Abstract
Freshwater representatives of Rhodophyta were sampled and the complete chloroplast and mitochondrial genomes were determined. Characteristics of the chloroplast and mitochondrial genomes were analyzed and phylogenetic relationship of marine and freshwater Rhodophyta were reconstructed based on the organelle genomes. The freshwater member Compsopogon caeruleus was determined for the largest chloroplast genome among multicellular Rhodophyta up to now. Expansion and subsequent reduction of both the genome size and GC content were observed in the Rhodophyta except for the freshwater Compsopogon caeruleus. It was inferred that the freshwater members of Rhodophyta occurred through diverse origins based on evidence of genome size, GC-content, phylogenomic analysis and divergence time estimation. The freshwater species Compsopogon caeruleus and Hildenbrandia rivularis originated and evolved independently at the inland water, whereas the Bangia atropurpurea, Batrachospermum arcuatum and Thorea hispida are derived from the marine relatives. The typical freshwater representatives Thoreales and Batrachospermales are probably derived from the marine relative Palmaria palmata at approximately 415–484 MYA. The origin and evolutionary history of freshwater Rhodophyta needs to be testified with more organelle genome sequences and wider global sampling.
Introduction
Rhodophyta is an anciently derived lineage, constituting one of the primary plastids-bearing hosts, and provides plastids for the secondary or tertiary endosymbiosis1. This phylum comprise two subphyla with seven classes2: the subphylum Cyanidiophytina composed of class Cyanidiophyceae, the other subphylum Rhodophytina composed of classes Florideophyceae, Bangiophyceae, Compsopogonophyceae, Stylonematophyceae, Rhodellophyceae and Porphyridiophyceae. Rhodophyta are primarily marine in distribution, with less than 3% of the over 6500 species occurring in truly freshwater habitats3, 4. Freshwater red algae are usually important constituents of stream floras, either in terms of abundance or distribution from local scale to biomes. Members of freshwater Rhodophyta cover six classes (Bangiophyceae, Compsopogonophyceae, Florideophyceae, Porphyridiophyceae, Stylonematophyceae and Cyanidiophyceae) of total seven in the Rhodophyta except for the Rhodellophyceae5, with some lineages exclusively inhabit in freshwater such as the Batrachospermales and Thoreales in Florideophyceae. Red algae in freshwater habitats tend to be macroscopic and benthic, meanwhile exhibiting a smaller size range than the marine members6. Their morphology varied from unicellular to multicellular forms including tufts, crusts and filaments. Reproductive types of freshwater red algae exhibit a diversity including cell division, monosporangia formation, carpogonium and spermatangium, and the last occurred during sexual reproduction and life history alternation5. The probable origin of these freshwater forms among the red algae, whether they were dwellers of inland waters or immigrants from the sea, is still in debate. Skuja considered the Cryptonemiales, Nemastomales, and especially of the Ceramiales representing higher types in freshwater Rhodophyta probably are migrants from the sea while the other members are primary types and have arisen from original inhabitants of inland waters based on the morphological and ecological features7. However, there are few molecular evidence for the origin of freshwater Rhodophyta up to now.
Chloroplast genomes of the red algae have high gene capacity and compact structure8, 9, and the mitochondria genomes of Florideophyceae are highly conserved despite a wide variety of morphological divergence10. Both chloroplast and mitochondrial genomes have been applied in the phylogenomic analysis of Rhodophyta10–12, and all have proved to capable to elucidate phylogenetic relationships at deep and terminal branches, thus reflecting evolutionary history between Rhodophyta and other eukaryotic lineages. The organelle genomes provide insights not only on phylogenetic relationship but also other features (genome size and structure, GC content, gene loss and genome synteny) involving in the genome evolution13. Other studies concerning organelle genomes of algae have also proved that organelle genomes can provide new insights into organelle function and evolution14. Furthermore the non-recombinant nature of organelles makes them good tools for inferring ancient phylogenetic relationships15. Considering the evolutionary status of Rhodophyta in plant kingdom, the chloroplast and mitochondrial genome evolution in Rhodophyta is of great importance to understand the organelle evolution in general.
Here we sequenced the organelle genomes of three members in freshwater Rhodophyta and analyzed their unique architecture and gene content features. Additionally, the available genome sequence of freshwater Rhodophyta in the GenBank were combined together to construct a phylogeny of Rhodophyta based on genome-wide information. We proposed a primary inference on the origin and evolutionary history of freshwater Rhodophyta, which needs to be testified with more sequence data and wider sampling.
Results
Organization and gene content of organelle genomes for freshwater Rhodophyta representatives
The resulting assembly generated complete circular genomes for organelles of all the three freshwater species. Batrachospermum arcuatum Kylin generated a contig of 187,354 bp with 449 coverage for chloroplast and 25,086 bp with 757 coverage for mitochondria (Fig. 1a for chloroplast, b for mitochondria). Thorea hispida Desvaux generated a contig of 175,278 bp with 700 coverage for chloroplast and 25,380 bp with 300 coverage for the mitochondria (Fig. 1c for chloroplast, d for mitochondria). Compsopogon caeruleus Montagne generated a contig of 221,013 bp with 880 coverage for chloroplast, 26,807 bp with 1130 coverage for mitochondria (Fig. 1e for chloroplast, f for mitochondria). GC contents of the chloroplast are 29.9%, 28.3% and 26.0%, and of mitochondria are 29.7%, 28.2% and 27.4%, respectively for Batrachospermum arcuatum, Thorea hispida and Compsopogon caeruleus.
Figure 1.
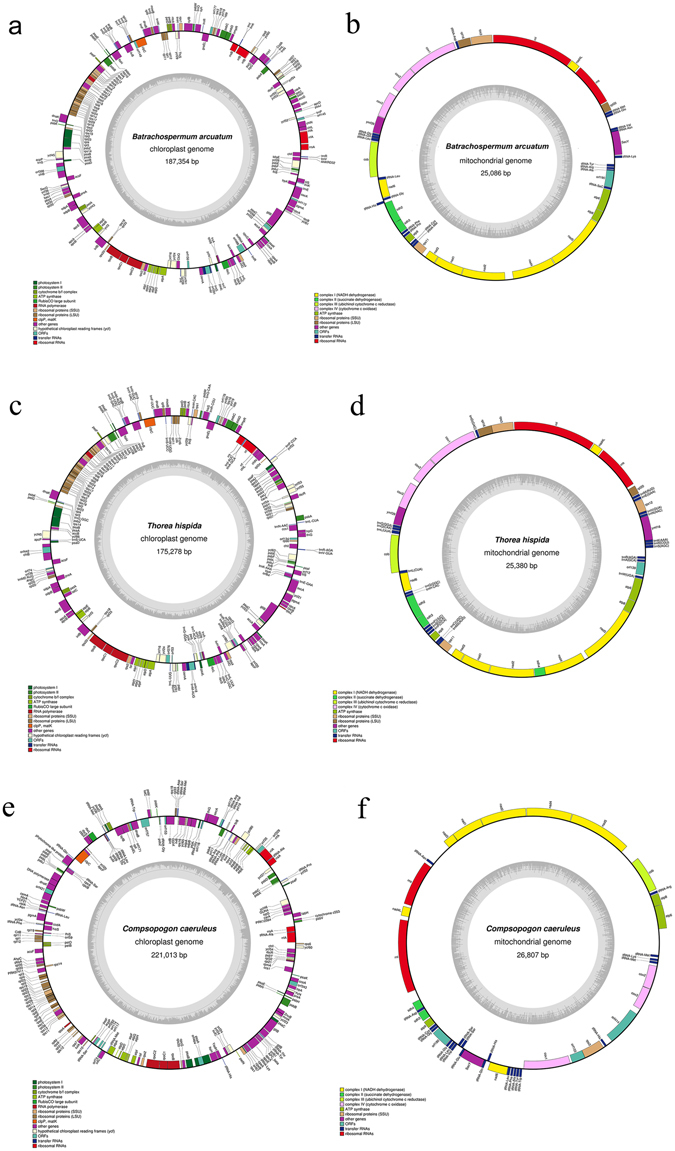
Organelle genome maps of freshwater Rhodophyta representatives. The genes inside and outside of the circles are transcribed in the clockwise and counterclockwise directions, respectively. Genes belonging to different functional groups are shown in different colors. (a) Chloroplast genome map of Batrachospermum arcuatum. (b) Mitochondrial genome map of Batrachospermum arcuatum. (c) Chloroplast genome map of Thorea hispida. (d) Mitochondrial genome map of Thorea hispida. (e) Chloroplast genome map of Compsopogon caeruleus. (f) Mitochondrial genome map of Compsopogon caeruleus.
The Batrachospermum arcuatum chloroplast genome encodes 221 genes including 185 protein coding genes, 31 tRNAs, 6 rRNAs, 1 ncRNA (rnpB) and 1 tmRNA (Suppl. Table S1), and the mitochondria genome encodes 43 genes including 23 protein coding genes, 20 tRNAs and 2 rRNAs (Suppl. Table S2). The Thorea hispida chloroplast genome encodes 228 genes including 193 protein coding genes, 30 tRNAS, 3 rRNAs, 1 ncRNA (rnpB) and 1 tmRNA (Suppl. Table S3), and the mitochondria genome encodes 49 genes including 25 protein coding genes, 22 tRNAs and 2 rRNAs (Suppl. Table S4). The Compsopogon caeruleus chloroplast genome encodes 229 genes including 211 protein coding genes, 20 tRNAs and 5 rRNAs (Suppl. Table S5), and the mitochondria genome encodes 45 genes including 21 protein coding genes, 22 tRNAs and 2 rRNAs (Suppl. Table S6).
Two introns were discovered in genes chlB and trnM for the chloroplast genome of Batrachospermum arcuatum and Thorea hispida. 15 introns were discovered in the chloroplast genome of Compsopogon caeruleus distributed in genes of odpB, clpC, phytochrome-like protein, pgmA, AhpC, rp12, rps3, infC, atpF, atpI, rpoC1, psaA, psaC, psbB and dnaK. No introns were found in the mitochondrial genomes of the three species.
Loss of tRNAs in the freshwater Rhodophyta members
Necessary tRNAs except trnN and trnP were found in the chloroplast genome of Batrachospermum arcuatum, whereas for the mitochondria genome absence of trnR, trnI, trnT, trnW and trnY was observed. For Thorea hispida, all necessary tRNAs were found in the chloroplast genomes whereas absence of trnI, trnT and trnY occurred in the mitochondria genome. In Compsopogon caeruleus, necessary tRNAs except trnE and trnI were found in the chloroplast genome and in the mitochondria genome the trnI and trnT have been lost.
Synteny analysis and structure comparison
The chloroplasts of the freshwater representatives (Compsopogon caeruleus, Bangia atropurpurea, Hildenbrandia rivularis, Batrachospermum arcuatum, Kumanoa americana) were poorly collinear, whereas the collinear alignment structure was observed for the mitochondrial genomes, particularly in the florideophyceae members (Crossing “X” pattern observed in lines connecting each colored bars was due to different annotation staring point) (Fig. 2). The mitochondrial genomes of Bangiophyceae and Compsopogonophyceae exhibited less collinear structure with that of the Florideophyceae. On the other hand, it was observed that freshwater and marine representatives from each Rhodophyta lineages shared high synteny when comparing their chloroplast genomes (Fig. 3). For the Compsopogonophyceae chloroplast, the freshwater and marine representatives all own 2 rRNA operons, which were identical in sequence while opposite in direction. A trnA was distributed in the region between 16 S rRNA and 23 S rRNA. The two marine members, Erythrotrichia carnea and Rhodochaete parvula share highly conservative structure, and the freshwater member Compsopogon caeruleus owned inverse SSC structure and partial inverse LSC rearrangement compared with the marine members (Fig. 3a). For the Bangiophyceae lineage, chloroplast genomes between freshwater and marine members had little structural rearrangements except for the loss of one rRNA operon in the freshwater member Bangia atropurpurea and the two rRNA operons in the marine members were arranged directly. Two tRNAs including trnI and trnA were distributed in the region of 16 S rRNA and 23 S rRNA (Fig. 3b). For the Hildenbrandiales in Florideophyceae, both freshwater and marine representatives own only one rRNA operon. Two rearrangements occurred between the marine members Hildenbrandia rubra and Apophlaea sinclairii and two larger blocks are rearranged between H. rubra and H. rivularis. One trnA was distributed in the region of 16 S rRNA and 23 S rRNA (Fig. 3c). For the Nemaliophycidae in Florideophceae, the marine representative owned two inverse rRNA operons and the freshwater members owned one or two rRNA operons. The freshwater member Thorea hispida shared highly collinear structure with the marine member Palmaria palmate except the loss of one rRNA operon. Chloroplast of Kumanoa americana owned one rRNA operon and was collinear with Thorea hispida except for the inverse arrangement of the SSC region. The Batrachospermum arcuatum were collinear with Kumanoa americana but gain one more rRNA operon and two rRNA operons were inverse arranged. Two tRNAs including trnI and trnA were distributed in the region between 16 S rRNA and 23 S rRNA (Fig. 3d).
Figure 2.
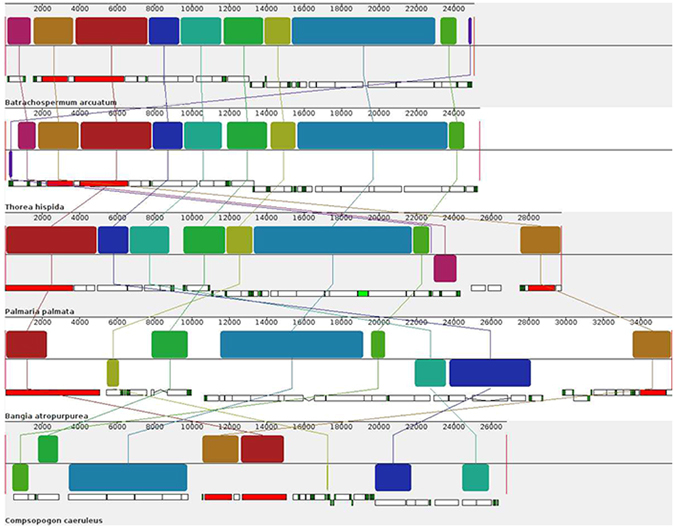
Synteny alignment of mitochondrial genomes between freshwater representatives. Each colored bar represents a locally collinear block and is assigned a unique color. White, red and green boxes represent annotated CDS (protein-coding sequence), rRNAs and tRNAs in the genomes respectively. Sequences each colored bar covered are homologous among the aligned genomes. The colored lines connect similar colored blocks and indicate which regions in each genome are homologous.
Figure 3.
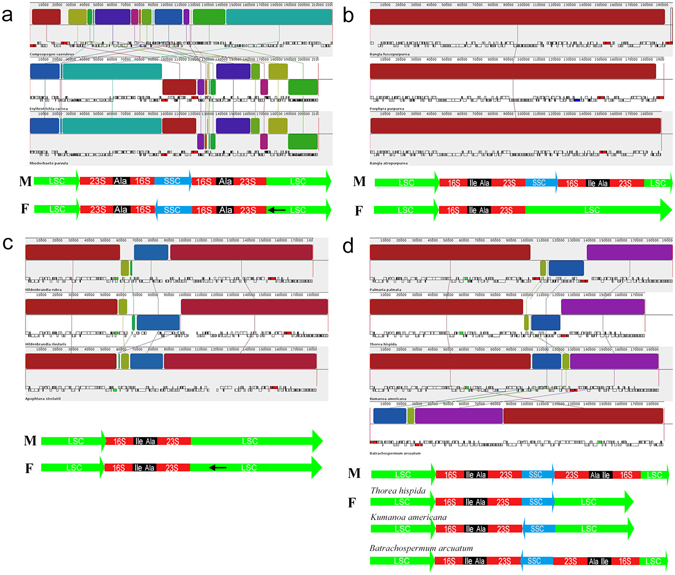
Synteny alignment of chloroplast genomes between freshwater and marine representatives. (a) Synteny alignment of chloroplast genomes between freshwater and marine Compsopogonophyceae. (b) Synteny alignment of chloroplast genomes between freshwater and marine Bangiophyceae. (c) Synteny alignment of chloroplast genomes between freshwater and marine Hildenbrandiophyceae. (d) Synteny alignment of chloroplast genomes between freshwater and marine Florideophyceae. Note: Lines with “M” and “F” mean types of chloroplast genome architecture for each Rhodophyta lineage. M represent marine species and F represent freshwater species; LSC and SSC represent genomic regions coding for large single copy and small single copy; 23 S and 16 S represent genes coding for 23 S rRNA and 16 S rRNA; Ala and Ile represent genes coding for tRNA-Ala and tRNA-Ile; Black arrows represent partial inverse of the LSC region. Lines connecting each colored bar with crossing “X” pattern represent genomic rearrangements or collinear structure annotated at different starting point.
Phylogenetic relationship of freshwater Rhodophyta
Both methods including Bayesian inference and maximum likelihood used for phylogenetic tree construction produced similar topologies, and only the Bayesian trees were showed with the Bayesian posterior probabilities and ML bootstrap supporting values labeled on the nodes (Fig. 4). Phylogeny based on chloroplast genome (Fig. 4a) revealed the freshwater members of each Rhodophyta lineage were distributed among the tree with robust supporting values. With the Cyanidiophyceae as the outgroup, the Porphyridiophyceae clustering together with the Compsopogonophyceae formed the basal branch, and the freshwater representative Compsopogon caeruleus was basal to the marine members Erythrotrichia carnea and Rhodochaete parvula. The second main branch contains classes of Bangiophyceae and Florideophyceae with the Bangiophyceae at the basal position. The freshwater representative in Bangiophyceae lineage, Bangia atropurpurea formed sister group with the marine member Porphyra purpurea. In the Hildenbrandiophycidae of Florideophyceae class, the freshwater representative Hildenbrandia rivularis was basal to the marine Hildenbrandia rubra and Apophlaea sinclairii, together formed the basal branch of Florideophyceae lineage. The typical freshwater members, Batrachospermales and Thoreales belonging to the Nemaliophycidae clustered together with the marine member Palmaria palmate. The Nemaliophycidae branch was basal to the other subclasses in Florideophyceae. As revealed by the genome size labeled on the branch, the Cyanidiophyceae lineage owned smallest chloroplast genome size, whereas the Porphyridiophyceae lineage owned the largest genome size. The genome size was decreasingly gradually from Porphyridiophyceae, Compsopogonophyceae, Bangiophyceae to Florideophyceae. In the Compsopogonophyceae lineage, the freshwater member Compsopogon caeruleus was slightly larger in chloroplast genome size than the marine members. In Bangiophyceae, freshwater representative owned smaller genome size than the marine representatives. In the Hildenbrandiophycidae, the freshwater H. rivularis are slightly larger than the marine member Hildenbrandia rubra and Apophlaea sinclairii in chloroplast genome size. In the Nemaliophycidae, the freshwater members Thorea hispida, Batrachospermum arcuatum and Kumanoa americana were slightly smaller than the marine member Palmaria palmata in chloroplast genome size. According to the results of the Spearman’s rank correlation tests, divergence of chloroplast genome size was principally caused by the variance of noncoding regions (r = 0.81, p = 0.000) and intron regions (r = 0.53, p = 0.000), whereas the length of protein-coding regions showed no significant difference in each Rhodophyta lineage. In Cyanidiophyceae, GC content of genus Galdieria was lower than other lineages. The GC content of Porphyridiophyceae, marine Compsopogonophyceae and Bangiophyceae were higher than the Florideophyceae. Totally, in all Rhodophyta lineages, freshwater representatives owns lower GC content than the marine representatives except for Hildenbrandiophycidae in Florideophyceae. Notably, the freshwater Compsopogonophyceae, Compsopogon caeruleus owned the lowest GC content in all the available Rhodophyta taxa.
Figure 4.
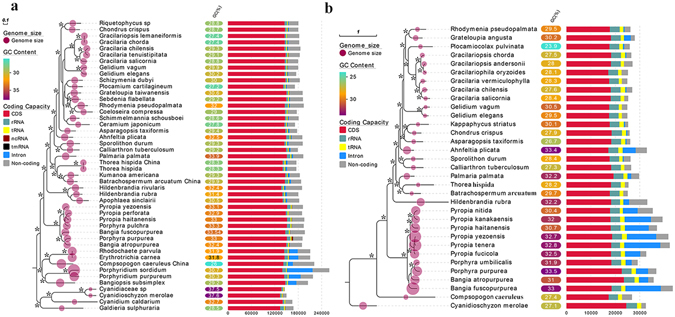
Bayesian phylogenetic trees based on organelle genomes, with each genomic features labeled on the tree. (a) Bayesian phylogenetic tree based on chloroplast genome. (b) Bayesian phylogenetic tree based on mitochondrial genome. Note: Asterisks on the nodes represent supporting values of Bayesian inference for 100% and Maximum Likelihood bootstrap more than 80%.
Phylogeny based on mitochondria genome produced similar relationship with the chloroplast revealed (Fig. 4b). With no Porphyridiophyceae lineage organelle genome information, the Compsopogon caeruleus was at the basal position. The Bangiophyceae and Florideophyceae formed two main classes with full supporting values. The freshwater Bangia atropurpurea was distributed between the marine members Bangia fuscopurpurea and genus Porphyra. In Florideophyceae lineage, the freshwater Thorea hispida and Batrachospermum arcuatum formed a branch with full supporting values. This branch again clustered together with Palmaria palmata. Mitochondrial genome size of Florideophyceae was significantly smaller than that of Bangiophyceae according to the Mann-Whitney U test (z = −4.268, p = 0.000), and the genome size of Bangiophyceae were larger than that of Compsopogonophyceae and Cyanidiophyceae. The typical freshwater representatives, Batrachospermum arcuatum and Thorea hispida were smaller in mitochondrial genome size than its sister taxa, the marine Palmaria palmata. The GC content of the three freshwater representatives were lower than the marine members of each lineage.
Divergence time estimation
Divergence time estimation based on chloroplast genome (Fig. 5a) showed that the Porphyridiophyceae, Stylonematophyceae and Compsopogonophyceae diverged first, following the divergence of Bangiophyceae and Florideophyceae. In the Florideophyceae, the Hildenbrandiophycidae diverged first and the Nemaliophycidae followed. The freshwater member Compsopogon caeruleus diverged with marine members in class Compsopogonophyceae at 547 MYA (95% HPD: 384–777 MYA). The class Bangiophyceae diverged at approximate 844 MYA (95% HPD: 704–1030 MYA), and freshwater Bangia atropurpurea diverged later (approximately 114 MYA) than the marine Bangia fuscopurpurea (approximately 130 MYA). In Hildenbrandiophycidae of the class Florideophyceae, the freshwater H. rivularis diverged earlier at 408 MYA (95% HPD: 305–489 MYA) and marine H. rubra diverged at 325 MYA (95% HPD: 251–406 MYA). The typical freshwater representative Thorea hispida diverged from Batrachospermales at approximately 484 MYA (95% HPD: 399–538 MYA) and Batrachospermum arcuatum diverged at 231 MYA (95% HPD: 152–373 MYA).
Figure 5.
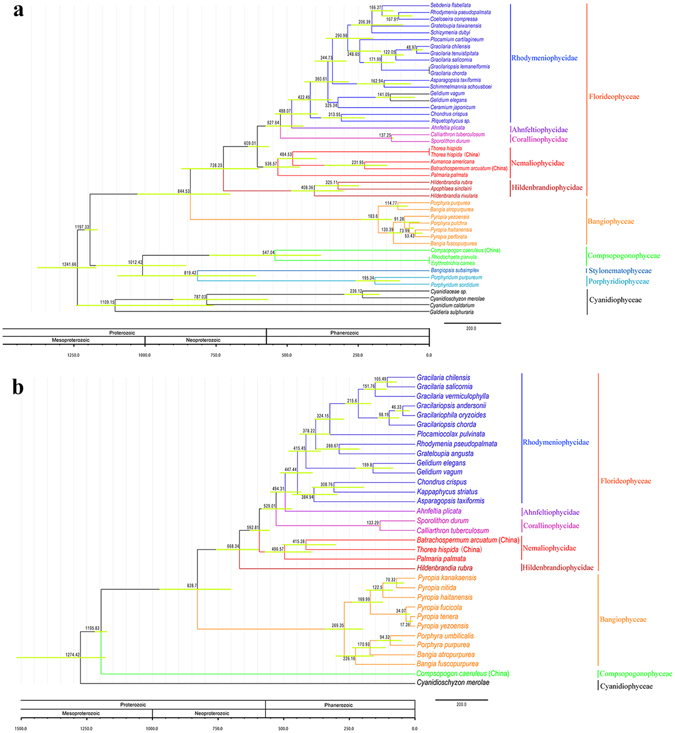
Divergence time estimation phylogram based on organelle genome sequence. Numbers at the nodes represent the mean estimated divergence time with the corresponding 95% highest posterior density (HPD) of the nodes indicated by horizontal bars. The geological timescale is given in million years ago (MYA). (a) Divergence time estimation tree based on the chloroplast genome sequence. (b) Divergence time estimation tree based on the mitochondrial genome sequence.
As illustrated by the divergence time estimation based on mitochondrial genome (Fig. 5b), the divergence time of lineages Bangiophyceae and Florideophyceae was approximately similar to the result revealed by chloroplast genome (828 MYA vs 844 MYA for Bangiophyceae). However, due to the absent information of Porphyridiophyceae and Stylonematophyceae mitochondrial genomes, the class Compsopogonophyceae was at the basal position, and divergence time estimation of Compsopogon caeruleus was evidently earlier (1195 MYA) than the result of chloroplast genome (547 MYA). The divergence time of freshwater Bangiophyceae taxa Bangia atropurpurea (170 MYA) was earlier than the result of chloroplast (114 MYA). The divergence between marine Palmaria palmata and freshwater Thorea hispida and Batrachospermum arcuatum was 496 MYA, slightly later than the result of chloroplast (536 MYA).
Gene loss between the freshwater and marine Rhodophyta
Unique chloroplast gene content of freshwater and marine representatives for each Rhodophyta lineage were illustrated in Fig. 6. Generally, ORF (open reading frame) and ycf (unknown reading frames) constituted most of the unique genes. Common genes loss, trx (thioredoxins) and grx (glutaredoxins) were noticed between the freshwater and marine members of Compsopogonophyceae and Florideophyceae (Fig. 6a,d). In Compsopogonophyceae, the freshwater representative Compsopogon caeruleus own unique protein-coding genes (GNU4, PRK12564) similar to cyanobacteria (Fig. 6a). For Bangiophyceae and Hildenbrandiophyceae, no special protein-coding genes were found in the freshwater members (Fig. 6b,c). Meanwhile, high conservation in mitochondrial gene contents were observed in Rhodophyta except that the Compsopogon caeruleus lost all the rpl genes (ribosomal protein large subunit).
Figure 6.
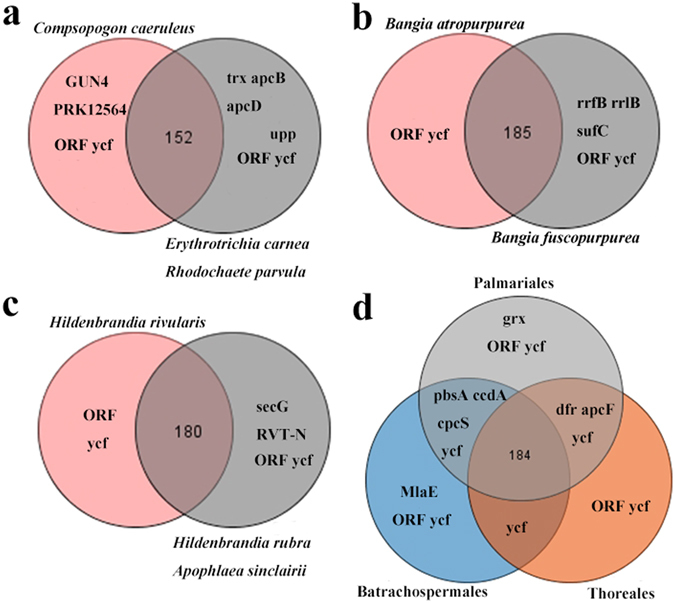
Venn diagram showing shared and unique gene content of each Rhodophyta class. (a) Venn diagram of gene content for the class Compsopogonophyceae. (b) Venn diagram of gene content for the class Bangiophyceae. (c) Venn diagram of gene content for the class Hildenbrandiophyceae. (d) Venn diagram of gene content for the class Florideophyceae. Note: Circles in gray represent marine members and circles in other colors represent freshwater members in each Rhodophyta lineage.
Substitution rates estimation of gene groups
Ratios of non-synonymous and synonymous substitutions of per gene group were calculated with the Galdieria sulphuraria and Cyanidioschyzon merolae as references (Suppl. Tables S7 and S8), as illustrated in Fig. 7. The ratios of non-synonymous and synonymous substitution for chloroplast gene groups including psa, psb and rbc were between 0 and 0.5, with psa the largest and psb the lowest. The ratios of apc and rpl were between 0.5 and 1.0 and the ratios of atp, rps and rpo were evidently higher with some species exceeding 1.0 (Fig. 7a). For mitochondrial gene groups, the ratio of cob was below 0.5, of the nad was around 0.5. Ratio of the cox was between 0.5 and 1.0 (except for genus Pyropia) and of the atp and sdh were higher than 1.0 (Fig. 7b). The difference of the nucleotide substitution rates among diverse gene groups were significant (χ 2 = 342.956, p = 0.000 for chloroplast gene groups; χ 2 = 147.493, p = 0.000 for mitochondria gene groups).
Figure 7.
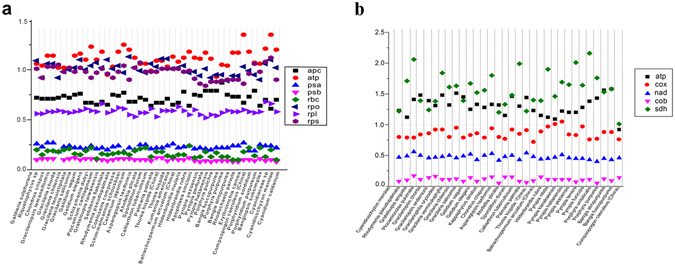
Substitution rates of organelle gene groups. (a) Substitution rates of chloroplast gene groups. (b) Substitution rates of mitochondrial gene groups.
Discussion
Of all the reported complete chloroplast genomes of multi-cellular freshwater Rhodophyta, Compsopogon caeruleus owned the largest genome size, and the chloroplast genome of Batrachospermum arcuatum and Thorea hispida were in the range of Florideophyceae9, 12, 16, 17. A possible history of expansion and subsequent contraction for the mitochondrial genome size has been discovered in green plant Cucurbitaceae18. Meanwhile, it has been proposed that all organelles undergo a genetic reduction, independently of their phylogenetic origin19. And the selection for small genomes contributes to the genome reduction19. Expansion and subsequent reduction of organelle genome size was observed in the red algae. The large-genome representatives Porphyridiophyceae, Stylonematophyceae and Compsopogonophyceae were more limited in species richness whereas the small-genome size representatives Bangiophyceae and Florideophyceae comprise greater than 98% of red algal diversity4. From the perspective of genomic size evolution, the freshwater members Compsopogon caeruleus and Hildenbrandia rivularis represented the primary forms in each class, and the Bangia atropurpurea, Batrachospermum arcuatum and Thorea hispida represented the evolved forms in each class.
Evidence have suggested that the pattern of spontaneous mutations is biased towards AT nucleotides in eukaryotes as well as Prokaryotes, thus decreasing the GC content20, 21. Šmarda et al. also argued the reduction process of genomic GC-content was probably ongoing and evolutionarily young22. The genomic GC content was decreasing with the evolution of Rhodophyta lineages, which was consistent with the general trend. However, the GC content of Compsopogon caeruleus was unexpectedly low contrasting its ancestral origin. Compsopogon caeruleus propagate by asexual reproduction and lack genetic recombination. Its low genetic variation has been found in previous record23. It is widely accepted that there exists positive correlation between genetic recombination and GC content24. Thus we speculated that the notably low GC content of Compsopogon caeruleus was caused by lack of genetic recombination. Considering the high GC content of Hildenbrandia rivularis comparing with its marine relatives, the freshwater Hildenbrandia rivularis represented the ancestral status. Whereas the contrary situation was observed in freshwater Bangia atropurpurea, Thorea hispida and Batrachoseprmum arcuatum, revealing their derived evolutionary stage.
Most of the early-diverged classes in Rhodophyta contained two copies of the ribosomal DNA operon, whereas single rDNA was found in most florideophycean species12. However, two inverted rDNAs were detected in the freshwater florideophycean taxa Batrachospermum arcuatum, whearas its sister genera Kumanoa americana and Thorea hispida owned single rDNA operon. It suggested that the loss and acquisition of rDNA in Rhodophyta were random and the structure of rDNA operon may be different even at closely-related taxa.
The mitochondria genome size of Bangiophyceae was significantly larger than that of the Florideophyceae and the GC content was higher than those of the Florideophyceae, which was consistent with previous report10. Unexpectedly, the mitochondrial genome of Compsopogon caeruleus exhibited an evidently small size compared with other ancient derived Rhodophyta lineages. The mitochondria genomes of freshwater Florideophycean representatives Thorea hispida and Batrachospermum arcuatum were highly conservative with its marine relative Palmaria palmata whereas extensive gene rearrangements occurred when comparing Compsopogonophycean, Bangiophyceae and Florideophyceae representatives. The similar phenomenon was also found in the marine Rhodophyta10. Additionally, introns were found in the marine Rhodophyta mitochondria genes, such as the cox1 and trnI10. Whereas no introns were found in all the freshwater representatives in this study, suggesting the independent evolution of freshwater and marine groups since their derivation.
Phylogenomic analysis in this study covered the freshwater and marine members of classes Compsopogonophyceae, Bangiophyceae and Florideophyceae, with the typical freshwater representatives Batrachospermales and Thoreales included. The class Compsopogonophyceae diverged at an earlier time followed by the sister group of Bangiophyceae and Florideophyceae, which was consistent with phylogenetic relationship in previous study12. Origin of freshwater Rhodophyta were lineage-specific based on phylogenomic data in this study, with the freshwater representatives Compsopogon caeruleus and Hildenbrandia rivularis diverging earlier than the marine relatives and probably originated and evolved independently at the inland water. Whereas the freshwater taxa Bangia atropurpurea in class Bangiophyceae and the typical freshwater representatives Thoreales and Batrachospermales were derived from the marine relatives. The results based on molecular data were partially consistent (on the origin of freshwater primary Rhodophyta like Compsopogon caeruleus and Hildenbrandia rivularis) with that proposed by Skuja based on morphological and ecological evidence7, whereas we expanded the scope of marine migrants origination to include freshwater Bangiales, Thoreales and Batrachospermales. Molecular-based inference on freshwater Rhodophyta origination and evolution were poorly studied before. The primary proposal in this study provided molecular evidence for further investigation of freshwater Rhodophyta.
It has proved difficult to resolve class-level relationships among red algae using multigene data2, 25, 26. Yoon et al. used seven plastid-encoded proteins and two rDNA sequences to construct phylogenetic tree of Rhodophyta and highlighted the inherent difficulties in resolving the intra-lineage relationship of the red algae except for the position of the Cyanidiales and the monophyly of Bangiales and Florideophyceae2. Phylogenetic trees generated in this study were robustly supported at most nodes, thus resolving the phylogenetic relationship at both the inter- and intra-class level for Rhodophyta and demonstrating the organelle genomes powerful tools for resolving the red algal phylogeny.
Based on the chloroplast phylogenomic analysis in this study, the Porphyridiophyceae, Stylonematophyceae and Compsopogonophyceae originated at 1012 MYA and the Bangiophyceae and Florideophyceae diverged 844 MYA (chloroplast genome based) vs 828 MYA (mitochondria genome based), which were similar to previous results based on multi-gene analysis. Yoon postulated the Porphyridiophyceae, Compsopogonophyceae, Stylonematophyceae, Rhodellophyceae and Bangiophyceae radiated around 1200 MYA, followed the divergence of Florideophyceae at around 800 MYA2, 27. Yang et al. suggested the Florideophyceae diverged 943 MYA beginning with the split of the Hildenbrandiophycidae followed by the divergence of Nemaliophycidae, Corallinophycidae, Ahnfeltiophycidae and Rhodymeniophycidae28. Phylogenomic data inferred the Compsopogonophyceae originated at Mesoproterozoic era and the Florideophyceae originated at Neoproterozoic era, which was consistent with the multi-gene evidence. The freshwater representatives Compsopogon caeruleus, Thorea hispida and Batrachospermum arcuatum diverged at 547 MYA, 484 MYA and 231 MYA respectively, all at the Phanerozoic era. Previous proposal based on morphological characterization highlighted the earlier divergence of Thoreales compared with other Nemaliophycidae lineages29. Genomic evidence was consistent with the morphological features in revealing the earlier divergence of Thoreales among Nemaliophycidae. Divergence of freshwater members occurred together with the radiation of the rhodymeniophycidaen algae28.
Thioredoxins (Trx) and glutaredoxins (grx) constituted families of thiol oxidoreductases and involved in disulphide/dithiol interchange. Marine algae produced large amounts of sulfated extracellular polysaccharides and sulfation accounted for a much greater proportion of the total assimilated sulfur30. Whereas polysaccharides of freshwater Rhodophyta characterized for Batrachospermum and Compsopogon species contained no ester sulfate31. Trx and grx genes have proved absent in the chloroplast genomes of Batrachospermum arcuatum and Compsopogon caeruleus in this study. Thus it was inferred that the trx and grx genes involved in the polysaccharides synthesis of Rhodophyta. No protein-coding unique genes were found in mitochondrial genomes of freshwater and marine Rhodophyta suggest the high conservation of gene content between these two groups.
Ka/Ks is significantly elevated for ribosomal protein (rpl and rps), RNA polymerase genes (rpo) and ATPase genes (atp) in the angiosperm plastid genomes32. Similarly, the three gene groups were proved increased in Rhodophyta chloroplast genomes, illustrating positive selection or relaxed selection in the Rhodophyta evolution. It was found that all ribosomal protein genes and sdh (succinate dehydrogenase) genes have been lost from the mitochondrial genome many times during angiosperm evolution33. In Rhodophyta, substitution rates are highly elevated for the sdh genes and all the rpl genes (ribosomal protein large subunits) have been lost in the mitochondrial genome of Compsopogon caeruleus. Given the ancient derivation of Rhodophyta, we inferred the features of high substitution rates for the chloroplast gene groups of ribosomal protein, RNA polymerase and ATPase, in combination with mitochondrial ribosomal protein genes and sdh genes, were shared by eukaryotic lineages produced through the primary and second endosymbiotic events. Variant substitution rates of each gene group can serve as candidate DNA barcoding at different taxonomic levels in the Rhodophyta systematics.
Methods
Taxon sampling
Algal specimens were collected from Nanlaoquan, Jinci Park and Niangziguan, Shanxi Province respectively (Table 1). Fresh thalli were first washed to eliminate the epiphytes and then desiccated in silica gel to be stored at −20 °C.
Table 1.
Specimen information used in this study.
| Species | Order and Class | Locality, latitude and longitude, collection date | GenBank accession numbers for chloroplast | GenBank accession numbers for mitochondria |
|---|---|---|---|---|
| Batrachospermum arcuatum | Batrachoserpmales, Florideophyceae | Nanlao Spring, Taiyuan, Shanxi province, China (37.71 N, 112.43E) March 2006. | KY033529 | KY083064 |
| Thorea hispida | Thoreales, Florideophyceae | Niangziguan, Pingding, Shanxi province, China (37.78 N, 113.52E) March 2009. | KY083065 | KY083066 |
| Compsopogon caeruleus | Compsopogonales, Compsopogonophyceae | Weizeguan Spring, Shanxi province, China (37.96 N; 113.88E) August 2013. | KY083067 | KY083068 |
Genome sequencing, Assembly and Annotation
Total DNA of algal specimens were sequenced using the Illumina Hiseq 2500 technology with 350 bp insertion fragments. The Illumina-generated reads were assembled with SPAdes 3.8.234. Contigs generated were blast against other Rhodophyta chloroplast and mitochondrial genomes. Matched contigs with similar coverage were screened out and extended by a baiting and iteration method using the Price software35. The resulting contigs were loaded as reference sequences in Bowtie 2.1.036 and the matched reads were used for another assembly under SPAdes 3.8.2. After iterative extension and assembly, the final circular structures were generated.
Annotation were conducted with Unipro UGENE for initial open reading frame (ORF) finding and blastp for annotation of protein-coding sequences (CDS)37. Large and small subunits of ribosomal RNA (rRNA) were identified using BLASTn with published red algal rRNAs as queries, and transfer RNAs (tRNA) and tmRNA were identified using using the tRNAscan-SE Search Server (http://lowelab.ucsc.edu/tRNAscan-SE/). The ribonuclease P gene (rnpB) was detected using Bcheck online web server38. Newly generated organelle genomes were deposited in GenBank.
Similarity and structure comparison of organelle genomes
Chloroplast and mitochondrial genomes of Batrachospermum arcuatum, Thorea hispida and Compsopogon caeruleus were aligned with other Rhodophyta organelle genomes (Suppl. Table S9) using Mauve ver. 2.3.1 under the progressive mode39. The alignment result file was used to extract syntenic alignments by customized Perl scripts.
Phylogenomic analysis and divergence time estimation
Sequence alignment at the genome-scale was used to construct phylogenetic trees. Optimal evolutionary models of the dataset were determined using Modeltest version 3.740. Maximum likelihood41 under the RaxML with 1000 bootstrap replicates42 were used to construct the trees. Bayesian inference43 was run for 1,100,000 generations with sampling every 100 generations under the temperature of 0.2. After discarding the first 25% of trees as burn-in, posterior probabilities were calculated under the MrBayes44. Organelle genome length, GC content and features (length of CDS, rRNA, tRNA, ncRNA, tmRNA, intron, non-coding region) of each taxa in the phylogenetic tree were labeled on the tree using online tools EvolView (http://www.evolgenius.info/evolview.html). SPSS ver. 16.0 (SPSS, Inc., Chicago, IL, USA) was used for statistical analyses. Appropriate statistical analysis were selected after testing for the normal distribution of the datasets. Primary components leading to overall genome size variation were analyzed using Spearman’s rank correlation analysis. The significance of difference in mitochondrial genome size was analyzed using Mann-Whitney U test. The alpha level for all the two-tailed tests was 0.01. Divergence time estimation were inferred based on a Bayesian tree using BEAST245. The analysis was performed under GTR nucleotide substitution model (determined by Modeltest in the phylogenetic analysis) with a Gamma distribution for four rate categories. Uncorrelated lognormal relaxed clock model was employed to account for the uncertainty in the divergence time estimation46. Calibration nodes were constrained based on three fossil records including the 1174–1222 MYA Bangiomorph multicellular red algae47, corallinalean algae from the Doushantuo Foamation at 635–551 MYA48, 49, and 130–136 MYA for the Sporolithales split50. Posterior distributions of parameters were approximated after 50,000,000 generations of MCMC runs, sampling every 5,000 generations with a 25% burn-in, and the divergence tree was visualized using FigTree51. The priors on the age of the nodes were set as a normal distribution as the normal approach has been proved more appropriate for red algae divergence time calculation28.
Gene loss and substitution rates
The coding content of organelle genome were compared between the freshwater Rhodophyta and marine members for each lineage using TBtools Venn analysis (www.omicshare.com/tools). Substitution rates of the genes were analyzed using DnaSP ver. 5 with the outgroup in the phylogenetic trees as reference52. Genes were grouped according to their function. Eight gene groups for chloroplast genome including allophycocyanin (apc), ATP synthase genes (atp), photosystem I genes (psa), photosystem II genes (psb), rubisco gene (rbc), ribosomal protein (rpo) and RNA polymerase (rpl and rps) were concatenated to estimate nonsynonymous substitutions (Ka), synonymous substitutions (Ks), and their ratios (Ka/Ks). And the gene groups selected for the mitochondrial genome were ATP synthase genes (atp), cytochrome b (cob), cytochrome c oxidase (cox), NADH dehydrogenase (nad), and cytochrome c oxidoreductase (sdh). The relative ratios were illustrated in scatter plot using OriginPro 2016 (OriginLab Corporation USA). The significance of difference in substitution rates for diverse gene groups was tested using Kruskal-Wallis H test.
Electronic supplementary material
Acknowledgements
This work was funded by the National Natural Science Foundation of China (No. 31370239 and 31670208 to S.L. Xie). We sincerely thank Y.X. Mao and G.Q. Bi for the technical help on assembly of organelle genomes.
Author Contributions
F.R.N. analyzed the data and wrote the manuscript. J.F., J.P.L. and Q.L. gave technological help. K.P.F. and C.Y.G. collected and analyzed data. S.L.X. designed the project and modified the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-03235-5
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yoon HS, Hackett JD, Pinto G, Bhattacharya D. The single, ancient origin of chromist plastids. P. Natl. Acad. Sci. USA. 2002;99:15507–15512. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoon HS, Müller KM, Sheath RG, Ott FD, Bhattacharya D. Defining the major lineages of red algae (Rhodophyta) J. Phycol. 2006;42:482–492. doi: 10.1111/j.1529-8817.2006.00210.x. [DOI] [Google Scholar]
- 3.Wehr, J. D. Introduction to Freshwater Algae (eds Sheath, R. G. & Wehr, J. D.) (Richmond Publishing, 1984).
- 4.Guiry, M. D. & Guiry, G. M. AlgaeBase world-wide electronic publication. In: Galway (Ireland): National University of Ireland. http://www.algaebase.org (2015) (Date of access 21/5/2015).
- 5.Wehr, J. D. & Sheath, R.G. Freshwater algae of North America. (Academic Press, 2003).
- 6.Sheath, R. G. & Hambrook, J. A. Freshwater ecology. In Biology of the Red Algae (eds Cole, K. M. et al.), 423–453 (Cambridge: Cambridge University Press, 1990).
- 7.Skuja H. Comments on freshwater Rhodophyceae. Bot. Rev. 1938;4:665–676. doi: 10.1007/BF02869845. [DOI] [Google Scholar]
- 8.Janouškovec J, et al. Evolution of red algal plastid genomes: ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PLoS One. 2013;8:e59001. doi: 10.1371/journal.pone.0059001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, et al. Complete sequence and analysis of plastid genomes of two economically important red algae: Pyropia haitanensis and Pyropia yezoensis. PLoS One. 2013;8:e65902. doi: 10.1371/journal.pone.0065902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang EC, et al. Highly conserved mitochondrial genomes among multicellular red algae of the Florideophyceae. Genome Biol. Evol. 2015;7:2394–2406. doi: 10.1093/gbe/evv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tajima N, et al. Analysis of the complete plastid genome of the unicellular red alga Porphyridium purpureum. J. Plant Res. 2014;127:389–397. doi: 10.1007/s10265-014-0627-1. [DOI] [PubMed] [Google Scholar]
- 12.Lee JM, et al. Parallel evolution of highly conserved plastid genome architecture in red seaweeds and seed plants. BMC Biol. 2016;14:75. doi: 10.1186/s12915-016-0299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa JF, Lin SM, Macaya EC, Fernández-García C, Verbruggen H. Chloroplast genomes as a tool to resolve red algal phylogenies: a case study in the Nemaliales. BMC Evol. Biol. 2016;16:205. doi: 10.1186/s12862-016-0772-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hovde BT, et al. The mitochondrial and chloroplast genomes of the haptophyte Chrysochromulina tobin, contain unique repeat structures and gene profiles. BMC Genom. 2014;15:1–23. doi: 10.1186/1471-2164-15-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leliaert F, et al. Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci. Rep. 2016;6:25367. doi: 10.1038/srep25367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagopian JC, Reis M, Kitajima JP, Bhattacharya D, de Oliveira MC. Comparative analysis of the complete plastid genome sequence of the red alga Gracilaria tenuistipitata var. liui provides insights into the evolution of rhodoplasts and their relationship to other plastids. J. Mol. Evol. 2004;59:464–477. doi: 10.1007/s00239-004-2638-3. [DOI] [PubMed] [Google Scholar]
- 17.Du Q, Bi G, Mao Y, Sui Z. The complete chloroplast genome of Gracilariopsis lemaneiformis (Rhodophyta) gives new insight into the evolution of family Gracilariaceae. J. Phycol. 2016;52:441–450. doi: 10.1111/jpy.12406. [DOI] [PubMed] [Google Scholar]
- 18.Alverson AJ, et al. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae) Mol. Biol. Evol. 2010;27:1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selosse MA, Albert B, Godelle B. Reducing the genome size of organelles favours gene transfer to the nucleus. Trends Ecol. Evol. 2001;16:135–141. doi: 10.1016/S0169-5347(00)02084-X. [DOI] [PubMed] [Google Scholar]
- 20.Hershberg R, Petrov DA. Evidence that mutation is universally biased towards AT in Bacteria. PLoS Genet. 2010;6:e1001115. doi: 10.1371/journal.pgen.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch M. Rate, molecular spectrum, and consequences of human mutation. P. Natl. Acad. Sci. USA. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Šmarda P, Bureš P, Horová L, Foggi B, Rossi G. Genome size and GC content evolution of Festuca: ancestral expansion and subsequent reduction. Ann Bot-London. 2008;101:421–433. doi: 10.1093/aob/mcm307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Necchi O, Jr, et al. Global sampling reveals low genetic diversity within Compsopogon (Compsopogonales, Rhodophyta) Eur. J. Phycol. 2013;48:152–162. doi: 10.1080/09670262.2013.783626. [DOI] [Google Scholar]
- 24.Meunier J, Duret L. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- 25.Le Gall L, Saunders GW. A nuclear phylogeny of the Florideophyceae (Rhodophyta) inferred from combined EF2, small subunit and large subunit ribosomal DNA: establishing the new red algal subclass Corallinophycidae. Mol. Phylogenet. Evol. 2007;43:1118–1130. doi: 10.1016/j.ympev.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Verbruggen H, et al. Data mining approach identifies research priorities and data requirements for resolving the red algal tree of life. BMC Evol. Biol. 2010;10:1. doi: 10.1186/1471-2148-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- 28.Yang EC, et al. Divergence time estimates and the evolution of major lineages in the florideophyte red algae. Sci. Rep. 2016;6:21361. doi: 10.1038/srep21361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi, Z. X. Flora algarum sinicarum aquae dulci, Tomus XIII, Rhodophyta, Phaeophyta (The Science Publishing Company, 2006).
- 30.Meyer Y, Belin C, Delorme-Hinoux V, Jean-Philippe R, Riondet C. Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid Redox. Sign. 2012;17:1124–1160. doi: 10.1089/ars.2011.4327. [DOI] [PubMed] [Google Scholar]
- 31.Gretz MR, Sommerfeld MR, Athey PV, Aronson JM. Chemical composition of the cell walls of the freshwater red alga Lemanea annulata (Batrachospermales) J. Phycol. 1991;27:232–240. doi: 10.1111/j.0022-3646.1991.00232.x. [DOI] [Google Scholar]
- 32.Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. P. Natl. Acad. Sci. USA. 2008;105:18424–18429. doi: 10.1073/pnas.0806759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams KL, Qiu YL, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. P. Natl. Acad. Sci. USA. 2002;99:9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bankevich A, et al. Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graham RJ, Priya B, Derisi JL. Price: software for the targeted assembly of components of (meta) genomic sequence data. G3-Genes Genom. Genet. 2013;3:865–880. doi: 10.1534/g3.113.005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okonechnikov K, Golosova O, Fursov M. & the UGENE team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 38.Yusuf D, Marz M, Stadler PF, Hofacker IL. Bcheck: a wrapper tool for detecting RNase P RNA genes. BMC Genome. 2010;11:432. doi: 10.1186/1471-2164-11-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 41.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 42.Stamatakis A. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rannala B, Yang Z. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 1996;43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 44.Ronquist F, et al. Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouckaert R, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. Plos Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butterfield NJ. Paleobiology of the late Mesoproterozoic (ca. 1200 Ma) hunting formation, Somerset Island, Arctic Canada. Precam. Res. 2001;111:235–256. doi: 10.1016/S0301-9268(01)00162-0. [DOI] [Google Scholar]
- 48.Xiao S, Zhang Y, Knoll AH. Tree-dimensional preservation of algae and animal embryos in a Neoproterozoic phosphorite. Nature. 1998;391:553–558. doi: 10.1038/35318. [DOI] [Google Scholar]
- 49.Condon D, et al. U-Pb ages from the Neoproterozoic Doushantuo Formation, China. Science. 2005;308:95–98. doi: 10.1126/science.1107765. [DOI] [PubMed] [Google Scholar]
- 50.Aguirre J, Perfecti F, Braga JC. Integrating phylogeny, molecular clocks, and the fossil record in the evolution of coralline algae (Corallinales and Sporolithales, Rhodophyta) Paleobiology. 2010;36:519–533. doi: 10.1666/09041.1. [DOI] [Google Scholar]
- 51.Rambaut, A. FigTree, a graphical viewer of phylogenetic trees. http://tree.bio.ed.ac.uk/software/figtree (2007) (Date of access: 28/7/2016).
- 52.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


