Abstract
Metabolic phenotypes reflect both the genetic and environmental factors which contribute to the development of varicose veins (VV). This study utilises analytical techniques to provide a comprehensive metabolic picture of VV disease, with the aim of identifying putative cellular pathways of disease pathogenesis. VV (n = 80) and non-VV (n = 35) aqueous and lipid metabolite extracts were analysed using 600 MHz 1H Nuclear Magnetic Resonance spectroscopy and Ultra-Performance Liquid Chromatography Mass Spectrometry. A subset of tissue samples (8 subjects and 8 controls) were analysed for microRNA expression and the data analysed with mirBase (www.mirbase.org). Using Multivariate statistical analysis, Ingenuity pathway analysis software, DIANALAB database and published literature, the association of significant metabolites with relevant cellular pathways were understood. Higher concentrations of glutamate, taurine, myo-inositol, creatine and inosine were present in aqueous extracts and phosphatidylcholine, phosphatidylethanolamine and sphingomyelin in lipid extracts in the VV group compared with non-VV group. Out of 7 differentially expressed miRNAs, spearman correlation testing highlighted correlation of hsa-miR-642a-3p, hsa-miR-4459 and hsa-miR-135a-3p expression with inosine in the vein tissue, while miR-216a-5p, conversely, was correlated with phosphatidylcholine and phosphatidylethanolamine. Pathway analysis revealed an association of phosphatidylcholine and sphingomyelin with inflammation and myo-inositol with cellular proliferation.
Introduction
Primary varicose vein disease, a vascular condition characterised by elongated, dilated and tortuous veins in the lower limbs, remains one of the most commonly reported chronic conditions in western medicine. While recent studies have identified familial clustering of disease, and a genetic component is generally accepted, age, occupation, pregnancy and obesity are still key risk factors. The determination of the existence of mechanistically different varicose vein “sub-phenotypes” would be important as this could reflect the different effects that various environmental factors have on disease progression. The morphological characteristics of the disease have been well documented, and known histological changes in the dilated vein include intimal hyperplasia and disruption of smooth muscle cells and the extracellular matrix1. However, how disease variants of risk loci disrupt the biochemistry of the cell and how these events translate into the formation of the varicose vein is still not known. Understanding how biochemical aberrations modulate metabolic pathways, enzymatic activity and normal cellular function is the natural next step in the investigation of the disease.
Metabonomics employs established and reproducible tools for the identification of the metabolic status of an individual and are highly appropriate for the evaluation of the biochemical components of a disease phenotype2, 3. The metabolic profiles of human varicose veins have been previously shown to be different from healthy vein tissue, when analysed using magic angle spinning (MAS) 1H- nuclear magnetic resonance (NMR) spectroscopy. 1H-MAS NMR spectroscopy allows for the profiling of intact whole tissue without the need for chemical extraction, and also provides information on metabolite mobility and metabolite localisation within the tissue4. Lactate, myo-inositol and creatine were found to be the differential biomarkers that distinguished between the healthy and diseased vein groups, and triglyceride moieties were also observed to be lower in concentration in varicose tissue5. However, the study in question only utilised one profiling technique and a very small sample size. Therefore, to be able to attain a more comprehensive metabolic picture, and move beyond phenotypic observation to identification of potential mechanisms, analysis of extracted metabolites from a larger number of participants is required. In addition to using multiple analytical platforms such as NMR and ultra performance liquid chromatography coupled to -mass spectrometry (UPLC-MS), microRNAs (miRNAs) have been profiled for a subset of patients to explore modulated pathways. miRNAs are small 22 nucleotide RNAs, that play an important role in cell regulation by silencing genes through targeting mRNAs for cleavage. miRNAs are involved in the regulation of many key cellular pathways and as such have been identified as potential markers for patient stratification and as therapeutic targets. Additionally, UPLC-MS, an inherently more sensitive analytical technology, can complement NMR analysis and provide further information of the perturbation of low concentration metabolites, and extend profiling coverage. This study uses three analytical techniques including NMR, UPLC-MS and miRNAs to provide a comprehensive metabolic picture of varicose vein disease, aiming to synthesise a metabolic map of the disease and identify putative cellular pathways of disease pathogenesis. Aqueous extracts from humans’ varicose vein samples and humans’ non-varicose vein control were analysed using NMR and UPLC-MS HILIC profiling. Lipid extracts from the diseased and controls veins samples were analysed using UPLC-MS. miRNAs profiling was performed on randomly selected diseases and long saphenous vein controls samples.
Results
Multivariate analysis of 1H NMR spectroscopic based profiling of vein tissue
Initial principal component (PC) modelling of the NMR data showed some stratification in the data (Supplementary Fig. 1) prompting further analysis with supervised methods.
The OPLS-DA models produced with the aqueous extract 1H NMR data showed clear metabolic differences between the diseased and control groups. The Q2Y and R2Y values were 0.379 and 0.202, respectively, and the test statistics were validated using permutation testing (p < 0.05). The significant peaks highlighted as driving the group differences corresponded to signatures from myo-inositol, taurine and glutamic acid (glutamate) which were higher in the disease group (Fig. 1). These metabolites were also found to be highly significant based on the univariate statistical analysis, after correction for multiple testing (Benjamini-Yekutieli correction) with corrected p-values < 0.0001 for each differential metabolite. In order to identify all the correlated peaks pertaining to a particular molecule, Statistical Total Correlation Spectroscopy (STOCSY) was carried out using a selected discriminatory peak as a driver for calculation of the correlation matrix6. Figure 2 shows the STOCSY output of glutamate (δ3.770), taurine (δ3.269), and myo-inositol (δ4.07), although at δ3.269, the taurine peaks and myo-inositol peaks are overlapping.
Figure 1.
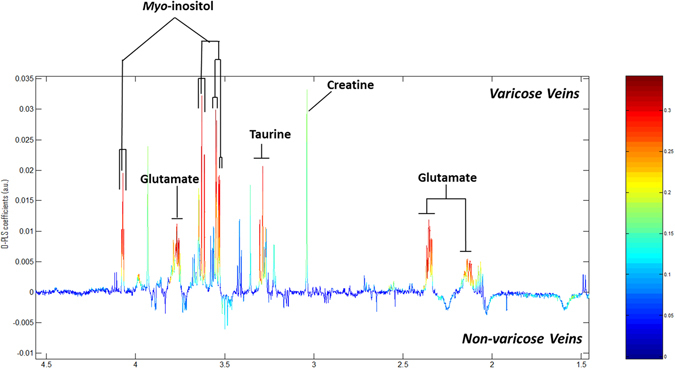
OPLS models for aqueous metabolites analysed by 1H NMR spectroscopy. Model is showing the differences between the varicose vein and non-varicose veins groups.
Figure 2.
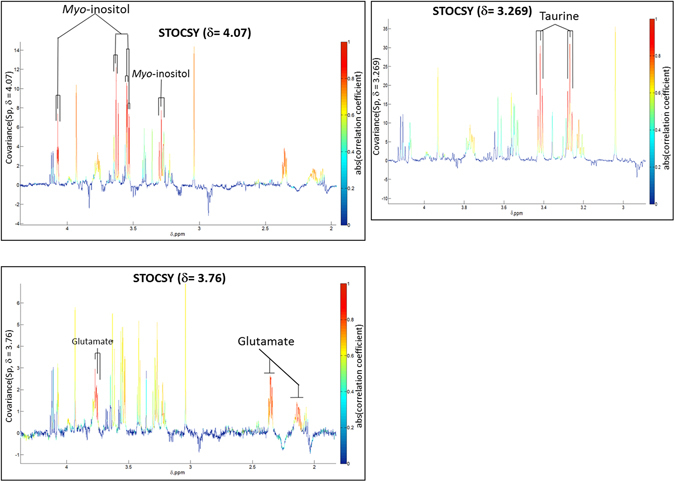
Statistical Total Correlation Spectroscopy (STOCSY). STOCSY showing intra-molecular correlation of peaks related to metabolites (taurine, myo-inositol and glutamate) associated with differentiation of disease from controls.
Multivariate analysis of UPLC-MS based lipid and HILIC profiling
Good instrumental stability during the lipid profiling (organic extracts) experimental run was indicated by the tight clustering of the QC samples near the middle of statistical space in the PCA scores plot from the multivariate analysis of both the electrospray ionization (ESI)+ and ESI- UPLC-MS data (Supplementary Figure 2). For the ESI+ UPLC-MS dataset, comparisons of the control and test group show some spatial trends. Subsequent orthogonal projection to latent structures discriminant analysis (OPLS-DA) modelling, after cross validation, showed a strong separation between the two groups, although there was some overlap, with the test statistics being Q2Y = 0.486 and R2Y = 0.693 (Fig. 3a). The loadings S- plot was produced for the model indicating which variables are correlating for each group. Variables clustered towards the bottom left of loadings S- plot are variables which represent metabolites more abundant in the disease group, whereas variables in the top right are down regulated in the disease group. The loadings S-plot identified triglycerides and ceramides as higher in the control group, while phosphatidylserines, phosphatidylcholines, sphingomyelins, and phosphatidylethanolamines were elevated in varicose veins group (Fig. 3b and Table 1). Details of these features are described in Table 1.
Figure 3.
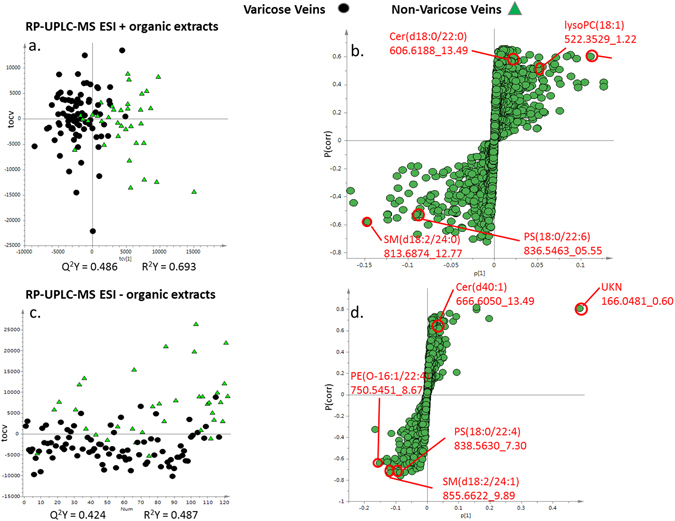
Multivariate OPLS –DA cross-validated scores and loadings S-plots for UPLC-MS based lipid profiling (ESI +/−) (a) and (c) comparing the varicose veins and non-varicose veins samples and (b) and (d) showing the S loadings plots for the OPLS-DA models. Metabolites at the outer most edges of the S plot are present in higher concentration in one or other group. Robustness of the OPLS-DA model was confirmed by permutations testing. TG Triglyceride, PC Phosphatidylcholine, SM Sphingomyelin, PE Phosphatidylethanolamine, PS phosphatidylserine.
Table 1.
Details of differential features identified between varicose and non-varicose veins groups on UP-LCMS ESI +/− and UP-LCMS HILIC +/− ESI analysis.
| Met Name | Mol Formula as Detected | Retention Time (min) | m/z (found) | m/z (theor) | Fold change | p-value | Δppm | Higher in group |
|---|---|---|---|---|---|---|---|---|
| UP-LCMS ESI +/− (Organic extracts) | ||||||||
| Cer(d18:0/22:0) | C40H80NO2+[M+H−H2O]+ | 13.49 | 606.6188 | 606.6189 | −2.1 | 0.01 | 0 | Non-VVs |
| lysoPC(18:1) | C26H53NO7P+[M+H]+ | 01.22 | 0522.3529 | 522.3554 | −1.7 | 0.03 | −5 | Non-VVs |
| TG(12:0/14:0/18:1) TG(12:0/16:0/16:1) | C47H92NO6+[M+NH4]+ | 14.77 | 0766.6937 | 766.6919 | −2.1 | 0.03 | 2 | Non-VVs |
| TG(12:0/16:0/18:1) TG(14:0/16:1/16:0) | C49H96NO6+[M+NH4]+ | 15.10 | 0794.7231 | 794.7232 | −1.8 | 0.007 | 0 | Non-VVs |
| TG(14:0/16:0/18:1) | C51H100NO6+[M+NH4]+ | 15.42 | 0822.7546 | 822.7545 | −1.6 | 0.002 | 0 | Non-VVs |
| TG(15:0/16:0/18:1) TG(14:0/17:0/18:1)* | C52H102NO6+[M+NH4]+ | 15.55 | 0837.7767 | 837.7741 | −1.5 | 0.02 | 3 | Non-VVs |
| TG(16:0/16:0/18:1)* | C53H104NO6+[M+NH4]+ | 15.71 | 0851.7901 | 851.7897 | −1.5 | 0.01 | 0 | Non-VVs |
| PC(18:0/20:3)* | C46H87NO8P+[M+H]+ | 08.61 | 0813.6229 | 813.6203 | 1.5 | 0.007 | 3 | VVs |
| PC(38:4)* | C46H85NO8P+[M+H]+ | 07.06 | 0811.6074 | 811.6047 | 1.4 | <0.0001 | 3 | VVs |
| PE(O-16:1/22:6) | C43H75NO7P+[M+H]+ | 06.49 | 0748.5290 | 748.5276 | 1.4 | 0.01 | 2 | VVs |
| PS(16:0/22:6) | C44H75NO10P+[M+H]+ | 04.29 | 0808.5168 | 808.5123 | 1.6 | 0.0002 | 6 | VVs |
| PS(18:0/20:1) | C44H85NO10P+[M+H]+ | 10.07 | 0818.5855 | 818.5906 | 1.5 | 0.0002 | −6 | VVs |
| PS(18:0/20:3) | C44H81NO10P+[M+H]+ | 06.65 | 0814.5615 | 814.5593 | 1.4 | 0.01 | 3 | VVs |
| PS(18:0/22:6) | C46H79NO10P+[M+H]+ | 05.55 | 0836.5463 | 836.5436 | 1.7 | 0.003 | 3 | VVs |
| SM(d18:2/24:0) | C47H94N2O6P+[M+H]+ | 12.77 | 0813.6874 | 813.6844 | 1.3 | 0.0007 | 4 | VVs |
| SM(d18:2/24:1) | C47H91N2O6PNa+[M+Na]+ | 09.85 | 0833.6557 | 833.6507 | 1.3 | 0.0008 | 6 | VVs |
| Cer(d40:1) | C41H80NO5 [M+FA−H]− | 13.49 | 0666.6050 | 666.6042 | −1.2 | 0.01 | 1 | Non-VVs |
| Alanine | C3H6NO2−[M−H]− | 00.54 | 0088.0398 | 88.0404 | 1.9 | <0.0001 | −7 | VVs |
| Creatine | C4H8N3O2−[M−H]− | 00.54 | 0130.0611 | 130.0622 | 1.6 | 0.003 | −8 | VVs |
| Glutamine | C5H8NO4−[M−H]− | 00.54 | 0146.0459 | 146.0459 | 1.8 | 0.0002 | 0 | VVs |
| Guanosine | C10H12N5O5−[M−H]− | 00.54 | 0282.0840 | 282.0844 | 1.4 | 0.005 | −1 | VVs |
| Inosine | C10H11N4O5−[M−H]− | 00.54 | 0267.0724 | 267.0735 | 1.6 | 0.001 | −4 | VVs |
| PC(18:0/20:3) | C47H87NO10P− [M+FA−H]− | 08.62 | 0856.6094 | 856.6073 | 1.4 | 0.008 | 2 | VVs |
| PE(O-16:1/22:4) | C43H77NO7P−[M−H]− | 08.67 | 0750.5451 | 750.5443 | 1.2 | 0.004 | 1 | VVs |
| PE(O-16:1/22:6) | C43H73NO7P−[M−H]− | 06.53 | 0746.5136 | 746.513 | 1.3 | 0.007 | 1 | VVs |
| PI(18:2/18:0) | C45H82O13P−[M−H]− | 06.01 | 0861.5527 | 861.5499 | 1.4 | <0.0001 | 3 | VVs |
| PI(36:1) | C45H84O13P−[M−H]− | 07.40 | 0863.5676 | 863.5655 | 1.4 | 0.001 | 2 | VVs |
| PS(16:0/22:6) | C44H73NO10P−[M−H]− | 04.33 | 0806.4999 | 806.4978 | 1.6 | 0.0005 | 3 | VVs |
| PS(18:0/20:1) | C44H83NO10P−[M−H]− | 10.15 | 0816.5787 | 816.576 | 1.4 | 0.01 | 3 | VVs |
| PS(18:0/20:3) | C44H79NO10P−[M−H]− | 06.72 | 0812.5485 | 812.5447 | 1.4 | 0.004 | 5 | VVs |
| PS(18:0/22:4) | C46H81NO10P−[M−H]− | 07.30 | 0838.5630 | 838.5604 | 1.5 | 0.0006 | 3 | VVs |
| PS(18:0/22:6) | C46H77NO10P−[M−H]− | 05.60 | 0834.5309 | 834.529 | 1.4 | 0.0007 | 2 | VVs |
| SM(d18:2/24:0)* | C47H93N2O6P−[M+FA−H]− | 12.79 | 0858.6876 | 858.6781 | 1.3 | 0.0005 | 11 | VVs |
| SM(d18:2/24:1) | C48H92N2O8P−[M+FA−H]− | 09.89 | 0855.6622 | 855.6597 | 1.4 | <0.0001 | 3 | VVs |
| Uridine | C9H11N2O6−[M−H]− | 00.55 | 0243.0616 | 243.0623 | 1.4 | 0.006 | −3 | VVs |
| UP-LCMS HILIC +/− Analysis (Aqueous extracts) | ||||||||
| Creatine | C4H10N3O2+[M+H]+ | 6.623 | 132.0773 | 132.0767 | 1.3 | <0.0001 | 4.5 | VVs |
| Inosine | C5H5N4O+[M+H]+ | 6.073 | 137.0463 | 137.0457 | 1.3 | 0.007 | 4.3 | VVs |
| PC (16:0/20:4) | C44H81NO8P+[M+H]+ | 4.737 | 782.5684 | 782.5694 | 1.4 | 0.009 | −1.2 | VVs |
| Glutamate | C5H8NO4−[M−H]− | 7.467 | 146.0455 | 146.0458 | 1.5 | <0.0001 | −2 | VVs |
| Inosine | C10H11N4O5−[M−H]− | 3.167 | 267.0729 | 267.0734 | 1.24 | 0.01 | −1.8 | VVs |
| Taurine | C2H6NO3S−[M−H]− | 5.808 | 124.0071 | 124.0073 | 1.17 | 0.07 | −1.6 | VVs |
| Uridine | C9H11N2O6−[M−H]− | 1.517 | 243.0617 | 243.0622 | 1.2 | 0.04 | −2 | VVs |
TG Triglyceride, PC Phosphatidylcholine, SM Sphingomyelin, PE Phosphatidylethanolamine, PS phosphatidylserine, PI phosphatidylinositols
Corrected p-values using Benjamini-Yakutieli multiple testing corrections. *Tentative assignments.
Similar distribution trends were also seen in the ESI- UPLC-MS dataset with the positions of the control and disease subjects within the model, with some separation between the classes on PC1 (Supplementary Figure 2). Further supervised analysis with OPLS-DA modelling show a better separation, with good test statistic values (Q2Y = 0.424 and R2Y = 0.487). The metabolites identified as driving the class differences, again using the loadings S-plot, included phosphatidylserines, phosphatidylcholines, sphingomyelins, ceramides, phosphatidylethanolamines and phosphatidylinositols (Fig. 3c and d, and Table 2). Additionally, several other small molecules demonstrated high correlations with the disease or control group and exhibited retention times at or close to the solvent-front. These were inosine, taurine, guanosine and uridine, which were elevated in patients with varicose veins. Furthermore, there were some differential unknown metabolites observed as well on UPLC MS ESI+/− modes. Details of these are listed in Supplementary Table 1. Future work may be required to identify these unknown metabolites and their relevance in veins.
Table 2.
Details of human vein tissue and Demographic details of patients.
| Non-varicose veins | Varicose veins | P value | |
|---|---|---|---|
| n = 35 | n = 80 | — | |
| Type of vein tissue | Cephalic = 3 | Truncal = 44 | — |
| Long saphenous = 15 | Varicosities = 36 | ||
| Inferior epigastric/mesenteric = 8 | |||
| Facial = 9 | |||
| CEAP clinical score | — | CEAP 2 = 4 | — |
| CEAP 3 = 66 | |||
| CEAP 4 = 9 | |||
| CEAP 5 = 1 | |||
| Recurrent varicose veins | — | 9 | — |
| Sex | Female = 6 | Female = 40 | — |
| Male = 29 | Male = 40 | ||
| Age, years | 32–85 (mean 62.7) | 18–82 (Mean 45.16) | <0.0001 |
| HTN | 21 | 14 | <0.0001 |
| Diabetes mellitus | 9 | 2 | 0.002 |
| PAD | 12 | 8 | 0.02 |
| IHD | 12 | 2 | 0.001 |
| Stroke | 6 | 2 | 0.03 |
| Cancer | 1 | 0 | 0.3 |
| Connective tissue disorders | 0 | 0 | — |
| Aspirin | 18 | 12 | 0.0002 |
| Simvastatin | 20 | 12 | <0.0001 |
| Lisinopril | 17 | 5 | <0.0001 |
| Amlodipine | 6 | 5 | 0.18 |
CEAP Clinical Aetiological Anatomical Pathophysiological. IHD ischemic heart disease, HTN hypertension, PAD peripheral arterial disease.
Principal component analysis of the aqueous extracts by UPLC-MS HILIC ESI+ also indicated that there were differences between the classes (Supplementary Figure 2). There was some separation of groups on PCA score plots with clustering of samples in varicose veins group. Therefore cross-validated OPLS-DA models were again run to identify differences between the groups (Fig. 4a). These identified inosine, creatine and phosphatidylcholine (PC) which were higher in varicose veins group (Fig. 4b). The same process was also repeated for the aqueous extracts analysed on UPLC-MS HILIC ESI-, and both the PCA analysis (Supplementary Figure 2) and the OPLS-DA modelling showed clear separation of varicose and non-varicose veins samples (Fig. 4c). Further interrogation of the loadings S-plot identified inosine, glutamate, uridine and taurine as the differential metabolites, and these were present in higher concentration in varicose veins group relative to non-varicose veins samples (Fig. 4d). Details of differentials features identified in HILIC+/− ESI are shown in Table 1.
Figure 4.
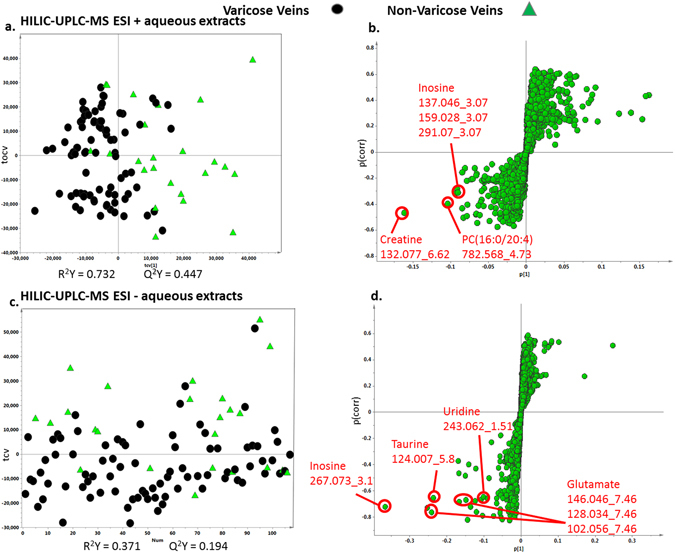
Multivariate OPLS –DA cross-validated scores and loadings S-plots for UPLC-MS HILIC based profiling (ESI +/−). (a) and (c) Comparing the varicose veins and non-varicose veins samples. (b) and (d) Showing the S loadings plots for the OPLS-DA models. Metabolites at the outer most edges of the S plot are present in higher concentration in one or other group. Robustness of the OPLS-DA model was confirmed by permutations testing. PC Phosphatidylcholine.
Patients participating in the test group, attending hospital for varicose vein intervention, were on average younger and with fewer co-morbid conditions than the controls; furthermore many were taking medications, such as aspirin, simvastatin and lisinopril, at the time of sample collection (Table 2). Multivariate partial least square (PLS) regression modelling approach showed that none of these secondary factors produced any significant model. The sole exception was a PLS-DA model for hypertension in UPLC-MS lipid profiling in negative ionisation, whereby the R2Y and the Q2Y for that model were 0.207 and 0.053 on first component with the p- value 0.05 for the first component. However, when the second orthogonal component was added to the model the Q2Y value was reduced to a negative value and p value changed to 1. This would indicate that the initial model was “over-fitted” and thus it is unlikely hypertension was a real bias which would skew the main comparisons (Supplementary Table 2).
The PCA scores plots for all the datasets showed some dispersal within the control group itself, therefore to evaluate if these within-group variations would affect the main models, PCA models containing only control samples were constructed, and the scores plots were coloured according to vein origin (Supplementary Figure 3). This highlighted that there were no significant metabolic differences between control veins of different anatomical locations. Furthermore, when models comparing the disease group samples with only control veins from the long saphenous vein (the most common control vein location) were created similar results to the main models were produced, albeit with lower significance values, which is to be expected when reducing sample size (Supplementary Figures 4, 5 and 6).
miRNA analysis and Pathway analysis
This experiment highlighted differential expression of 7 miRNAs in varicose vein disease which are listed in Table 3, together with their expected regulated pathways. The elevated levels of hsa-miR-4459, hsa-miR-135a-3p and hsa-miR-216a-5p in varicose vein tissue are of particular interest as they regulate signal cascades that directly influence cell proliferation and survival.
Table 3.
Differentially expressed miRNAs in the varicose vein cells, together with the fold change ratio, overexpression analysis p-value and the relevant target processes which each miRNA regulates.
| miRNA | Fold change | P value | Target Genes | Target Processes |
|---|---|---|---|---|
| hsa-miR-642a-3p | −2.913896 | 4.14 × 10−3 | 285 | Biological regulation, transcription, nucleobase-containing compound metabolism, fatty acid metabolism |
| hsa-miR-718 | −2.582323 | — | 11 | Pentose Phosphate pathway |
| hsa-miR-4459 | −2.5259194 | 2.72 × 10−2 | 349 | EGF receptor signalling pathway, primary and secondary bile acid synthesis |
| hsa-miR-135a-3p | −2.2473247 | 1.02 × 10−3 | 718 | PI3K pathway, EGF receptor pathway, TGF-beta signalling pathway, Thiamine metabolism |
| hsa-miR-363-3p | −2.2425704 | 1.5 × 10−3 | 893 | GRH receptor pathway, Leucine and Isoleucine biosynthesis |
| hsa-miR-216a-5p | 8.4616585 | 8.79 × 10−3 | 287 | Wnt signalling pathway, TGF signalling pathway |
| hsa-miR-136-5p | 4.2504244 | — | 270 | glycosaminoglycan biosynthesis |
A Spearman correlation matrix was constructed to investigate the correlation between miRNA expression and the levels of the significant metabolites identified through the analytical chemistry. The resultant heatmap and dendrogram are provided (Fig. 5). This highlighted that hsa-miR-642a-3p expression levels were highly correlated with levels of inosine within the vein tissue, as are expression levels of hsa-miR-4459 and hsa-miR-135a-3p. Expression of miR-216a-5p, known to regulate the Transforming Growth Factor beta (TGF-β) signalling pathway, conversely showed high levels of correlation with a variety of metabolites including phosphatidylcholine, phosphatidylethanolamine, and uridine. Finally, hsa-miR-142-5p showed a very strong correlation with sphingomyeline levels. IPA analysis included the identified miRNAs in addition to the previously identified differential metabolites. IPA provides a putative network based on statistical probability. The analysis used an IPA core analysis with a miRNA target filter, and this was continued to identify a causal network. This process is similar to gene-set enrichment, but with the additional power of incorporating cause-effect relationships. Figure 6, the output of the IPA, shows the association between the highlighted miRNAs, hsa-miR-642a-3p (representing the miRNAs with seed GACACAU) and hsa-miR-142-5p (and other miRNAs with seed UUGGCUU) and levels of sphingomyeline, phosphatidylinositol, glutamate and myo-inositol. The analysis revealed that the miRNAs regulate production of a variety of receptors which converge on the activation of a number of enzymes involved in phospholipid homeostasis and activation of MAP kinase. ABCA1, an ATP-binding cassette transporter involved in phospholipid homeostasis, and PTEN, a phosphatase and tensin homolog which catalyses the conversion phosphatidylinositol (3,4,5)-trisphosphate (PIP3) to phosphatidylinositol 4,5-bisphosphate (PIP2), are predicted to contribute to the regulation of both the PI3K/AKt and MAPK pathways, which are known to be involved in cell proliferation. The identities of remaining nodes of the pathway are defined in the supplementary section.
Figure 5.
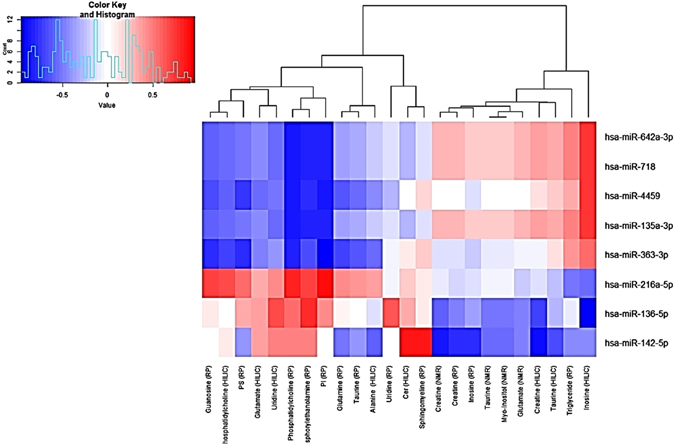
Correlation heatmap and a dendrogram. A correlation heatmap and a dendrogram of the spearman correlation mapping between the significant metabolites (across the bottom) and the differentially expressed miRNAs (along the right). Areas of intense red colour signify a high level of correlation between the expression of that miRNA and levels of the matching metabolite, whereas blue areas signify anti-correlation. The dendrogram at the top shows correlation distances by clustering the metabolites hierarchically.
Figure 6.
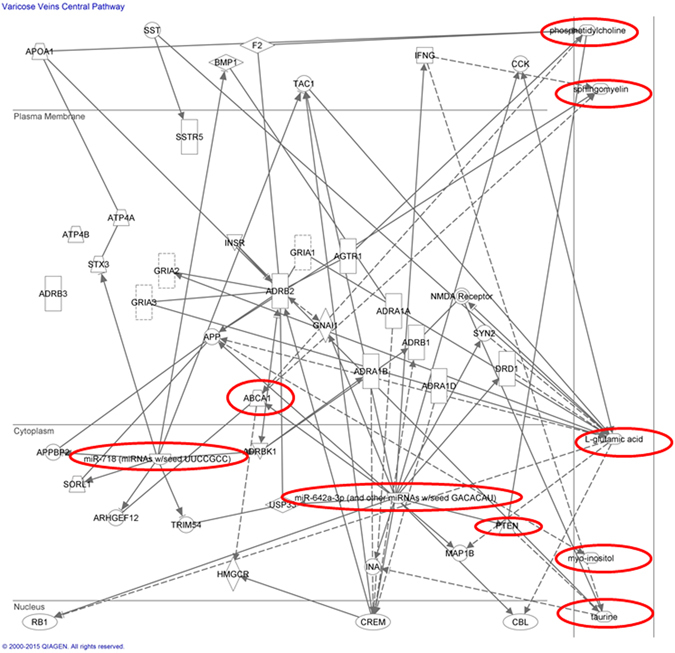
Ingenuity Pathway Analysis showing a network biology system for the interaction between both the miRNAs and the metabolites identified from the metabonomic analysis.
Discussion
The aim of this study was to provide a comprehensive characterisation of the metabolic profiles of varicose and non-varicose veins, and to identify any metabolic differences between the two. Chemometric analysis of the tissue aqueous extracts revealed that the polar metabolites creatine, glutamate, myo-inositol, taurine and inosine were present in higher concentrations in varicose veins. The validity of these results was reenforced by the agreement of both MS-based analytical techniques and the results of the NMR spectroscopy in identification of the differential metabolites in varicose vein disease. In addition the non-polar metabolites sphingomyelin and several phospholipids were also found to be elevated in the disease group. Pathway analysis provided a potential network of molecular interactions between these metabolites and identified miRNAs within VV; this network paves the way for further investigation of disease etiology. Variations in age, sex, differences in past medical history and medications had no correlation with any metabolic differences noticed between the groups.
Glutamate, one of the key metabolites identified by the multivariate modeling, is an alpha-amino acid, a key metabolite in many aspects of cellular metabolism, and plays a central role in hepatic amino acid metabolism, glycolysis, gluconeogenesis and the tricarboxylic acid cycle7. In adition glutamate is also involved in the production of gamma-aminobutyric acid (GABA) in the GABA-shunt and also part of glutathione synthesis8. Glutathione, being one of the primary conjugative molecules of phase two detoxification, is an important factor in the cellular defence mechanism and also helps regulate cell proliferation and apoptosis9. However there were no discernable changes in glutathione levels detected in the samples.
Another of the significantly changed metabolites is taurine. Taurine, a ubiquitous organic osmolyte, is one of the most abundant amino acids within the body, although it is not involved in protein synthesis. Metabolically derived from cysteine, taurine is involved in the formation of bile salts, regulation of cell volume and modulation of intracellular calcium concentration, essential for the function of signal transduction pathways10. Administration of taurine has also been shown to protect from stress-induced vascular endothelial cell apoptosis10, 11, and high levels of taurine are thought to promote cell survival.
Myo-inositol, one of the more abundant inositol isomers, is a carbocyclic polyol and is also a structural component of a number of secondary cellular messengers such as inositol phosphates, phosphatidylinositol and phosphatidylinositol phosphate lipids. Myo-inositol, like taurine, predominately operates as an intercellular osmolyte, and is also involved in the Akt signalling pathway, membrane trafficking and regulating apoptosis. Cytosolic myo-inositol is essential for cell survival and promotes growth and proliferation12.
Sphingomyelin was elevated in VV tissue, and correlation analysis showed its concentration is closesly linked to the hsa-miR-142-5p part of the predicted VV network (Fig. 5). Sphingomyelin has also previously been observed in varicose veins by Tanaka et al. using matrix assisted laser desorption ionization imaging13. Sphingomyelin is the most abundant type of sphingolipid and constitutes a key structural component of cellular membranes and vesicles. Sphingomyelin acts as a platform for the regulation of transmembrane signalling, and also intracellular vesicle trafficking as bilayer sphingomyelin levels are involved in chemotaxis-based migration. This modulation extends to transferrin-mediated cell proliferation triggering cell growth and division14.
The statistical analysis showed that there is an increase of cellular concentrations of a wide range of phospholipids which are the key component of the lipid bilayer and are essential for the viability of eukaryotic cells. While both phosphatidylserine and phosphatidylethanolamine, have been shown to be involved in intracellular signal transduction and cell proliferation, of most interest is phosphatidylcholine. Phosphatidylcholine, a glycerophospholipid, is synthesised by the CDP-choline pathway or through the hepatic phosphatidylethanolamine methyltransferase. As a key component of membranes and lipid messengers, phosphatidylcholine is directly involved in cell cycle progression and cell proliferation15. Aberrant activation of phosphatidylcholine production is usually through induction of the normal PI3K/Akt pathway, and is a key feature of many forms of cancer. Furthermore, it has been shown that increased phosphatidylcholine due to the increased expression of choline kinase A (CKa) is related to hypoxia, as the gene encoding for CKa includes a hypoxia induced element in the promoter region16; this means that during hypoxic conditions, cells will increase choline uptake potentially triggering downstream cell cycle progression. Phosphatidylcholine is also a substrate for phospholipase A2 (PLA2). PLA2 causes hydrolysis of phosphatidylcholine into free fatty acids such as arachidonic acid (AA) and lysophosphatidylcholine. AA is a precursor molecule for prostaglandins and leukotriene, both responsible for chronic inflammation13. We didn’t detect phosphatidylcholine with AA incorporated, however, we detected phosphatidylcholine (20:3).
Finally the two other metabolites of note are inosine and creatine, which both follow the theme of hypoxia-induced metabolic changes; inosine is known to be released during hypoxia17 and increased creatine has been seen in dilated cardiomyopathy18. Correlation matrix analysis (Figs 5 and 6) showed that inosine levels may be highly regulated by miR-4459 and mIR-216a-5p, in addition to miR-642a-3p the key miRNA in the presented VV network, with taurine levels also showing a strong correlation with the same miRNAs predicted to be under the direct regulation of PTEN.
Myo-inositol, found to be elevated in varicose vein tissue, both in this study and previous work by Anwar et al.5, is also related to this process, as it constitutes the precursor molecule for phosphatidylinositol, a substrate for the PI3K signalling pathway. In fact, PI3K constitutes a key component of the suggested varicose vein pathway from the IPA (Fig. 6). Identified miRNAs mIR-216a-5p (upregulated in VVs) and miR-4459 (downregulated in VVs) are predicted to regulate expression of membrane receptors TFG-beta receptor 1 and EGF receptor, respectively. These receptors play a key role in the activity of PI3K. Furthermore expression of PI3K itself is predicted to be controlled by miR-135a-3p. The altered activies of these particular miRNAs, based upon our putative interaction network, seem to indicate that there is a phenotypic switch in the expression profile of the varicose vein cells, likely in smooth muscle cells, with dysregulation of the systems controlling cell growth and proliferation.
A similar narrative can be discerned from evaluating the results while focusing on sphingolipid metabolism. Sphingolipids share a common 18 carbon backbone, with different accessory structures giving rise to the various members of the sphingolipid family. The metabolic network could be thought of a web of interconnected pathways, branching out from a common progenitor molecule and coming together to terminate in a common catabolic end-point. The various intermediate molecules on each branch of the network, however, have highly diverse functions, and as such the metabolic trajectory each molecule is highly regulated to ensure correct cellular function. Sphyngomyelin, mentioned above and found to be raised in varicose veins, is synthesised by the transfer of a phosphocholine group from phosphatidylcholine (also shown to be elevated in varicose veins) to ceramide, by sphingomyelin synthases (SMSs)19; this reaction also produces diaclyglycerol. Diaclyglycerol is typically converted to phosphatidic acid by the diacylglycerol kinases; a process that has been shown to protect myocardium cells from hypertrophy when subjected to abnormally high pressure and stretch forces20. However, sphingomyelin is catabolised by the hydrolysis of the phosphocholine group by sphingomyelinases (SMase), the activity of which is closely linked with local pH and oxygen levels21; Indeed studies investigating the effect of sphingomyelinase deficiency has shown that cells that do not catabolise sphingomyelin efficiently are protected from stress-induced apoptosis22. Each player in this complex set of interactions has been shown to be a bioactive lipid, with sphingomyelin, phosphocholine, phosphatidylcholine, ceramide and DAG all exerting an influence on the survivability of the cell, albeit an opposing one. The first three seemingly are encouraging growth and survival, whereas the latter two promoting cell death and quiescence23. Taken together this could indicate that the local chemical environment and physical duress which cells are exposed to following chronic blood stasis could be triggering changes in the balance of metabolism and disrupting lipid homeostasis, leading to the production of certain pro-survival molecules with the relevant downstream effects.
This study has presented a metabolic characterisation of the VV disease, highlighting the molecular processes underlying VV pathogenesis and progression. It has been well established that such a phenotypic switch will affect vein wall contraction and result in vein wall relaxation and weakness, which may be the cause of the tortuous and loose appearance of the diseased vein. The identified metabolites suggest that the processes of cell proliferation and survival should be further investigated with a view to identifying potential therapeutic drug targets. Development of drug therapy in venous disease may help in halting the disease progression as well as a preventative measure for individuals with a genetic propensity for the development of the disease.
One of the key limitations of the work presented here is the heterogeneity of the control group. The control group did not accurately match with the varicose veins group in terms of demographic features, past medical history and the medications. Factors such as age, medications and past medical history may influence the metabolic profile of vein tissue. However, to our knowledge, there has been no demonstration of the effect of age on metabolic profile of vein tissue so far. The control group included great saphenous vein samples from patients undergoing lower limb bypass operations or amputations in addition to vein samples from other anatomical locations such as the neck and lower abdomen, which are subject to different biomechanical stresses than the GSV. While statins and most other cardiovascular medications have a short half-life (less than 24 hours)24 and neither parent drugs nor their metabolites were detected in our NMR or LCMS based profiling, it is possible that the above factors confounded the presented results by pharmacological effect. It is, nonetheless, important to stress that despite these issues the varicose vein samples were consistently different from the control groups in the statistical modelling, suggesting that any variability within the non-varicose veins groups was far outweighed by the differences between the two groups. Ultimately due to the ethical issues surrounding vein sample retrieval from often young and otherwise healthy individuals, it may prove challenging if not impossible to correctly age match groups in future varicose veins studies.
Another consideration for the future is CEAP classification, the scheme by which the disease is classified into different grades based on severity and type of presentation25, and how CEAP grading relates to metabolic phenotypes. Future studies should potentially explore any differences of metabolites in tissue, serum and urine between patients of different disease grades. This could aid clinicians in following disease progression and help tailor therapy accordingly. Varicose vein disease is a multifactorial disease influenced by both environmental factors such as occupation, obesity, pregnancy and also has a strong familial predisposition. Environmental stimuli in the patient exposes the vein wall to high pressure and hypoxia, which in combination trigger molecular changes in the vein wall leading to vein wall relaxation and dilatation. This work has shown that the metabolic profiles of human varicose veins are systematically different from non-varicose veins tissues, as well as exemplifying the utility of the metabonomic approach in understanding the molecular changes at cellular level of vascular disease.
Methods
Ethical approval was obtained for the research from the Riverside Research Ethics Committee (RREC 3092). The research has also been registered to the Imperial College London Research Services, and the Imperial College Healthcare NHS Trust Research and Development department and all the experiments were performed in accordance with the department’s guidelines and regulations. All the specimens collected were anonymised according to the rules set out in the Human Tissue Act (2004). Informed consent was obtained from all the patients involved in the study. Varicose veins were retrieved from patients who were treated with ligation of sapheno-femoral junction and stripping of great saphenous vein (GSV) or phlebectomies within Imperial College London Healthcare NHS Trust. Initial assessment of the patient was performed in outpatient department. Duplex ultrasound imaging of the lower limb venous system was performed for all varicose veins patients by accredited vascular scientists where the presence of venous reflux was assessed. Control veins were obtained from patients undergoing any surgery which involved the routine removal of non-varicose veins that would otherwise be discarded, including residual vein following vein harvest for extremity arterial bypass. Patients with a current or past history of varicose veins were not included in the non-varicose vein control group. Varicose and non-varicose veins of the affected limbs of patients with a history of deep vein thrombosis (DVT) or superficial thrombophlebitis were excluded from the study. Patients with congenital diseases associated with varicose veins, including Klippel-Trenaunay Syndrome, arterio-venous malformations, secondary varicose veins, or human immunodeficiency virus (HIV) infection were not included in the study.
In total, 80 participants with varicose veins and 35 with non-varicose vein samples were included in the study. Tissue and demographic details are provided in Supplementary Table 2. Samples were cut and prepared on dry ice in the class II vacuum hood in the Division of Computational and Systems Biology at Imperial College London. Samples were cut circumferentially and an average sample weight of 140+/− 5 mg was used. However, some non-varicose veins were of low weight especially inferior epigastric veins retrieved from hernia surgery and facial veins retrieved from carotid endarterectomy. The effect of different sample weight was compensated by respectively suspending the metabolites in different concentrations of solvents during their extractions process, such that the tissue to solvent ratio remained constant.
Chemicals used
The organic solvents used were chloroform, acetonitrile, DCM, ISP, hexane and MTBE (Sigma-Aldrich Gillingham, UK). Methanol (Fisher) and water were (Fluka) both LC-MS grade. Additionally, formic acid, leucine enkephalin, ammonium formate (HPLC grade) and sodium formate, were obtained from Sigma-Aldrich, Gillingham, UK. Deuteriated chloroform with 0.05% tetramethylsilane (TMS) was obtained from Goss Scientific, UK. Biological tissue was handled in Class II biological cabinet and local laboratory protocols were strictly followed. Chloroform was handled in safety cabinet due to hazardous risks involved with chloroform. Local laboratory safety precautions were followed while handling liquid nitrogen.
Extraction of metabolites
Metabolite extraction was performed as previously optimised in human vein tissue26. Aqueous and organic extractions were performed sequentially on the tissue samples. Organic extraction was performed first followed by aqueous extraction26 (please see Supplementary Information for the details of extraction protocol).
1H-NMR spectroscopic analysis of aqueous extract
Two aliquots holding dried aqueous extracts from 80 varicose vein and 35 non-varicose vein samples were prepared for NMR spectroscopy using the method previously by Anwar et al.26 (please see Supplementary information for the details of aqueous extracts preparation). Aqueous vein extracts were analysed using 1H NMR spectroscopy at a field strength of 14.1 T (1H 600.29 MHz) using a 5 mm broadband inverse configuration probe with a z axis magnetic field-gradient capability. The spectrometer was equipped with a Bruker Sample Jet system set to 5 mm shuttle mode with a cooling rack of refrigerated tubes at 6 °C and controlled via a Bruker Avance III console (Bruker, Rheinstetten, Germany). A standard 1D pulse sequence [recycle delay (RD)-90°-t1-90°-tm-90°-acquire free induction decay (FID)) with water suppression applied during RD of 2 s and tm was used to acquire data for each sample. A total of 512 scans were acquired into 64 K data points. The spectral width of δ1H 20.00 and acquisition time of 1.36 sec were employed for all aqueous experiments. Aqueous Spectra acquired from NMR spectroscopy were phased and calibrated using chemical shift of sodium 3-trimethylsilyl-1-[2,2,3,3,-2H4] propionate (TSP) at δ1H 0.00 in TOPSPIN 3.0 software (Bruker BioSpin, Rheinstetten, Germany). The baseline was corrected manually. Spectra were imported into MATLAB R2009b (Mathworks™, 2009) using an in-house algorithm built in MATLAB. The spectral region containing the water resonance (from δ1H 4.68 to 5.24) was removed from all spectra. In addition, the resonance of TSP (from δ1H −1 to 0.2) was removed from all spectra. All spectra were subsequently aligned and normalised using probabilistic quotient normalisation27. NMR assignments were performed by observing signal peak, chemical shift and comparing this to a list of standards available on central database HMDB, and an internal database. Statistical Total Correlation Spectroscopy (STOCSY)6, which computes the correlation between variables in the spectrum, by taking advantage of the multicollinear nature of NMR data, was also employed for the identification of related peaks, aiding the metabolite identification process.
UPLC-MS analysis of aqueous and organic extracts
UPLC-MS analysis was performed with the protocols previously published by our group26, 28. A total of 50 µl from each sample was pooled to make a quality control (QC) sample29. QC samples were injected every 10 samples throughout the run to monitor the stability of the UPLC-MS instrument. Details of samples preparation for UPLC-MS analysis is given in supporting information section. Organic extracts (lipid profiling) from 80 varicose vein and 35 non-varicose vein samples were analyzed as previously described28 using an Acquity UPLC system (Waters Ltd Elstree, UK) coupled to a Q-TOF Premier mass spectrometer (Waters Technologies, Ltd. Manchester, UK). For chromatography of organic extracts, an Acquity UPLC column CSH (1.7 µm, 2.1 × 100 mm, Waters, USA) was used. Analysis of aqueous extracts from 80 varicose vein and 24 non-varicose vein samples with hydrophilic interaction liquid chromatography (HILIC) was performed as previously described28 using an Acquity UPLC System (Waters, Ltd. Elstree, UK), coupled with LCT Premier mass spectrometer (Waters MS Technologies, Ltd., Manchester, U.K.). An Acquity UPLC BEH HILIC column (1.7 μm, 2.1 × 100 mm, Waters, USA) was used and maintained at 35 °C. Details of instruments settings (for organic and HILIC UPLC-MS analysis are provided in supplementary information section)28.
For UPLC-MS, data were processed using the XCMS package30 (version 1.46.0) in the R programing language (version 3.2.0). After peak-picking, retention time correction and grouping of the chromatographic peaks, an algorithm was used for filling zero values using the adjacent background intensity. The three-dimensional table produced consisted of features characterized by their m/z, retention time and signal intensity. The dataset was then subjected to total area normalization.
Metabolite structural assignments for UPLC-MS were conducted by: 1) matching accurate mass measurements to theoretical values from on-line databases including METLIN (http://metlin.scripps.edu/metabolites), HMDB (http://www.hmdb.ca) and LIPID MAPS, (http://www.lipidmaps.org), 2) isotopic patterns, 3) in-house developed libraries of standards28 and 4) MSE and/or MS/MS spectra, by matching to tandem MS experiments from online databases.
Multivariate statistical analysis
SIMCA-P+ 12.5 statistical software (UMETRICS™, Sweden) was used to analyse the data acquired from UPLC-MS, whereas the NMR data was analysed using MATLAB R2009b (Mathworks™, 2009) with scripts developed in-house. Both principal components analysis (PCA) and orthogonal projection to latent structures discriminant analysis (OPLS-DA) were performed. The validity of the OPLS models were assessed by performing 1000 permutations with randomized scrambling of class labels. OPLS loadings S-plots derived from the OPLS-DA models were used to discern the variables driving group differences. Further univariate analysis of variance (one-way ANOVA) was performed on the complete set of data to identify the individual effects of each variable; Benjamini-Yekutieli correction was used to adjust the significance statistics to account for multiple testing31. Only the biomarkers which were identified by both the validated multivariate OPLS-DA models and adjusted univariate statistics were considered to be significant overall.
A multivariate partial least square (PLS) regression modelling approach was used to identify any influences of demographic and clinical confounders (Table 2) on the both aqueous NMR and non-polar UPLC-MS metabolic vein tissue profiles. For each factor (age, sex, diabetes, etc.) a PLS model was constructed whereby the y matrix was replaced with the factor under investigation instead of the sample class. Q2Y, R2X values and p values (relating to the predictive variability, variance explained and significance of the model respectively) for each model were calculated.
miRNA analysis
A subset of tissue samples (8 test subjects and 8 controls) were analysed for microRNA expression, by Miltenyi Biotec (Bergisch Gladbach, Germany). These were chosen randomly from the cohort of 115 samples prepared for metabonomic analysis. Samples underwent RNA extraction using TRIzol (Invitrogen) under RNase-free conditions. Details of experimental protocol are given in supplementary information section. The quantity and quality of the RNA extraction was investigated by bioanalyzer and the generation of an RNA Integrity Number (RIN). A RIN value of greater than 6 was considered to be acceptable and based on this six samples (3 test and 3 control) of the original 16 samples were rejected. Electropherograms of the accepted samples indicated that there was good signal intensity in each of the sample runs. The fold change ratios of expression were calculated based on light intensity (light units) after hybridisation of the extracted RNA onto microarrays.
Functional analysis of the identified miRNA targets was performed using computational methods. Initial confirmation of miRNA nomenclature was achieved using miRNA database mirBase (www.mirbase.org), after which the miRNAs were ranked according to experimental versus control expression ratios. The 10 most correlated and the 10 most anti-correlated miRNAs were then examined for their expected regulated cellular processes using the DIANALAB database, which provided a list of potential KEGG pathways, ranked by statistical confidence. Prediction modelling tool TargetScan was then used to produce a list of mRNAs under the regulation each of the target miRNAs, which was subsequently imported into the PANTHER classification system to produce a list of pathways containing the mRNAs in question. These pathways were analysed using a statistical overrepresentation analysis to produce a p-value to identify which ones were statistical significant for the experiment. All processes that were statistically significant were incorporated into Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com/products/ipa) to create a network biology system for the interaction between both the miRNAs and the metabolites identified from the metabonomic analysis. A Pearson correlation matrix was constructed to identify which miRNA expression levels correlated with concentrations of the previously identified significant metabolites. Finally, the results from all the above analyses were combined into a single IPA pathway mapping analysis, which mapped the biological functions of the differential metabolites and miRNAs.
Electronic supplementary material
Acknowledgements
We would like to acknowledge the support of the European Venous Forum Research Grant and the Graham Dixon Charitable Trust Grant. This research was supported by the National Institute Health Research Centre based at Imperial College Healthcare Trust and Imperial College London. The views expressed are those of the authors and not necessarily those of the NHS, NIHR and department of Health. We also acknowledge Mr Tristan Lane and Mr Chung Sim Lim for their support in providing vein tissues for the research.
Author Contributions
M.A. conceive the study design with E.H., A.H.D. and P.A.V. M.A. and K.A.-G. contribute equally in conducting the experiments, analysis of the data and writing the manuscript and preparing the figures. K.S., P.A.V. and J.L. conducted some of the experiments and perform data analysis. J.S. recruited patients for the study and collected tissue samples. E.H. and A.H.D. critically appraise the study design and results. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Muzaffar A. Anwar and Kyrillos N. Adesina-Georgiadis contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02529-y
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lim CS, Davies AH. Pathogenesis of primary varicose veins. Br J Surg. 2009;96(11):1231–1242. doi: 10.1002/bjs.6798. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 3.Clayton TA, et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 4.Beckonert O, et al. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc. 2010;5:1019–1032. doi: 10.1038/nprot.2010.45. [DOI] [PubMed] [Google Scholar]
- 5.Anwar MA, et al. In-vitro Identification of Distinctive Metabolic Signatures of Intact Varicose Vein Tissue via Magic Angle Spinning Nuclear Magnetic Resonance Spectroscopy. Eur J Vasc Endovasc Surg. 2012;44:442–450. doi: 10.1016/j.ejvs.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Coen M, et al. Heteronuclear 1H-31P statistical total correlation NMR spectroscopy of intact liver for metabolic biomarker assignment: application to galactosamine-induced hepatotoxicity. Anal Chem. 2007;79:8956–8966. doi: 10.1021/ac0713961. [DOI] [PubMed] [Google Scholar]
- 7.Newsholme P, Procopio J, Lima MM, Pithon-Curi TC, Curi R. Glutamine and glutamate–their central role in cell metabolism and function. Cell biochemistry and function. 2003;21:1–9. doi: 10.1002/cbf.1003. [DOI] [PubMed] [Google Scholar]
- 8.Newsholme P, Costa Rosa LF, Newsholme EA, Curi R. The importance of fuel metabolism to macrophage function. Cell biochemistry and function. 1996;14:1–10. doi: 10.1002/cbf.644. [DOI] [PubMed] [Google Scholar]
- 9.O’Neill AJ, et al. Glutathione depletion-induced neutrophil apoptosis is caspase 3 dependent. Shock. 2000;14:605–609. doi: 10.1097/00024382-200014060-00006. [DOI] [PubMed] [Google Scholar]
- 10.Ripps H, Shen W. Review: taurine: a “very essential” amino acid. Molecular vision. 2012;18:2673–2686. [PMC free article] [PubMed] [Google Scholar]
- 11.Das J, Sil PC. Taurine ameliorates alloxan-induced diabetic renal injury, oxidative stress-related signaling pathways and apoptosis in rats. Amino Acids. 2012;43:1509–1523. doi: 10.1007/s00726-012-1225-y. [DOI] [PubMed] [Google Scholar]
- 12.Eagle H, Oyama VI, Levy M, Freeman A. Myo-inositol as an essential growth factor for normal and malignant human cells in tissue culture. Science. 1956;123:845–847. doi: 10.1126/science.123.3202.845-a. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka H, et al. Imaging mass spectrometry reveals unique lipid distribution in primary varicose veins. Eur J Vasc Endovasc Surg. 2010;40:657–663. doi: 10.1016/j.ejvs.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Shakor AB, et al. Sphingomyelin synthase 1-generated sphingomyelin plays an important role in transferrin trafficking and cell proliferation. J Biol Chem. 2011;286:36053–36062. doi: 10.1074/jbc.M111.228593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ratnam S, Kent C. Early increase in choline kinase activity upon induction of the H-ras oncogene in mouse fibroblast cell lines. Archives of biochemistry and biophysics. 1995;323:313–322. doi: 10.1006/abbi.1995.9959. [DOI] [PubMed] [Google Scholar]
- 16.Glunde K, et al. Hypoxia regulates choline kinase expression through hypoxia-inducible factor-1 alpha signaling in a human prostate cancer model. Cancer Res. 2008;68:172–180. doi: 10.1158/0008-5472.CAN-07-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schrader J, Haddy FJ, Gerlach E. Release of adenosine, inosine and hypoxanthine from the isolated guinea pig heart during hypoxia, flow-autoregulation and reactive hyperemia. Pflugers Archiv: European journal of physiology. 1977;369:1–6. doi: 10.1007/BF00580802. [DOI] [PubMed] [Google Scholar]
- 18.Diguet N, et al. Muscle creatine kinase deficiency triggers both actin depolymerization and desmin disorganization by advanced glycation end products in dilated cardiomyopathy. J Biol Chem. 2011;286:35007–35019. doi: 10.1074/jbc.M111.252395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huitema K, van den Dikkenberg J, Brouwers JF, Holthuis JC. Identification of a family of animal sphingomyelin synthases. The EMBO journal. 2004;23:33–44. doi: 10.1038/sj.emboj.7600034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niizeki T, et al. Diacylglycerol kinase-epsilon restores cardiac dysfunction under chronic pressure overload: a new specific regulator of Galpha(q) signaling cascade. Am J Physiol Heart Circ Physiol. 2008;295:H245–255. doi: 10.1152/ajpheart.00066.2008. [DOI] [PubMed] [Google Scholar]
- 21.Gatt S. Enzymic Hydrolysis and Synthesis of Ceramides. J Biol Chem. 1963;238:3131–3133. [PubMed] [Google Scholar]
- 22.Horinouchi K, et al. Acid sphingomyelinase deficient mice: a model of types A and B Niemann-Pick disease. Nat Genet. 1995;10:288–293. doi: 10.1038/ng0795-288. [DOI] [PubMed] [Google Scholar]
- 23.Chao R, Khan W, Hannun YA. Retinoblastoma protein dephosphorylation induced by D-erythro-sphingosine. J Biol Chem. 1992;267:23459–23462. [PubMed] [Google Scholar]
- 24.Raffetto JD, et al. Functional adaptation of venous smooth muscle response to vasoconstriction in proximal, distal, and varix segments of varicose veins. J Vasc Surg. 2010;51:962–971. doi: 10.1016/j.jvs.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eklof B, et al. Revision of the CEAP classification for chronic venous disorders: consensus statement. J Vasc Surg. 2004;40:1248–1252. doi: 10.1016/j.jvs.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 26.Anwar MA, et al. Optimization of metabolite extraction of human vein tissue for ultra performance liquid chromatography-mass spectrometry and nuclear magnetic resonance-based untargeted metabolic profiling. The Analyst. 2015;140:7586–7597. doi: 10.1039/C5AN01041A. [DOI] [PubMed] [Google Scholar]
- 27.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 28.Vorkas PA, et al. Untargeted UPLC-MS profiling pipeline to expand tissue metabolome coverage: application to cardiovascular disease. Anal Chem. 2015;87:4184–4193. doi: 10.1021/ac503775m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spagou K, et al. HILIC-UPLC-MS for exploratory urinary metabolic profiling in toxicological studies. Anal Chem. 2011;83:382–390. doi: 10.1021/ac102523q. [DOI] [PubMed] [Google Scholar]
- 30.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Yoav YD. The control of the false discovery rate in multiple testing under dependency. Ann. Statist. 2001;29:1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


