Abstract
Colorectal cancer (CRC) is a leading cause of death, yet facile preclinical models that mimic the natural stages of CRC progression are lacking. Through the orthotopic engraftment of colon organoids we describe a broadly usable immunocompetent CRC model that recapitulates the entire adenoma-adenocarcinoma-metastasis axis in vivo. The engraftment procedure takes less than 5 minutes, shows efficient tumor engraftment in 2/3 mice, and can be achieved using organoids derived from GEMMs, wild type organoids engineered ex vivo, or from patient-derived human CRC organoids. In this model, we describe the genotype and time-dependent progression of CRCs from adenocarcinoma (6 weeks), to local disseminated disease (11–12 weeks) and spontaneous metastasis (>20 weeks). Further, we use the system to show that loss of dysregulated Wnt signaling is critical for the progression of disseminated CRCs. Thus, our approach provides a fast and flexible means to produce tailored CRC mouse models for genetic studies and pre-clinical investigation.
Colorectal cancer (CRC) is the second leading cause of cancer related deaths in developed countries, yet there remain few effective therapies for treating advanced disease. CRC develops in a step-wise fashion, whereby loss of function mutations in the Adenomatous Polyposis Coli (APC) tumor suppressor drive the formation of precancerous benign adenomas, and subsequent mutations in KRAS and TP53 support malignant progression1, 2.
Unlike many other cancer types, it has been difficult to develop genetically engineered mouse models (GEMMs) of CRC that accurately recapitulate advanced staged disease in the correct anatomical location. This is because traditional GEMMs harboring germline Apc mutations frequently develop small intestinal, rather than colonic lesions, and overall tumor burden limits the time whereby malignant progression can occur3–5. Tissue restricted Cre/LoxP-based strategies can drive cancer-predisposing lesions in the colon, but require multi-allelic intercrossing and still rarely show disease progression, thus limiting their utility for genetic and pre-clinical studies6–9. Although tumors produced by transplantation of human colon organoid cultures10, 11 provide one platform to produce more efficient CRC models, to date, these models have only been used to study tumors at ectopic sites and in immunocompromised recipients Thus, no one system accurately recapitulates human CRC progression, yet is simple, fast, and flexible enough to produce tailored preclinical models at a reasonable cost.
We set out to build a modular and rapid approach to generate CRC preclinical models in the context of a physiologically accurate tissue environment. We envisaged an ideal model would: (1) develop focal tumors in the colon; (2) enable longitudinal analysis of disease progression and regression using endoscopy; (3) permit all stages of CRC progression; (4) allow rapid and iterative genetic manipulation; and (5) be broadly adaptable to laboratories that lack advanced surgical expertise. Given recent success in the isolation, culture, and genetic manipulation of colon organoids in vitro10–17, we hypothesized that genetically engineered organoids engrafted into the mucosal layer of the mouse colon would satisfy the above requirements.
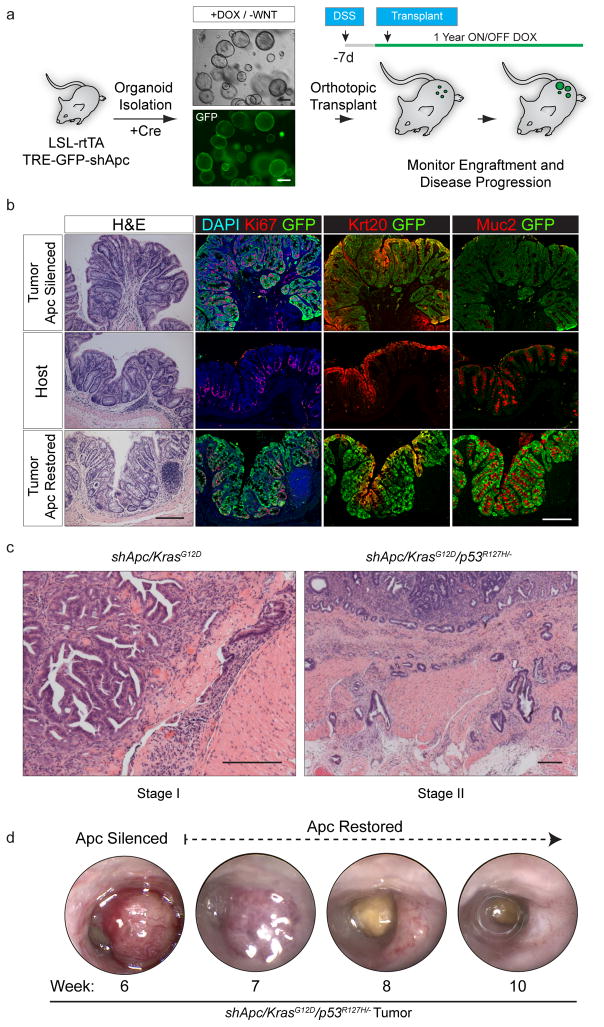
First, to examine the feasibility this approach, we isolated organoids from an ‘shApc’ GEMM that allows shRNA-mediated Apc silencing linked to GFP expression18. In this system, expression of the reverse tet-transactivator (rtTA) enables doxycycline-dependent Apc silencing and polyp growth, which is reversed upon dox withdrawal (Apc restoration)18. Initial use of this model facilitated tracking of engrafted cells by GFP fluorescence and allowed us to benchmark the fidelity of the orthotopic engraftment approach against a well-characterized transgenic system18. We therefore generated colon organoid cultures from CAGs-LSL-rtTA3/TRE-GFP-shApc (shApc) transgenic mice, induced rtTA3 expression via adenoviral Cre, and drove GFP expression and Apc silencing by doxycycline (dox) treatment (Fig. 1a). Consistent with previous work10, 11, 16, 18–20, Apc suppression disrupted the hallmark crypt-villus structure of wildtype organoids, producing spheroid structures that could survive and proliferate in the absence of exogenous Wnt ligand.
Figure 1.
Transplanted organoids can recapitulate autochthonous GEMMs a: Schematic depiction of the protocol used to generate organoid engraftments to model Apc loss in the colon. b. Histochemical (H&E) and immunofluorescent (Ki67, Krt20, Muc2, GFP) stains of an engrafted shApc adenoma (top row) and neighboring normal host mucosa (middle row), and long-term (29 weeks) Apc-restored (bottom row). c. H&E stain of an shApc/KrasG12D tumor 8 weeks post transplant (left), H&E stain of an shApc/KrasG12D/p53R172H/− tumor 16.5 weeks post transplant (right). d. Serial endoscopies of an shApc/KrasG12D/p53R172H/− tumor that was maintained ON Dox (Apc Silenced) for 6 weeks, and then longitudinally followed OFF Dox (Apc restored). Scale bars are 200 μm.
To implant shApc organoids in the colon, we adapted a protocol established for the engraftment of wildtype colon organoids21 (Online Methods, Supplemental Movie 1, Supplemental Fig. 1, and Fig. 1a). This approach relies on short- term treatment with dextran sodium sulfate (DSS) to induce transient colonic injury and establish a niche for cell engraftment. Following transplant, macroscopic GFP+ lesions were first identifiable using fluorescence endoscopy at 10 days and persisted up to a year after transplantation (Supplementary Fig. 1). As occurs in Apc mutant GEMMs, mice receiving shApc organoids also develop benign tumors – however in this setting they are restricted to the colon, rather than the small intestine. One-year post-engraftment, polyps remained embedded within normal host mucosa as benign tubular adenomas (Fig. 1b). That these single-mutant (shApc only) lesions never show submucosal invasion up to a year after engraftment indicates that the DSS procedure does not in itself enable tumor dissemination.
Oncogenic KRAS and TP53 mutations are associated with malignant disease progression in human CRC2. Accordingly, transplantation of shApc organoid cultures harboring oncogenic KrasG12D and Trp53R172H/− mutations produced frank carcinomas withsubmucosal invasion (Fig. 1c). Consistent with studies in germline transgenic animals18, we showed that restoration of Apc expression in benign shApc and invasive shApc/KrasG12D/p53mut engrafted tumors induced cell differentiation, tumor regression, and the reestablishment of crypt-villus homeostasis (Fig. 1b and d, Supplementary Figs. 1–3). Hence, orthotopic engraftment of colonic organoids enables the rapid production of focal colorectal tumors while retaining the histopathology and physiology of ‘gold-standard’ autochthonous models.
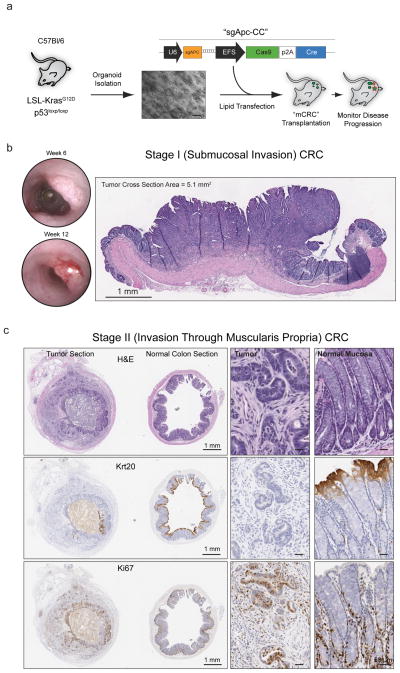
In principle, the organoid engraftment approach described above could be used to produce cohorts of mice from pre-existing engineered organoids in a fraction of the time and expense of traditional GEMMs. Having credentialed the first generation model, we set out to establish a generalizable platform in which the tumor genotype or tissue context could be easily manipulated and the resulting tumors studied in a physiologically accurate microenvironment. To do this, we generated organoid cultures harboring the most common CRC mutant alleles in a C57Bl/6 background, which would allow subsequent engraftment into syngeneic recipient mice. We reasoned that studying CRCs with Apc, Kras and Trp53 mutations was the most relevant context because they represent the three most frequent genetic alterations in human CRC22. To engineer organoids with this configuration, we isolated colon stem cells from LSL-KrasG12D/p53flox/flox C57Bl/6 mice and engineered biallelic loss-of-function mutations in Apc by CRISPR/Cas9-mediated genome editing with an sgApc-Cas9-Cre (sgApc-CC) vector (Fig. 2a)16. This construct also expresses Cre recombinase to simultaneously activate Kras and delete Trp53, which we confirmed by genomic PCR, DNA sequencing, and functional selection (Supplementary Fig. 4). Of note, although the genes we modified permit rapid selection by altering growth factor dependencies (RSPO/Wnt) or sensitivity to pathway inhibitors (Nutlin-3), it is possible to manually isolate edited clones for those mutational events that do not confer an in vitro or in vivo selective advantage11. Additionally, the approach can also be multiplexed, enabling the production of tailored genetic models by simply combining additional sgRNAs (Supplementary Fig. 4).
Figure 2.
Transplanted organoids create focal colorectal tumors. a. Schematic depiction of the protocol used to generate Apcmut/KrasG12D/p53mut organoids for syngeneic orthotopic engraftment. b. Serial endoscopies of an Apcmut/KrasG12D/p53mut transplant growing as a polypoid mass in the mucosal layer of an engrafted host (left), and H&E stained cross-section of an Apcmut/KrasG12D/p53mut tumor (7 weeks post transplant). c. Histochemical (H&E) and immunohistochemical (Krt20, Ki67) stained sections of tumor (left) and normal (right) tissue of a Stage II CRC tumor collected 75 days post transplant. Whole slide scan images are presented on the left (Scale Bar, 1mm), with higher resolution insets on the right (Scale Bar, 50 μm).
For engraftment in syngeneic mice, we treated cohorts of C57BL/6J animals with a 3% DSS and transplanted 300,000 Apcmut/KrasG12D/p53mut cells per mouse, as disassociated organoid fragments. This treatment regimen had a 97% survival rate, associated with minimal and transient weight loss (Table 1). Further, comprehensive immunophenotyping and CBC analysis of non-engrafted animals confirmed that this DSS pretreatment regimen had no significant long-term effects on the immune system or peripheral blood counts (Supplementary Fig. 5). We observed tumor engraftment in 62% of viable DSS-treated mice, and an average of one engrafted tumor per mouse (median = 1, mean = 1.2 +/− 0.41) (Fig. 2b and Table 1). Individual engrafted tumors arose from the mucosal layer of the colon, ranged in size from 1–5 mm2, and appeared histologically as localized or invasive adenocarcinoma (Fig. 2b and Supplementary Fig. 6). All tumors examined during the first 4–9 weeks post transplant were classified as Stage I (T1N0M0), mirroring the early stages of colorectal cancer tumorigenesis in vivo.
Table 1.
Summary of the orthotopic engraftment approaches.
| Cell Type | Recipient | Total Mice | # Surviving DSS | % of Starting Weight On D7 | Stdev Weight Loss | # Engrafted | Transplant Efficiency (#Engrafted/Surviving Mice) |
|---|---|---|---|---|---|---|---|
| shAPC.8745e | Nude | 18 | 16 | 73% | 6% | 8 | 50% |
| shAPC.3374 | Nude | 15 | 15 | 87% | 9% | 9 | 60% |
| shAPC.3374/Kras | Nude | 15 | 13 | 78% | 26% | 6 | 46% |
| shAPC.3374/Kras/p53−/− | Nude | 14 | 12 | 89% | 9% | 6 | 50% |
| shAPC.3374/Kras/p53 R172H/− | Nude | 15 | 15 | 94% | 13% | 7 | 47% |
| shAPC.3374/Kras/p53 R172H/− MNIL-shRenilla | Nude | 25 | 21 | 80% | 10% | 18 | 86% |
| shAPC.3374/Kras/p53 R172H/− MNIL-shSmad4 | Nude | 20 | 13 | 81% | 9% | 8 | 62% |
| APC (mut)/Kras/p53 −/− (3D Organoids) | C57Bl/6J | 35 | 34 | 95% | 10% | 21 | 62% |
| APC (mut)/Kras/p53 −/− (2D Cell Line) | C57Bl/6J | 20 | 19 | 92% | 5% | 19 | 100% |
| Total Transplanted | Total Engrafted | Total Transplant Efficiency | |||||
| 158 | 102 | 65% |
Engrafted tumors progressed to high-grade adenocarcinomas that invaded the muscle wall and pericolorectal tissue (Fig. 2c), and displayed advanced signs of disease, including dysplasia, loss of tissue architecture, and lymphovascular invasion (Supplementary Fig. 7). We were unable to detect any signs of dysplasia or aberrant tissue architecture in host mucosa examined at these later time points (Fig. 2c). At 16 weeks post transplant, 2 of 3 mice that were staged radiographically by MRI showed locally invasive disease with tumor extending past the serosal lining and infiltrating the pericolorectal adipose tissue (Stage II (T3)) (Supplementary Fig. 7). Notably, we never observed disease progression past Stage I in transgenic mice of a similar genotype18, likely owing to the reduced survival of GEMMs harboring multiple primary lesions throughout the small and large intestine (Supplementary Fig. 7).
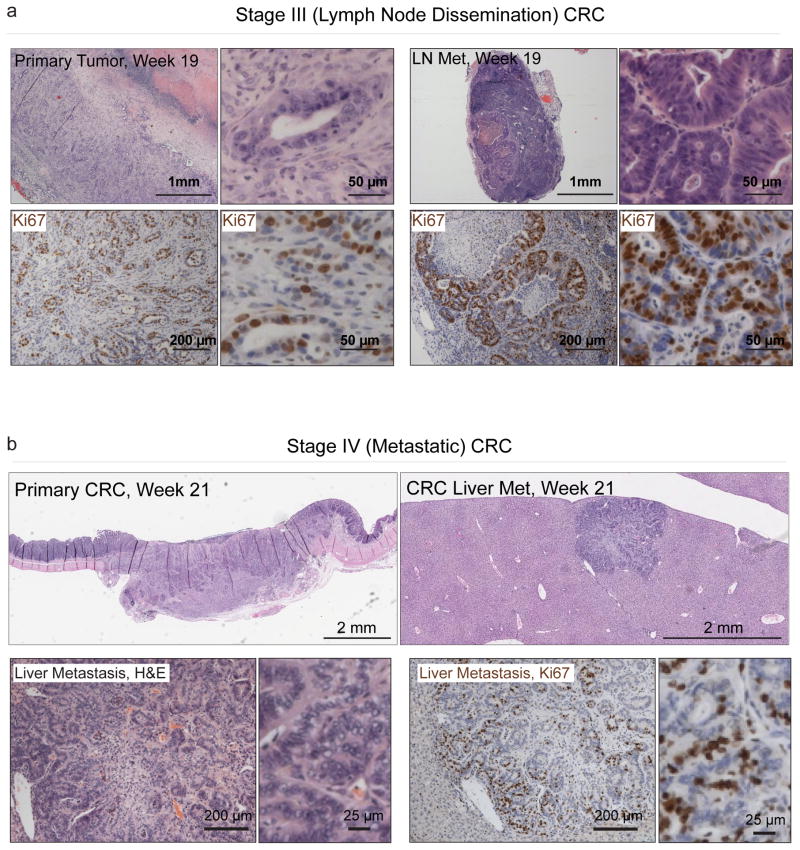
Transplanted C57Bl/6 mice had a maximum survival of 21.4 weeks (compared to 10.1 weeks for the GEMM model18), providing an expanded time window for local dissemination to regional lymph nodes, and metastatic progression (Fig. 3a and Supplementary Fig. 7–8). We identified macroscopic liver metastasis in 1/6 animals examined 21 weeks post transplant (Fig. 3b, Supplementary Figs. 8–9). Consistent with metastatic human CRC, this metastasis contained moderately differentiated gland forming adenocarcinoma, regions of stratified nuclei, and intracellular Mucin. Immunohistochemical staining for colon specific markers (Krt20 and Villin) and PCR genotyping confirmed the lesion was derived from transplanted organoids (Supplementary Fig. 8–9). These studies demonstrate that orthotopic engraftment of Apcmut/KrasG12D/p53mut organoids in syngeneic recipients can recapitulate the entire adenoma-carcinoma-metastasis progression sequence in vivo (Supplementary Fig. 10). Since distant metastasis does not occur in every animal, this platform provides an ideal context for interrogating the additional genetic and environmental factors that enhance metastatic progression from the primary site.
Figure 3.
Orthotopically transplanted organoids progress to metastatic CRC. a. Histochemical (H&E, top) and immunohistochemical (Ki67, bottom) of a T3 primary tumor, 137 days post transplant, and a caudal lymph node metastasis (right). b. H&E stained sections (top) of a primary T3 CRC tumor 21 weeks post transplant, and a liver metastasis harvested from the same animal (low res slide scan, top right, higher magnification, bottom left), and immunohistochemical (Ki67) stained section of the liver met (bottom right).
Previous efforts to develop orthotopic transplant models have relied on tumor-derived cell lines cultured on plastic, rather than as organoids in a 3D protein matrix5, 23. To assess any differences between these approaches, we derived isogenic 2D cultures from our engineered organoids and transplanted cells exactly as described for 3D cultures. In contrast to the glandular and well differentiated tumors derived from organoids, 2D cells engrafted as poorly differentiated, flat lesions that invaded the muscle wall and formed distant metastases in as little as five weeks (Supplementary Fig 11). Thus, although the model is aggressive, the engraftment of 2D cell lines in this context does not recapitulate the histopathology or stepwise tumor progression characteristic of organoid engraftment and human CRC.
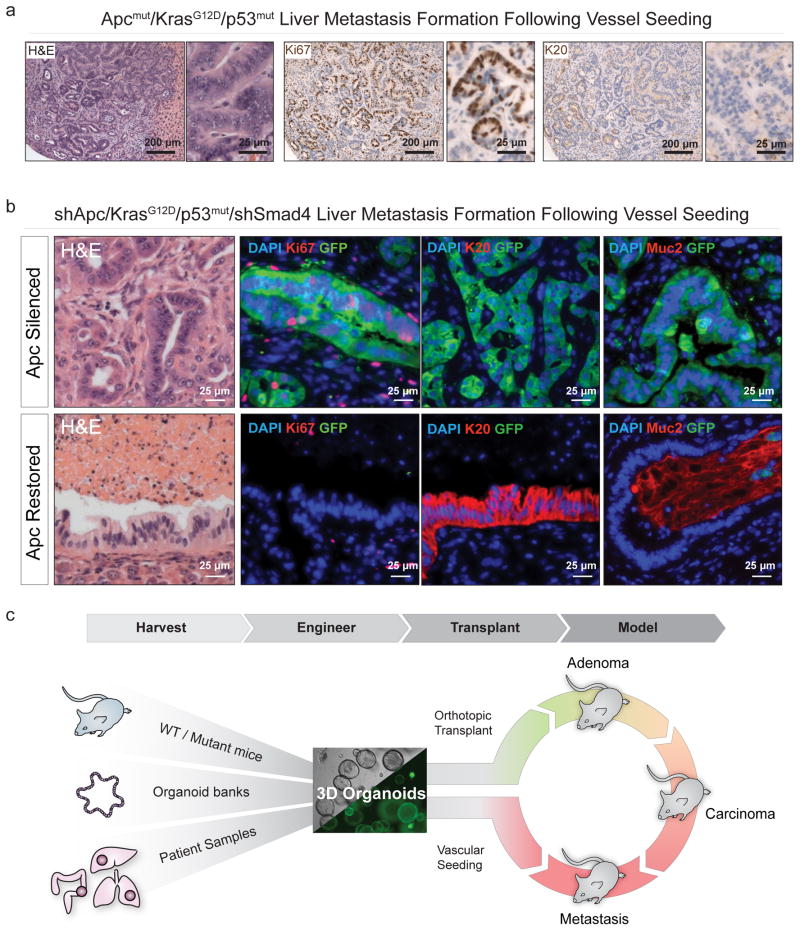
The step-wise tumor progression model described above effectively mimics all stages of human CRC, including the stochastic timing of metastatic spread. However, robust pre-clinical studies for treatment of metastatic disease rely on the generation of large cohorts of mice with synchronous disease onset and progression. Given that engineered organoids can accurately model disease following orthotopic engraftment, we asked whether it was possible to produce a histologically accurate model of ‘experimental metastasis’ using splenic, intravenous, or direct liver injection methods. We observed tumor formation using either triple mutant C57Bl/6 organoids (Fig. 4a and Supplementary Figs. 12–13) or organoids that had been further engineered to model Smad4 loss, which has been shown to promote more aggressive disease progression in animal models24, 25 (Supplemental Figure 14. Tumor foci in the liver were histologically indistinguishable from spontaneous metastases in orthotopically engrafted mice, containing cells with stratified nuclei, high Ki67 positivity and little to no expression of Krt20 (Fig. 4a). Similarly, organoids showed robust colonization and growth of histologically accurate metastatic CRC foci in the lungs following intravenous injection (Supplementary Figs. 12–13). Thus, engineered organoids can be used to rapidly derive genotypically-defined and histologically accurate models of metastatic CRC, either by orthotopic transplantation to faithfully recapitulate disease progression, or by surrogate ‘vessel injection’ to create large cohorts of predictable size- and age-matched tumors for interventional studies.
Figure 4.
Dysregulated Wnt signaling is required to sustain advanced CRC.a. Histochemical (H&E, left) and immunohistochemical (Ki67, middle, Krt20, right) stains of sections from an Apcmut/KrasG12D/p53mut liver metastasis that formed 10 weeks after intra-splenic injection. b. Histochemical (H&E) and immunofluorescent (GFP, Ki67, Krt20, Muc2, Dapi) stains of liver tumors in animals maintained on dox for 10 weeks (“Apc silenced”),” see also Supplementary Fig. 19f), and of liver tumors maintained on dox for 10 weeks, then off dox for 3 weeks (“Apc Restoration,” bottom). c. A schematic depicting the two-tiered approach for ex vivo manipulation of organoids, sourced from murine models, tissue banks or human samples, engineered to further alter genotype, and transplanted into the colon mucosa by DSS treatment/pipette enema. In parallel organoids can be seeded in the vasculature to model metastatic CRC growth.
To illustrate the use of the organoid models to evaluate therapeutic strategies, we examined the requirement for sustained Apc loss and Wnt hyperactivation in metastatic lesions, which had not been possible in previous GEMM models due to early morbidity and limited disease progression. To test if triple mutant (TRE-shApc/Kras/p53/MNIL-Control – ‘AKP’) or quadruple mutant (TRE-shApc/Kras/p53/MNIL-shSmad4 – ‘AKPS’) murine CRC tumors rely on hyperactive Wnt signaling when growing at distant tissue sites, we utilized the splenic injection method. Ten weeks following liver seeding of Luciferase expressing tumors via splenic injection, histological examination of livers revealed multiple metastatic foci ranging in size from <1 – 7mm at their widest point (Fig. 4b, top panel, and Supplementary Fig. 14).
In both AKP and AKPS tumors, restoration of Apc expression during this period induced a marked drop in Luciferase signal (Supplementary Fig. 14). Similar to what we observed in either transgenic or transplanted primary tumors, Apc restoration drove Wnt pathway suppression and tumor cell differentiation, but now also formed large areas of fibrosis or necrosis in the liver (Fig. 4b, bottom panel). We also observed, large glands of post-mitotic colon epithelium that appeared atrophic and flattened. Notably, some glandular structures stained positive for the enterocyte marker Keratin 20, while others expressed a canonical marker of differentiated intestinal goblet cells, Muc2 (Fig. 4b, bottom panel), which we also confirmed by QRTPCR analysis on tumor derived organoids (Supplemental Fig. 14). Thus, aggressive CRC tumor cells growing in the liver remain dependent on Wnt hyperactivation for survival, and retain their intrinsic differentiation program in the absence of local microenvironmental cues. Long-term follow-up of ‘Apc Restored’ tumors was complicated by the development of severe ascites, perhaps due to the presence of ectopic, functional colonic epithelium in the liver of these mice. Similarly, restoration of Apc expression in CRC lung lesions led to labored breathing and endobronchial accumulation of colonic Mucin (data not shown). From a translational perspective, this implies that Wnt pathway inhibitors may be effective in treating metastatic CRC, but additional consideration needs to be paid to the effects of colonic cell differentiation in distant tissue sites.
Our studies illustrate that use of ex vivo engineered mouse organoids provides a flexible, fast and low cost platform to model all stages of CRC (Fig. 4c). The approach is also easily adapted for the orthotopic engraftment of patient-derived CRC organoids for the production of histopathologically accurate pre-clinical human cancer models (Supplementary Fig. 15). In this context, our work is complementary to previous efforts to build genetically defined human organoid models10, 11 and ‘living biobank’ repositories26. Combined, these tools provide an opportunity platform to create genetically tractable and orthotopic human CRC models. Moreover, resulting mice are “mosaic” such that engrafted cells are surrounded by genetically normal tissue as occurs during human cancer disease progression.
This broadly adaptable strategy does not require complicated surgical expertise or special equipment (see Supplementary Movie 1), with a throughput and versatility that substantially exceeds previous efforts to model this disease. It facilitates the rapid production of immunocompetent mice harboring primary or metastatic CRC, allows longitudinal studies on the natural process of CRC progression, and enables the production of large cohorts of mice with metastatic disease for preclinical studies. As the system produces genetic chimeras, it is possible to engraft benign or malignant organoid models into genetically modified hosts, enabling an examination of tumor-stroma interactions likely to be important in disease progression and therapy response. Collectively these models will foster an expanded understanding of the genetic factors mediating CRC behavior, and help drive the development of targeted small molecules, immunotherapies, and/or diagnostic tests for detection and treatment of metastatic CRC.
Online Methods
Animals
Production of mice, treatments described, and criteria for euthanasia were approved by the Institutional Animal Care and Use Committee (IACUC) at Memorial Sloan Kettering Cancer Center (NY), under protocol number 11-06-012. Foxn1nu and C57Bl/6NHsd female mice, age 4–6 weeks, were purchased from Envigo. C57Bl6/J female mice, age 4–6 weeks, were purchased from Jackson Labs. TG-Apc.3374 and TG-Apc.8745e animals were made as previously described18. Doxycycline was administered via food pellets (625mg/kg) (Harlan Teklad). 4-hydroxytamoxifen (4-OHT, Sigma Aldrich, 70% Z-isomer) was delivered by a single intraperitoneal injection (0.5mg/mouse) at 5–6 weeks of age. Differences in animal survival were calculated using a Log-Rank (Mantel-Cox) test performed using GraphPad Prism version 6.00 for Mac OS X, (GraphPad Software, La Jolla California USA). Animal randomization was performed by randomly assigning animal ID numbers to treatment arms after tumor formation was observed; investigators were not blinded to group allocation. No statistical methods were used to predetermine sample size. Animal and organoid genotyping, as well as necropsy, tissue fixation, immunohistochemistry, and immunofluorescence were performed as previously described18. Non-phosphorylated (active) beta catenin levels were measured by immunofluorescent staining with Non-phospho (Active) β-Catenin (Ser33/37/Thr41) (4270, Cell Signaling).
Organoids
Organoid isolation, passage and cryopreservation were performed as previously described13, 18. Cre recombination in vitro was induced by adding 3ul of 2×1010 PFU/mL Adenoviral-Cre (University of Iowa Gene Transfer Vector Core) to 250ul of Matrigel containing freshly isolated crypts and plated as above. Mouse Tgfb1 (eBioscience, 14-8342-62) was used at 10ng/ml. Nutlin (Selleck Chemicals, S1061) was used at 10 μM. DNA, Protein and RNA isolation, and qRT-PCR analysis of organoids was performed as previously described16, 18. Prior to transplantation, organoids were amplified in 200 μl of Matrigel plated in one well each of a 6-well dish and supplemented with 3 milliliters of colon organoid growth media13. For conversion of organoids into 2-D tissue monolayer conditions, organoids were passaged as previously described but resuspended into DMEM (+Pen/Strep +10% Fetal Bovine Serum) and plated onto a 10 cm sterile tissue culture dish (Falcon, 353003), and passaged 1:5 every 3–5 days.
Primers
qRT-PCR Primers in this study include:
| Axin2: | F: GCAGCTCAGCAAAAAGGGAAAT |
| R: TACATGGGGAGCACTGTCTCGT | |
| b2mBM: | F: ACCCCCACTGAGACTGATAC |
| R: ATCTTCAGAGCATCATGATG | |
| Fzd7: | F: CCATCCTCTTCATGGTGCTT |
| R: ATGGCCAAAATGGTGATTGT | |
| MycMYC: | F: CTCAGTGGTCTTTCCCTACCCG |
| R: TGTCCAACTTGGCCCTCTTGGC | |
| Krt20: | F: CCCAGAAGAACCTGCAAGAG |
| R: CCTCCGTGTTCACTGTGACT | |
| GFP: | F: GGACGACGGCAACTACAAGA |
| R: AAGTCGATGCCCTTCAGCTC |
PCR of the sgApc target region in sgApc-CC treated cells was performed with the following primers:
| Apc_SVR_F: | GCCATCCCTTCACGTTAG |
| Apc_SVR_R: | TTCCACTTTGGCATAAGGC |
Genotyping of organoids before and after sApc-CC transfection was performed with the following primers:
-
Y116-kras2-common: TCCGAATTCAGTGACTACAGATG
Y117-kras2-LSL: CTAGCCACCATGGCTTGAGT
Y118-kras2-wt: ATGTCTTTCCCCAGCACAG
-
P53-A: CACAAAAACAGGTTAAACCCAG
P53-B: AGCACATAGGAGGCAGAGAC
P53-C: GAAGACAGAAAAGGGGAGGG
Organoid Transfection
Prior to transfection, organoids were cultured in complete growth media (ENRW) supplemented with 10 μM Y-27632 (EMD Millipore, 688001) for two days. 2 μg of vector DNA was mixed with 6 μL of Lipofectamine 2000 (Thermo-Fisher Scientific, 11668-027) in 50 μL of Opti-MEM Reduced Serum Medium (Thermo-Fisher Scientific, 31985070) and allowed to incubate for 20 minutes at room temperature. Organoids, containing approximately 100,000 cells, were digested in .25% Trypsin/.02% EDTA in PBS at 37°C for exactly 7 minutes, and then mechanically disassociated using a p200 pipette, then 10ml of DMEM (+Pen/Strep +10% Fetal Bovine Serum) was mixed with the solution and centrifuged at 1500 RPM for 5 minutes. The pellet was resuspended in 450 μL of ENRW + Y-27632 and mixed with the 50 μL of lipid:DNA complex, plated in a 24-well plate and spun at 600g for 60 minutes at 32°C, and then placed in an incubator for 4 hours at 37°C. The cells were then collected, spun at 1500 RPM for 5 minutes, and replated in matrigel as normal. pMaxGFP (Lonza) was used as a positive control and cells were checked for viability and GFP expression 24h post transfection. Transfected organoids were allowed to recover for 5 days in ENRW+Y-27632 before being passaged into selective media conditions.
Copy number profiling
Tumor DNA was isolated using the AllPrep DNA/RNA Micro Kit, (Qiagen, 80284) according to instructions provided with the kit. 1 μg of DNA was sonicated (17W, 75 sec) using the E220 sonicator instrument (Covaris, Woburn, MA). Samples were subsequently prepared and indexed using Illumina TruSeq library prep system. Libraries were purified using AMPure XP magnetic beads (Beckman Coulter, Brea, CA), PCR enriched, and sequenced on an Illumina HiSeq instrument. Sequence reads were mapped using Bowtie with PCR duplicates removed. Uniquely mappable reads were further processed for copy number determination using the ‘varbin’ algorithm using 5000 bins, allowing for a median resolution of ~600 kb. GC content normalization, segmentation and copy number estimation was determined as described previously27. Tumor ploidy was confirmed via propidium iodide staining and cell cycle FACS analysis.
Human Organoid Specimen Procurement
All patient derived fresh tissue samples were collected with informed patient consent according to protocols approved by the institutional review board of Weill Cornell Medicine (IRB # 1305013903). Fresh tissue biopsies or resection specimens were directly taken from the procedure rooms to the frozen section area in pathology. A board certified pathologist collected the tumor tissue for sequencing and tumor organoid development. Time between harvesting fresh tissue specimens to placing them in transport media [Dulbecco’s modified Eagle medium (DMEM, Invitrogen) with Glutamax (1x, Invitrogen), 100U/ml penicillin, 100ug/ml streptomycin (Gibco), primocin 100ug/ml (InvivoGen), 10 uM Rock inhibitor Y-27632 (Selleck Chemical Inc)] varied from 10 to 45 minutes. Sample size varied between biopsies (<0.2 to 0.3 mm2) and resection specimens (<0.5 mm2 to 0.5 cm2).
Human Organoid Tissue Processing and Cell Culture Conditions
Tissue samples were washed at least three times with transport media and placed in a sterile 3 cm petri dish (Falcon) for either total mechanical dissociation or dissection into smaller pieces (<2 mm diameter) prior to enzymatic digestion. Enzymatic digestion was done with 2/3rd of 250 U/mL collagenase IV (Life Technologies) in combination with 1/3rd of 0.05% Trypsin-EDTA (Invitrogen) in a volume of at least 20 times the tissue volume. Tubes were incubated on a shaker at 200 rpm at a temperature of 37°C until the digestion solution turned cloudy. The suspension was mixed with 10% FBS (Denville) enriched DMEM to inactivate the enzymes and centrifuged at an average of 1000 rpm for 5 min to pellet cells. The cell pellets were washed once with 10% FBS enriched DMEM and once washed with DMEM only to wash away FBS residues. The cells were resuspended in a small volume of growth media [Advanced DMEM/F-12 (Invitrogen) with B27 supplement (Invitrogen), Glutamax (Invitrogen), Penicillin/Streptomycin (Invitrogen), HEPES (Invitrogen), N-Acetylcysteine (Sigma-Aldrich), [Leu15]-Gastrin I (Sigma-Aldrich), Y-27632 (VWR), A-83-01 (Tocris), SB202190 (Selleck Chemical Inc), Nicotinamide (Sigma-Aldrich), Recombinant EGF, and Noggin, Rspondin, WNT (all derived from in house conditioned media)] and mixed in a 1:2 volume of growth factor reduced Matrigel (Corning). Up to ten, 50–125uL drops of Matrigel/cell suspension were distributed into a 6-well cell suspension culture plate (SARSTEDT Ltd, Leicester, UK). The drops were solidified by a 15–30 minute incubation in the cell culture incubator at 37°C and 5% CO2. After solid drops formed 3–4 mL primary culture media was added. Each well was replaced with fresh culture media every 5 days until the organoid culture started growing and then every 2–4 days. Regular mycoplasma screening was performed using the MycoAlert kit (Lonza Inc).
Human Organoid Maintenance
Organoid media was changed every 2–3 days and they were passaged approximately 1:4 every 6–8 days. To passage, the growth media was removed and the Matrigel was resuspended in 3ml cold PBS, then transferred to a 15ml falcon tube. The organoids were mechanically disrupted using a p1000 pipette by repeatedly pipetting, approximately 50 times. 8ml of cold PBS was then added to the tube and the tubes were again pipetted, approx. 20 times, for further mechanical disruption. The cells were centrifuged at 1000RPM for 4 minutes and the supernatant was aspirated. They were then resuspended in Matrigel and replated as above. For freezing down organoid cells lines, the cells were resuspended in growth media containing 10% DMSO and 10% FBS after centrifugation above. They were stored in liquid nitrogen indefinitely.
Organoid Transplantation
‘shApc’ organoids were engrafted into recipient Athymic-Nude-Foxn1nu hosts (Envigo), which were pretreated with 2.5%–3% DSS in their drinking water for 5 days and daily body weights were recorded on Day 0, 3, 5 and 7. Notably, we observed no successful engraftment in Foxn1nu hosts treated with 1% or 2% DSS, and we also observed an increased rate of lethality when we treated males with 2.5%–3% DSS compared to females. After the animals were allowed to recover for 2 days, mice were anesthetized with isofluorane (2%) and the colon was flushed with room temperature sterile PBS. After flushing, 2000 shApc organoid fragments in 50ul of 5% Matrigel-PBS were instilled into the lumen of the colon by a p200 pipette enema, coated sparingly with petroleum jelly (Vaseline), over the course of 30 seconds. Organoid fragments were prepared identical as to passaging13 except that after centrifugation they were resuspended in 5% Matrigel-PBS and kept on ice until infusion. After infusion, the anal verge was sealed with 4 μl of Vetbond Tissue Adhesive (3M, 1469SB) to prevent luminal contents from being immediately excreted. They were then maintained on anesthesia for 5 minutes, and then allowed to recover in room air underneath a heat lamp. Six hours after the procedure the anal canal was inspected to make sure it was patent. For syngeneic transplants, C57Bl/6NHsd (Envigo) and C57Bl/6J (Jackson Labs) animals were treated with 2.5–3% DSS for 5 days as above. Importantly, we noticed significant differences in DSS toxicity, as measured by weight loss and animal survival, amongst C57Bl/6 strains ordered from different vendors, and so we proceeded forward with C57Bl/6J animals ordered from Jackson Labs. Prior to transplantation, organoids were digested in .25% Trypsin/.02% EDTA in PBS at 37°C for exactly 7 minutes, and then mechanically disassociated using a p200 pipette, then 10ml of DMEM (+Pen/Strep +10% Fetal Bovine Serum) was mixed with the solution and centrifuged at 1500 RPM for 5 minutes. They were inspected underneath the microscope to determine if they remained in clumps, and then resuspended to a final concentration of ~300K Cells/50ul in 5% Matrigel-PBS. Importantly, Apcmut/KrasG12D/p53mut cells were grown from freshly thawed stocks of cells that were cryogenically preserved at the same time they were characterized to be chromosomally stable and thus, genetically defined.
Splenic Injections
Organoid fragments were prepared as single cells following 10 minutes of digestion in .25% Trypsin/.02% EDTA at 37°C, followed by mechanical disassociation, and centrifugation with DMEM (+ Pen/Strep, +10% FBS), and then resuspension in 100% PBS. After induction of anesthesia, under sterile conditions, animal abdomens were shaved and sterilized. A 1 cm flank incision was made to expose the tip of the spleen, which was gently withdrawn from the abdomen and held in place with a sterile Q-tip. Using a 26-1/2 gauge needle, 0.1 mL total volume, containing 300,000 cells were injected underneath the splenic capsule over the course of 30 seconds. After needle withdrawal, pressure was held for 3–5 minutes to assure no bleeding, and then the incision was closed.
Mouse Tumor Organoid Derivation
Tumors were dissected and washed in sterile PBS. Under sterile conditions, a razor blade was used to mince the tumor into a slurry, and then collected in sterile PBS and pelleted. The pellet was resuspended in 1mg/ml Type IV Collagenase (Worthington) at 37°C for 30 minutes with occasional mixing, and then pelleted and washed in PBS twice. It was then resuspended in 5ml .25% Trypsin/.02% EDTA in PBS at 37°C for 5 minutes, and then washed with DMEM+FBS and pelleted, followed by two additional washes with DMEM+FBS. The supernatant was removed, leaving about .2ml, and the resuspended cells were added at 1:50 to matrigel and plated as organoids. The cultures were checked for 100% GFP positivity (ON Dox) following multiple passages to ensure a pure population of tumor derived organoids, as contaminating host stroma cells did not contain GFP.
Small Animal Imaging
Noninvasive, optical in vivo imaging was conducted using a Xenogen IVIS Spectrum. The IVIS Imaging System was prepared for use according to the IACUC Guidelines for Animal Use at MSKCC. Mouse colon MRI scans were performed on a 300 MHz Bruker 7T Biospec scanners (Bruker Biospin MRI GmbH, Ettlingen, Germany) equipped with 640 mT/m ID 115 mm gradients (Resonance Research, Inc., Billerica, MA). RF excitation and acquisition was achieved by a custom-built quadrature birdcage resonator with ID of 32 mm (Stark Contrast MRI Coils Research Inc., Erlangen, Germany). The mice were anesthetized with 1% isoflurane (Baxter Healthcare Corp., Deerfield, IL) gas in oxygen. Animal respiration was monitored with a small animal physiological monitoring system (SA Instruments, Inc., Stony Brook, New York). Scout images along three orthogonal orientations were first acquired for animal positioning. For colon imaging, axial T2-weighted images using fast spin-echo RARE sequence (Rapid Acquisition with Relaxation Enhancement) was acquired with TR 5.1, TE 47 ms, RARE factor of 8, slice thickness of 0.8 mm, FOV 30 mm, in-plane resolution of 117 × 156 um, and 8 averages.
Immunophenotyping and CBC Analysis
Wildtype female C57Bl6/J mice, aged 4–6 weeks, were treated with 3% DSS for 5 days per the transplantation protocol and harvested at Day 7, Day 35 and Day 70 post transplant. Untreated mice were collected as controls. Single cell suspensions were derived from spleen through filtration (40μM) and peripheral blood, while red blood cells were lysed with ACK buffer. Cells were incubated with antibodies for 1 hour on ice. All antibodies were purchased from eBioscience or BioLegend. We used the following fluorochrome anti-mouse antibodies: CD11b (M1/70), Gr-1 (RB6-8C5), CD3 (17A2), CD4 (GK1.5), CD8 (53-6.7), B220 (B114968) (Should you include the concentrations or dilutions used?). Stained cells were quantified using a BD Fortessa analyzer. FlowJo software (Tree Star) was used to generate FACS plots, tables and calculate the percentages of each population. The bar graphs were generated using Prism 6 and significance was determined using unpaired two-tailed t-test, without Welch correction as the variance was not significantly different. For the CBC analysis we used the Hemavet 950FS (Drew) hematology analyzer after collecting the peripheral blood in EDTA tubes (Sarstedt 20.1278.100). The CBC graphs were generated as described above using Prism 6.
Vector Generation
The U6-sgRNA-EFS-Cas9-P2A-Cre (sgRNA-CC) lentiviral vector was constructed based on the lentiCRISPR backbone28. We replaced PuroR with a Cre (PCR amplified from Puro.Cre empty vector29) and, subsequently, we destroyed the BsmB1 restriction sites inside the Cre cDNA using Quick-Change II Site Directed Mutagenesis (200523, from Agilent Technologies) according to instructions provided with the kit. The primers used were NheI-Fw-Cre: 5′TCAGCGCTAGCGGCAGCGGCGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGTCCAATTTACTGACCGTACACC 3′ and MluI-Rv-Cre: 5′ GATCGACGCGTCTAATCGCCATCTTCCAGCAGGCGCACC 3′. For sgRNA cloning, the sgRNA-CC vector was digested with BsmBI and ligated with BsmB1-compatible annealed APC sgRNA oligos made previously16.
Supplementary Material
Acknowledgments
We thank D Grace, S Tian and M Taylor for technical assistance with animal colonies, and other members of the Lowe laboratory for advice and discussions, and J Shia for assistance with histopathology, M Gollub for assistance with interpreting MRI studies, as well as C LeKaye, M Lupu and D Winkleman for their technical support. We also thank members of the Englander Institute for Precision Medicine Organoid Platform, T McNary, Y Churakova and C Cheung. This work was supported by grants from the Starr Cancer Consortium (I7-A771, to Mark A. Rubin and Himisha Beltran; and I8-A8-030 to Norbert Perrimon, Scott Lowe, and Lukas Dow), the Department of Defense (PC121341; to Himisha Beltran), and a Damon Runyon Cancer Research Foundation-Gordon Family Clinical Investigator Award (CI-67-13; to Himisha Beltran). This work was supported by grants from the NIH (U54 OD020355-01, R01 CA195787-01 and P30 CA008748). KPO is supported by an F30 Award from the NIH/NCI (1CA200110-01A1). TB was supported by the MSKCC Single-Cell Sequencing Initiative, The William and Joyce O’Neil Research Fund. KPO and EMS were supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM07739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. PBR is supported by a K12 Paul Calebresi Career Development Award for Clinical Oncology (CA 187069). LED was supported by a K22 Career Development Award from the NCI/NIH (CA 181280-01). Animal imaging studies were supported by the NIH Small-Animal Imaging Research Program (SAIRP), R24 CA83084; NIH Center Grant, P30 CA08748; NIH Prostate SPORE, P50-CA92629. SWL is the Geoffrey Beene Chair of Cancer Biology and an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions: K.P.O., L.E.D. and S.W.L. conceived the project. K.P.O, designed, performed and analyzed experiments, and wrote the paper. E.L, G.L, E.M.S, T. B., E.M, J.S., P.R., B.L, T.H, C.P, H.B., and M.A.R provided reagents, performed or analyzed experiments. L.E.D and S.W.L supervised experiments, analyzed data and wrote the paper.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. New England Journal of Medicine. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taketo MM, Edelmann W. Mouse Models of Colon Cancer. YGAST. 2009;136:780–798. doi: 10.1053/j.gastro.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 4.Heijstek MW, Kranenburg O, Borel Rinkes IHM. Mouse Models of Colorectal Cancer and Liver Metastases. Digestive Surgery. 2005;22:16–25. doi: 10.1159/000085342. [DOI] [PubMed] [Google Scholar]
- 5.Oh BY, Hong HK, Lee WY, Cho YB. Animal models of colorectal cancer with liver metastasis. Cancer Lett. 2016 doi: 10.1016/j.canlet.2016.01.048. [DOI] [PubMed] [Google Scholar]
- 6.Hinoi T, et al. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 2007;67:9721–9730. doi: 10.1158/0008-5472.CAN-07-2735. [DOI] [PubMed] [Google Scholar]
- 7.Byun AJ, et al. Colon-specific tumorigenesis in mice driven by Cre-mediated inactivation of Apc and activation of mutant Kras. Cancer Lett. 2014;347:191–195. doi: 10.1016/j.canlet.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue Y, Johnson R, Desmet M, Snyder PW, Fleet JC. Generation of a transgenic mouse for colorectal cancer research with intestinal cre expression limited to the large intestine. Mol Cancer Res. 2010;8:1095–1104. doi: 10.1158/1541-7786.MCR-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tetteh PW, et al. Generation of an inducible colon-specific Cre enzyme mouse line for colon cancer research. Proc Natl Acad Sci U S A. 2016;113:11859–11864. doi: 10.1073/pnas.1614057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matano M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 11.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 12.O’Rourke KP, Dow LE, Lowe SW. Immunofluorescent Staining of Mouse Intestinal Stem Cells. bio-protocol.org. 2016;6:e1732. doi: 10.21769/bioprotoc.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Rourke KP, Ackerman S, Dow LE, Lowe SW. Isolation, Culture, and Maintenance of Mouse Intestinal Stem Cells. bio-protocol.org. 2016;6:e1733. doi: 10.21769/bioprotoc.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 15.Sato T, et al. Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett's Epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 16.Dow LE, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015;33:390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koo BK, et al. Controlled gene expression in primary Lgr5 organoid cultures. Nature methods. 2012;9:81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- 18.Dow LE, et al. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell. 2015;161:1539–1552. doi: 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Onuma K, et al. Genetic reconstitution of tumorigenesis in primary intestinal cells. Proc Natl Acad Sci U S A. 2013;110:11127–11132. doi: 10.1073/pnas.1221926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yui S, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nature Medicine. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- 22.Brannon AR, et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome biology. 2014;15:454. doi: 10.1186/s13059-014-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin ES, et al. Development of a colon cancer GEMM-derived orthotopic transplant model for drug discovery and validation. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:2929–2940. doi: 10.1158/1078-0432.CCR-12-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura T, et al. SMAD4-deficient intestinal tumors recruit CCR1+ myeloid cells that promote invasion. Nat Genet. 2007;39:467–475. doi: 10.1038/ng1997. [DOI] [PubMed] [Google Scholar]
- 25.Takaku K, et al. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 26.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baslan T, et al. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res. 2015;25:714–724. doi: 10.1101/gr.188060.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar MS, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.