser extent by AS. This could potentially explain and be one of the reasons for a compromised splicing fidelity in cancer. Of note, we did not find any association between the aberrations of spliceosomal machinery and patients’ age.
Discussion
Here, we provide the first comprehensive tissue-based AS landscape of PDAC, with the report of a number of newly identified AS events. We show that the most common alterations in the protein-coding AS genes in PDAC are skipped exon and alternative 1st exon, followed by intron retention. Exon skipping was previously highlighted as being the most common AS form involved in shaping eukaryotic evolution22, and we show here that the same mechanism is also utilised in PDAC development. In contrast, intron retention, a rare event in normal eukaryotic tissues (most commonly seen in plants, fungi and protozoa)1 seemed to be also hijacked in PDAC evolution. This inclusion of intronic sequences within the mRNA (sometimes referred to as the phenomenon of exonization) has been reported in cancer23, suggesting that aberrant AS is not merely a side effect of cancer, but a bona fide regulator of cancer development22.
In addition to our validation, a number of genes affected by AS described here have already been reported, providing an independent validation of our data. A total of 121 genes (8.9%) found to have AS signatures in our PDAC cohort were also found to have skipped exons across different tissues4 (Supplementary Table S6). The AS of CALD1, COL6A3, FN1, MAST2, LGR5 and ITGB4 were previously validated in colon cancer using RT-PCR24, while CLSTN1, AUP1, CTNND1, CALD1 and COL6A3 were shown to exhibit tumour-specific splice variants in colon, bladder and prostate cancers8. ADAM12, DKK3, GSN, TNFSF11, CDH3, CXCL5, HCK, ITGA5 and VEGFC have also been shown to possess known or novel splice variants in lung cancer25. The BIRC5 (survivin) gene and its spliced isoforms have been shown to be associated with prostate cancer cell proliferation and aggressive phenotypes26; and tumour-associated splice variants of MACF1, ITGB3, TLE3, SHC1, ETS1 and BCAS1 have been reported in lung, prostate, breast cancer and glioblastoma27–32.
In PDAC, AS in PSMD2, PTPN18, SUPT16H, CUL4A, NIN, SLK, and ABCC3 were also reported previously11, and increased AS of the KLF6 tumour suppressor gene was shown to correlate with prognosis and tumour grade33. Interestingly, in our data, four other members of the Krüppel-like family, KLF5, KLF7, KLF12 and KLF16, also showed significant AS events (Supplementary Table S2A). The AS in human tissue factor (TF or F3) was shown to promote tumour growth in an orthotopic pancreatic cancer model34, 35, and our data also show a highly skipped event for this gene (within the first exon) in PDAC (probe-set: 2423935; FIRMA log2FC = −4.69, adj. p = 1.99e-08).
Overall, collagen genes showed the highest versatility in AS in PDAC; interestingly, a switch in alternate promoters in collagen IX during fracture healing36, and a dynamic process of tumour-specific AS in several exons of COL6A3 were previously reported37. While in evolutionary terms increased splicing presents novel opportunities for expansion of gene families, its functional implications are far-reaching, particularly in PDAC, where the abundant stroma, predominantly composed of collagens, is a characteristic and pronounced feature. It is highly plausible that alternatively spliced collagen genes play a critical role in intense stromal remodelling with important repercussions to mechanical and stiffness properties, shown recently to be implicated in progression and invasive properties of PDAC38.
Of note, while our samples were pre-selected to predominantly comprise tumour cells, it is possible that a large number of AS signatures seen here in collagen and other ECM genes are derived from the desmoplastic stromal compartment still present in our samples. Therefore, future studies that involve the microdissection and AS profiling of both tumour and stromal cells will further enrich our understanding of this phenomenon, and inform on tumour and stroma specific AS events in PDAC. Furthermore, to compare the AS/DE profile of chronic pancreatitis to that of normal and/or tumour tissues would also be an important further step in understanding the degree to which differences described here are tumour-specific.
Around a third of the significant genes in our study were shown to be affected by both DE and AS (Fig. 4), and they were particularly enriched for ECM-receptor interaction and focal adhesion pathways as well as complement and coagulation cascades. Of these genes, 135 encode cell surface proteins. Although it still remains to be fully established if these events are PDAC-specific, such surface proteins might represent an as yet untapped source of both potential imaging tools or drugable targets. Furthermore, the discovery of PDAC-associated splice variants in body fluids might present a novel diagnostic opportunity, as at least 32 AS gene products were found as circulating proteins in mouse plasma20. These represent a rich pool of non-invasive diagnostic candidates that now need to be explored and validated directly in human biofluids.
Interestingly, we describe here a group of hypersensitive genes, which showed predilection to be affected by mutations, copy number aberrations, DE and AS signatures. While the detailed mechanistic annotation for such a multidimensional regulation of this set of genes remains to be further established, their functionality in the ECM-receptor interactions, focal adhesion, collagen fibril organisation and actin cytoskeleton provides a versatility of options in communication networks between cancer cells and their ever-changing microenvironment.
Finally, we also looked at the spliceosomal machinery itself. A deregulation of around a third of genes belonging to this dynamic ribonucleoprotein complex was found, with upregulation being more prevalent than downregulation; this has also been established for the majority of other cancers. Based on the percentage of upregulated vs. downregulated splicing factor genes, PDAC appeared to be more similar to renal cell carcinoma and lung adenocarcinoma than to prostate, colorectal and breast cancers5. Of note, the spliceosomal signature itself was sufficient to clearly separate the normal and cancer specimens, suggesting its cancer-specificity; curiously, we also show that several spliceosomal genes can undergo alternative splicing themselves.
It was previously shown that spliced isoforms follow the principle of parsimony and adopt the simplest structural folds, with the most pronounced changes seen in the exposed surface of the affected proteins39. Integration of our data with the RNA-Seq data and development and refinement of the computational prediction methods to inform on the resultant protein sequence and corresponding 3D structural models on a global scale is now needed to gain full benefit from the available AS data.
In summary, we provide the most comprehensive landscape of AS events in PDAC to date, with underlying changes in the spliceosome and its regulators. We also report a group of alternatively spliced genes that encode surface and circulating proteins. These represent novel candidates of translational relevance as potential diagnostic and therapeutic targets in pancreatic adenocarcinoma.
Materials and Methods
Samples and RNA isolation
A total of 43 freshly-frozen pancreatic samples (37 PDAC, four histologically normal samples adjacent to cancer and two normal donor tissue samples) were analysed in this study. Samples were obtained from the Department of Pathology, Verona, Italy, after informed consent with full ethical approval from the Institutional Review Boards of The University of Verona. The experiment and methods were conducted in accordance with the Declaration of Helsinki. The demographic and clinical information of the patients and samples are summarised in Table 1. Of note, the samples have been selected based on tumour cellularity, so over 80% of PDAC specimens had >60% cancer cell content.
Total RNA was isolated using TRIzol (Invitrogen); 1.5 μg of total RNA was further processed (depleted of ribosomal RNA and labelled) according to supplied protocols (Affymetrix, Santa Clara, CA, USA).
Affymetrix Exon array expression analysis
Affymetrix GeneChip® Human Exon 1.0 ST Arrays, comprising ~1.4 million probe-sets consisting of over 5 million individual probes and >300,000 transcript clusters (group of probe-sets targeting individual exons in genes and noncoding transcripts) were used for gene-level expression profiling and AS analysis. Labelling using Affymetrix GeneChip Whole Transcript (WT) ST Labeling Assay and subsequent hybridization were performed according to the manufacturer’s instructions. After scanning, CEL files were checked for quality and analysed following the pipeline described in Rodrigo-Domingo et al.40. Exon array data files have been submitted to Gene Expression Omnibus (GEO) under the accession number of GSE63158, a SuperSeries consisting of the gene-level data GSE56560 and the exon-level data GSE63111.
Within the pipeline, the “aroma. affymetrix” R package41 was used for data preprocessing, normalisation and summarisation to produce transcript, probe-set and probe-level intensities. Here, only the “core” probe-sets, supported by the most reliable evidence from RefSeq and full-length mRNA GenBank records containing complete coding DNA sequence (CDS) information and transcript clusters were used. This was followed by intensity filtering across samples as recommended by the Affymetrix White Paper10. Data obtained from one replicate of the two donor samples were of low quality and were subsequently removed. Using the limma R package42, transcript cluster expression intensities were further analysed to identify differentially expressed (DE) transcripts between PDAC and control groups. The raw p-values were adjusted using the Benjamini-Hochberg (BH) model43. Differentially expressed transcript clusters were identified using a double threshold of adjusted p-value < 1e-04 and absolute fold change (FC) > 2. The transcript clusters were further matched to Ensembl genes and gene symbols.
Alternative splicing (AS) analysis
Differential alternative splicing events were detected using FIRMA (Finding Isoforms using Robust Multichip Analysis)44. FIRMA scores for all filtered probe-sets were calculated using “aroma. affymetrix”. After log2 transformation, the differential splicing analysis was conducted using limma. Alternatively spliced probe-sets were identified using a double threshold of adjusted p-value < 1e-06 and absolute log2 FC > 2. The ANalysis Of Splice VAriation (ANOSVA) method45 was also used as an additional filter with an adjusted p-value < 1e-06 at both the probe and probe-set levels. Only protein-coding genes were finally selected according to the Ensembl gene annotation. Identified AS events in PDAC were categorised into common patterns of alternatively spliced exonic regions according to the Ensembl alternative splicing event set4, 46, i.e. alternative 5′ and 3′ sites, alternative first and last exon, skipped and consecutive exon, intron retention and isoform and mutually exclusive exons.
Gene set enrichment tests
The gene set enrichment tests were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID)12 to inspect overrepresented Gene Ontology (GO) Biological Process terms and KEGG pathways. The p-values were adjusted using the BH model. The functional and pathway analyses were also conducted using Ingenuity Pathway Analysis (IPA, Ingenuity® Systems, www.ingenuity.com). To identify surface protein coding genes, the GO Cellular Component terms for genes were explored using DAVID to determine those coding for plasma membrane proteins.
Real-time PCR validation
First strand cDNA was prepared from 1 µg of total human pancreatic RNA using Quantitect Reverse Transcription kit (Qiagen, Crawley, UK). Real-time PCR was performed on a 7500 Real Time PCR System (Applied Biosystems, Warrington, UK) using SYBR Green dye (Qiagen) according to the manufacturer’s instruction. Specific primers were designed and evaluated for amplification efficiency with the use of Universal Human Reference RNA (Agilent technologies, Stockport, UK) (Supplementary Table S7). To confirm the exon array data, relative changes of expression were shown for each target after normalization to the reference genes HPRT1, RPLP0 and S16, according to the formula: 2ΔΔCt47. In addition to samples used for Affymetrix experiments, eight more PDAC and four more pancreatic normal tissues were used; in total, the validation was performed on 20 PDAC and six normal samples.
Molecular subtype analysis
In addition to our exon array dataset, an analogous dataset from Zhang et al.15, was also used for clinical inferences. The two data sets (gene expression and clinical follow-up) were compiled, processed and merged as previously reported48. To identify PDAC molecular subtypes using the Collisson gene signature14, non-negative matrix factorisation (NMF) consensus clustering49 was employed to identify stable sample clusters based on normalised or z-score standardised expression values for each dataset. The R package ‘ConsensusClusterPlus”50 was also used to verify sample clustering. After sample clustering and grouping, Kaplan-Meier (KM) analysis and Log Rank test as well as Cox proportional hazards model were undertaken for survival analyses examining the impacts of grouping on overall survival using the R ‘survival’ package (https://cran.r-project.org/web/packages/survival/).
Electronic supplementary material
Acknowledgements
The study was funded by the MolDiagPaCa European Union Framework Programme and CR-UK Programme grant A12008 from CR-UK (C. Chelala, T. Crnogorac-Jurcevic, and N.R. Lemoine). Italian Cancer Genome Project – Ministry of University [FIRB RBAP10AHJB]; Associazione Italiana Ricerca Cancro [grant number: 12182]; FP7 European Community Grant Cam-Pac [no: 602783]; Italian Ministry of Health [FIMP- CUP_J33G13000210001]. The funders were not involved in the design of the study, collection, analysis, and interpretation of data and in writing of the manuscript. We thank Tracy Chaplin-Perkins for help with running the Affymetrix experiments.
Author Contributions
Study concept and design: C.C., A.S., N.R.L., T.C.J.; Acquisition of data: P.U., L.D., N.A.A., T.P.R.; Analysis and interpretation of data: J.W., L.D., A.S., A.M., C.C., T.C.J.; Drafting of the manuscript: J.W., L.D., T.C.J.; Critical revision of the manuscript for important intellectual content: J.W., C.C., T.C.J.; Material and clinical data support: R.T.L., R.S., A.S.; study supervision: C.C., T.C.J. All authors have reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Claude Chelala and Tatjana Crnogorac-Jurcevic jointly supervised this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-03354-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jun Wang, Email: j.a.wang@qmul.ac.uk.
Tatjana Crnogorac-Jurcevic, Email: t.c.jurcevic@qmul.ac.uk.
References
- 1.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 2.Skotheim RI, Nees M. Alternative splicing in cancer: noise, functional, or systematic? Int J Biochem Cell Biol. 2007;39:1432–1449. doi: 10.1016/j.biocel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 4.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2016;35:2413–2427. doi: 10.1038/onc.2015.318. [DOI] [PubMed] [Google Scholar]
- 6.Pagliarini V, Naro C, Sette C. Splicing Regulation: A Molecular Device to Enhance Cancer Cell Adaptation. Biomed Res Int. 2015;2015:543067. doi: 10.1155/2015/543067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ladomery M. Aberrant alternative splicing is another hallmark of cancer. Int J Cell Biol. 2013;2013:463786. doi: 10.1155/2013/463786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorsen K, et al. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Molecular & cellular proteomics: MCP. 2008;7:1214–1224. doi: 10.1074/mcp.M700590-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, et al. Computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. Cancer Res. 2003;63:655–657. [PubMed] [Google Scholar]
- 10.Affymetrix. Alternative transcript analysis methods for exon arrays http://www.affymetrix.com/support/technical/whitepapers/exon_alt_transcript_analysis_whitepaper.pdf. The White Paper (2005).
- 11.Carrigan PE, Bingham JL, Srinvasan S, Brentnall TA, Miller LJ. Characterization of alternative spliceoforms and the RNA splicing machinery in pancreatic cancer. Pancreas. 2011;40:281–288. doi: 10.1097/MPA.0b013e31820128d2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 13.Dayem Ullah, A. Z. et al. The pancreatic expression database: recent extensions and updates. Nucleic Acids Res42 (2014). [DOI] [PMC free article] [PubMed]
- 14.Collisson EA, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nature medicine. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One. 2012;7:e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moffitt RA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 18.Lapuk A, et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res. 2010;8:961–974. doi: 10.1158/1541-7786.MCR-09-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menon R, et al. Identification of novel alternative splice isoforms of circulating proteins in a mouse model of human pancreatic cancer. Cancer Res. 2009;69:300–309. doi: 10.1158/0008-5472.CAN-08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 22.Kim E, Goren A, Ast G. Insights into the connection between cancer and alternative splicing. Trends Genet. 2008;24:7–10. doi: 10.1016/j.tig.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Dvinge H, Bradley RK. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7:45. doi: 10.1186/s13073-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardina PJ, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi L, et al. Whole genome exon arrays identify differential expression of alternatively spliced, cancer-related genes in lung cancer. Nucleic acids research. 2008;36:6535–6547. doi: 10.1093/nar/gkn697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koike H, Sekine Y, Kamiya M, Nakazato H, Suzuki K. Gene expression of survivin and its spliced isoforms associated with proliferation and aggressive phenotypes of prostate cancer. Urology. 2008;72:1229–1233. doi: 10.1016/j.urology.2007.12.064. [DOI] [PubMed] [Google Scholar]
- 27.Jin R, Trikha M, Cai Y, Grignon D, Honn KV. A naturally occurring truncated beta3 integrin in tumor cells: native anti-integrin involved in tumor cell motility. Cancer biology & therapy. 2007;6:1559–1568. doi: 10.4161/cbt.6.10.4710. [DOI] [PubMed] [Google Scholar]
- 28.Laitem C, et al. Ets-1 p27: a novel Ets-1 isoform with dominant-negative effects on the transcriptional properties and the subcellular localization of Ets-1 p51. Oncogene. 2009;28:2087–2099. doi: 10.1038/onc.2009.72. [DOI] [PubMed] [Google Scholar]
- 29.Misquitta-Ali CM, et al. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Molecular and cellular biology. 2011;31:138–150. doi: 10.1128/MCB.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakaya HI, et al. Splice variants of TLE family genes and up-regulation of a TLE3 isoform in prostate tumors. Biochemical and biophysical research communications. 2007;364:918–923. doi: 10.1016/j.bbrc.2007.10.097. [DOI] [PubMed] [Google Scholar]
- 31.Venables JP, et al. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008;68:9525–9531. doi: 10.1158/0008-5472.CAN-08-1769. [DOI] [PubMed] [Google Scholar]
- 32.Cheung HC, et al. Global analysis of aberrant pre-mRNA splicing in glioblastoma using exon expression arrays. BMC genomics. 2008;9:216. doi: 10.1186/1471-2164-9-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartel M, et al. Increased alternative splicing of the KLF6 tumour suppressor gene correlates with prognosis and tumour grade in patients with pancreatic cancer. European journal of cancer. 2008;44:1895–1903. doi: 10.1016/j.ejca.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hobbs JE, et al. Alternatively spliced human tissue factor promotes tumor growth and angiogenesis in a pancreatic cancer tumor model. Thrombosis research. 2007;120(Suppl 2):S13–21. doi: 10.1016/S0049-3848(07)70126-3. [DOI] [PubMed] [Google Scholar]
- 35.Unruh D, et al. Alternatively spliced tissue factor contributes to tumor spread and activation of coagulation in pancreatic ductal adenocarcinoma. Int J Cancer. 2014;134:9–20. doi: 10.1002/ijc.28327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiltunen A, et al. Expression of type VI, IX and XI collagen genes and alternative splicing of type II collagen transcripts in fracture callus tissue in mice. FEBS Lett. 1995;364:171–174. doi: 10.1016/0014-5793(95)00384-L. [DOI] [PubMed] [Google Scholar]
- 37.Arafat H, et al. Tumor-specific expression and alternative splicing of the COL6A3 gene in pancreatic cancer. Surgery. 2011;150:306–315. doi: 10.1016/j.surg.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laklai H, et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat Med. 2016;22:497–505. doi: 10.1038/nm.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P, Yan B, Guo JT, Hicks C, Xu Y. Structural genomics analysis of alternative splicing and application to isoform structure modeling. Proc Natl Acad Sci USA. 2005;102:18920–18925. doi: 10.1073/pnas.0506770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodrigo-Domingo, M. et al. Reproducible probe-level analysis of the Affymetrix Exon 1.0 ST array with R/Bioconductor. Briefings in bioinformatics (2013). [DOI] [PMC free article] [PubMed]
- 41.Bengtsson, H., Simpson, K., Bullard, J. & Hansen, K. aroma.affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. Tech Report #745, Department of Statistics University of California, Berkeley (2008).
- 42.Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology3, Article3 (2004). [DOI] [PubMed]
- 43.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 44.Purdom E, et al. FIRMA: a method for detection of alternative splicing from exon array data. Bioinformatics. 2008;24:1707–1714. doi: 10.1093/bioinformatics/btn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cline MS, et al. ANOSVA: a statistical method for detecting splice variation from expression data. Bioinformatics. 2005;21(Suppl 1):i107–115. doi: 10.1093/bioinformatics/bti1010. [DOI] [PubMed] [Google Scholar]
- 46.Koscielny G, et al. ASTD: The Alternative Splicing and Transcript Diversity database. Genomics. 2009;93:213–220. doi: 10.1016/j.ygeno.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 47.Dumartin L, et al. AGR2 is a novel surface antigen that promotes the dissemination of pancreatic cancer cells through regulation of cathepsins B and D. Cancer Res. 2011;71:7091–7102. doi: 10.1158/0008-5472.CAN-11-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haider S, et al. A multi-gene signature predicts outcome in patients with pancreatic ductal adenocarcinoma. Genome Med. 2014;6:105. doi: 10.1186/s13073-014-0105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci USA. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


