Abstract
Large genome-mapping consortia and thousands of genome-wide association studies have identified non-protein coding elements in the genome as a having a central role in various biological processes. However, decoding the function of the millions of putative regulatory elements discovered in these studies remains challenging. CRISPR–Cas9-based epigenome editing technologies have enabled precise perturbation of the activity of specific regulatory elements. Here we describe CRISPR–Cas9-based epigenomic regulatory element screening (CERES) for improved high-throughput screening of regulatory element activity within the native genomic context. Using dCas9KRAB repressor and dCas9p300 activator constructs and lentiviral sgRNA libraries targeting DNase I hypersensitive sites surrounding a gene of interest, we perform both loss- and gain-of-function screens to identify regulatory elements for the β-globin and the HER2 loci in human cells. CERES readily identified known and novel regulatory elements, some of which were dependent on cell type or direction of perturbation. This technology allows the high-throughput functional annotation of putative regulatory elements in their native chromosomal context.
Understanding how the human genome transforms dynamic biological signals into complex patterns of gene expression remains a fundamental challenge for biological research. Overcoming that challenge is especially important for advancing our ability to treat human disease, since >90% of the genetic variation associated with complex disease falls within the non-coding genome1, 2. Large annotation efforts such as ENCODE3 and the NIH Roadmap Epigenomics Program4 have revealed millions of putative regulatory elements across >100 human cell types that contribute to the transformation of biological signals into cellular responses and phenotypes1. The potential function of any one of those elements is typically unknown, however, because there are not sufficiently high-throughput assays to measure endogenous regulatory element activity or to link that activity to specific functions and target genes. High-throughput massively parallel reporter assays have been effective in quantifying the activity of regulatory elements in particular cell types5–7, but do not recapitulate regulatory element interactions at the native chromosomal locus and cannot identify target genes of each element. Until recently, directly interrogating these critical regulatory regions in their native chromosomal position has not been possible.
The type II clustered, regularly interspaced, short palindromic repeat/CRISPR-associated protein (CRISPR–Cas9) system is a versatile tool for genome engineering8–12. Genome editing tools, including Cas9 and pooled guide RNA (gRNA) libraries, have also been used for high-throughput loss-of-function screens of genes13–15 and regulatory elements16–21. Such CRISPR–Cas9 screens rely on ablating the activity of critical regions, such as transcription factor binding sites, by introducing short insertions or deletions (indels) of DNA following Cas9-induced cleavage of the genome and repair by non-homologous end joining (NHEJ). Consequently, a high density of gRNAs is needed to saturate all possible transcription factor binding sites in each regulatory element. In many cases, this degree of saturation is not possible because a protospacer adjacent motif (PAM) sequence is not located within the transcription factor binding sequence. Finally, screening regulatory elements with genome editing only examines loss-of-function by genomic disruption, and does not permit gain-of-function analyses22.
Nuclease-deactivated Cas9 (dCas9) can be fused to epigenome-modifying protein domains to precisely modulate gene expression from gene promoters and both proximal and distal genomic enhancer regions23–37. Additionally, repressor- or activator-domain fusions to dCas9 combined with pools of gRNAs targeting gene promoters have been used to perform genome-wide CRISPRi and CRISPRa screens28, 29. Here, we focus on the dCas9-based transcriptional repressor, dCas9KRAB, and activator, dCas9p300. Fusion of the KRAB (Krüppel-associated box) domain to dCas9 and targeting it to a promoter or enhancer causes highly specific gene repression by recruiting a host of factors that deposit H3K9 trimethylation (H3K9me3) that ultimately results in heterochromatin formation24, 32. Conversely, fusion of the E1A-associated protein p300 histone acetyltransferase (HAT) core domain to dCas9 and targeting it to either promoters or enhancers induces target gene activation concomitant with deposition of H3K27 acetylation (H3K27ac)30. Because screening with epigenome editing perturbs regulatory element activity directly rather than via DNA mutation, the approach offers several advantages over genome editing screens. Epigenome editing can modulate regulatory element activity even if there is not a PAM located precisely within a critical transcription factor binding motif. Fewer gRNAs may be needed to achieve robust modulation of each candidate regulatory element, enabling the screening of a larger number of genomic regions with a fixed gRNA library size (Supplementary Table 1). Finally, the dCas9p300 tool provides an opportunity to screen for gain of regulatory element function that is not possible with NHEJ-mediated genome editing, even if the specific transcription factors that bind the element when active are not present in the cell type being studied. The use of dCas9KRAB and dCas9p300 in parallel screens around the same loci uniquely facilitates the identification of elements that are necessary and sufficient, respectively, for target gene expression.
DNase I hypersensitivity sequencing (DNase-Seq) is a genome-wide, transcription factor-agnostic measure of chromatin accessibility, corresponding to genomic loci where proteins are bound to DNA38, 39. Many of the ~2 million DNase I hypersensitive sites (DHSs) that have been identified across many human cell and tissue types are likely regulatory elements which influence gene expression, and many of these are cell-type-specific1. However, systematically identifying and quantifying the impact that these regulatory elements have on gene expression levels has not been possible. Here we demonstrate the utility of CRISPR–Cas9-based Epigenomic Regulatory Element Screening (CERES) by targeting DHSs surrounding genes of interest to identify endogenous regulatory elements through loss- and gain-of-function epigenome editing.
Results
Epigenetic Repression Recovers Known Regulatory Elements
We first designed pools of gRNAs to specifically target all DHSs in a particular segment of the genome. Protospacer sequences are identified within each DHS surrounding the gene of interest and ranked by possible off-target alignments (see Methods). The gRNA library pool is synthesized and cloned into a lentiviral expression vector used to generate a pool of lentiviral particles that deliver individual gRNAs to the target cells (Fig. 1a) 16, 17.
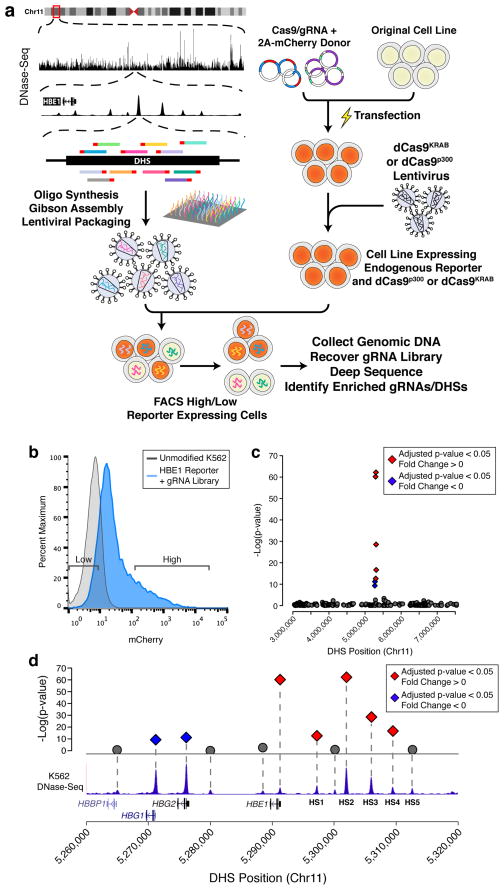
Figure 1.
CRISPR–Cas9-based Epigenetic Regulatory Element Screening (CERES) identifies regulatory elements of the β-globin locus in a loss-of-function screen. (a) CERES involves the design and synthesis of libraries of gRNAs targeted to all candidate gene regulatory elements in a genomic region, in this case as defined by DNase I hypersensitive sites (DHS) identified by DNase-seq. Lentiviral vectors encoding the gRNA library are delivered to cell lines expressing the dCas9KRAB repressor, for loss-of-function screens, or the dCas9p300 activator, for gain-of-function screens. The cells can then be selected for changes in phenotype, such as gain or loss of expression of a target gene. Sequencing the gRNAs in the selected cell subpopulations and mapping them back to the genome reveals regulatory elements involved in controlling the selected phenotype. In the example shown here, a gRNA library was designed to all DHSs in a 4.5 Mb region surrounding the β-globin locus, and introduced into human K562 cells expressing dCas9KRAB and containing an mCherry reporter at the HBE1 gene. (b) Representative flow cytometry data of the HBE1 reporter cells containing the gRNA library, and expression levels of cells sorted for gRNA enrichment. (c) Manhattan plot of a high-throughput screen for regulatory elements in the 4.5 Mb surrounding the globin locus using the dCas9KRAB repressor. (d) Enriched DHSs following selection for decreased HBE1 expression were found only in the HBE1 promoter and enhancers (HS1-4), while the promoters of HBG1/2 were enriched in cells with increased HBE1 expression. Diamonds indicate adjusted p-value < 0.05 and gray circles represent adjusted p-value > 0.05. Red indicates DHS fold change < 0 and blue indicates DHS fold change > 0.
We screened for regulatory elements within the well-characterized β-globin locus. The β-globin locus control region (LCR) contains five DHSs (HS1-5) upstream of five globin genes (HBE1, HBG1/2, HBD, HBB)40. This LCR controls the expression of each gene at different times throughout development in erythroid cells. The K562 myelogenous leukemia cell line highly expresses HBE1 and HBG1/2 and has accessible chromatin at all five DHSs in the β-globin LCR42. We previously demonstrated robust modulation of HBE1 expression by epigenome editing of the HS2 enhancer30, 32. To more easily monitor transcriptional modulation of HBE1, we generated an endogenous reporter in K562 cells by replacing the stop codon of HBE1 with a P2A-mCherry sequence via CRISPR–Cas9-mediated genome editing. These reporter cells were then transduced with a lentivirus encoding dCas9KRAB and clonal isogenic populations were derived from single cells (Fig. 1a). To test for proper reporter activity, cells were transduced individually with four different gRNAs targeting the HS2 enhancer and flow cytometry was used to verify reduced mCherry expression (Supplementary Fig. 1 and Supplementary Table 2).
DNase-seq data from K562 cells43 was used to design a library of 10,739 gRNAs targeting 281 DHSs in a 4.5 megabase region surrounding the β-globin locus and 1,733 control gRNAs targeting regions between DHSs in this region. The HBE1 reporter cell line was transduced at a low MOI to introduce one gRNA per cell and after two days blasticidin S was added for 12 days of culture to select for gRNA-containing cells. We performed fluorescence-activated cell sorting (FACS) to isolate the cells expressing the lowest and highest 10% of mCherry levels (Fig. 1b). As a control, we also collected unsorted cells. We then used high-throughput sequencing to estimate the relative abundance of each gRNA in the genomic DNA of each sample. We identified DHSs involved in HBE1 regulation by grouping gRNAs within a DHS as replicates and using linear regression to identify DHSs with significant differences in gRNA abundance between the high and low mCherry cell populations. Notably, the only DHSs significantly enriched within the low HBE1 library were four of the β-globin LCR enhancers, HS1-4, and the promoter of HBE1 (Figs. 1c and 1d). The HBG1/2 promoters were significantly (FDR < 0.05) enriched in the high HBE1 population, indicating that repression of the HBG1/2 promoters leads to upregulation of HBE1, potentially by relieving competition between the promoters for the HS1-5 enhancers.
As an alternative analysis strategy, we determined enrichment of individual gRNAs and detected similar trends, with the strongest enrichment in the HBE1, HBG1/2 promoter regions and HS1-4 enhancer regions (Supplementary Fig. 2). Control gRNAs outside of the regions of DNase I signal in K562 cells, including gRNAs interspersed between HS1-4, were not significantly enriched. We validated our results by individually delivering the most differentially enriched gRNAs of each significant DHS under the same conditions as used in the screen. We then measured the fold-change of mCherry expression and HBE1 mRNA levels by flow cytometry and RT-qPCR, respectively. We observed a high correlation of both measurements (Spearman ρ = 0.9429 and ρ = 0.8857, respectively) with the log2(fold-change) of the individual gRNAs from the screen (Supplementary Fig. 3).
Epigenetic Repression Reveals Gene Regulatory Elements
We next extended this approach to identify regulatory elements of HER2 (ERBB2/Neu), an oncogene overexpressed or amplified in approximately 20–30% of breast cancers44. HER2-positive breast cancer has the second-poorest prognosis rate of all subtypes. The HER2 humanized monoclonal antibody trastuzumab is part of the standard of care treatment and can improve survival 20–25 months45. However, less than 35% of patients initially respond to treatment and 70% of those who first respond can develop resistance to the drug with resistance often arising due to protein regulation44, 46. This provides an attractive target to screen for the regulatory regions that may be controlling HER2 expression.
To design the gRNA library, we generated DNase-seq data for the SKBR3 HER2-amplified breast cancer cell line and used the DHS coordinates as input for identifying gRNAs. The pooled gRNA library was generated as described above with the final library containing 12,189 gRNAs targeting 433 DHSs in a 4 megabase region surrounding HER2 and 283 control gRNAs within that same region not targeting DHSs. As HER2 is a transmembrane protein, we used immunofluorescence staining to monitor expression as an alternative to generating an endogenous reporter cell line. A431 epidermoid carcinoma cells, which express moderate levels of HER2, were transduced with dCas9KRAB lentivirus and selected for stable expression. The selected, polyclonal A431-dCas9KRAB cells were then transduced with the gRNA library at low MOI and selected for the gRNA vector with antibiotic. Next, flow cytometry was used to sort cells with the lowest and highest 10% of HER2 (Fig. 2a). gRNAs were sequenced and enrichment analysis was performed. When comparing the highest to lowest HER2-expressing samples, we identified several DHSs containing individual gRNAs that were differentially represented between the two groups (Figs. 2b and 2c), as well as DHS that were called when using individual gRNAs as replicates within a DHS (Supp. Fig. 4). Enrichment in the low HER2 population included the HER2 promoter, several DHSs in the first intron of HER2 including one previously identified regulatory element47, and several DHSs downstream of HER2 including one DHS in the first intron of GRB7. In high HER2 cells, we detected enrichment of gRNAs in two DHSs near the promoter of GRB7. We also showed that GRB7 mRNA was significantly (p < 0.05) reduced when targeting the promoter or first intron of GRB7 (Supplementary Fig. 5c). Therefore the regulation of HER2 levels via the DHSs in the GRB7 promoter may be due to relieving competition of distal enhancers or possibly due to post-transcriptional secondary effects, as GRB7 is involved in the phosphorylation of HER248. To validate the screen, several of the most enriched gRNAs were delivered individually to the same A431-dCas9KRAB cells. We again found a high degree of correlation between the fold-change of HER2 mRNA or protein levels, determined by RT-qPCR (Spearman ρ = 0.5175) or immunofluorescence staining (ρ =0.5701), respectively, and the log2(fold-change) of gRNA representation in the screen (Figs. 2e and Supplementary Figs. 5b, p < 0.05).
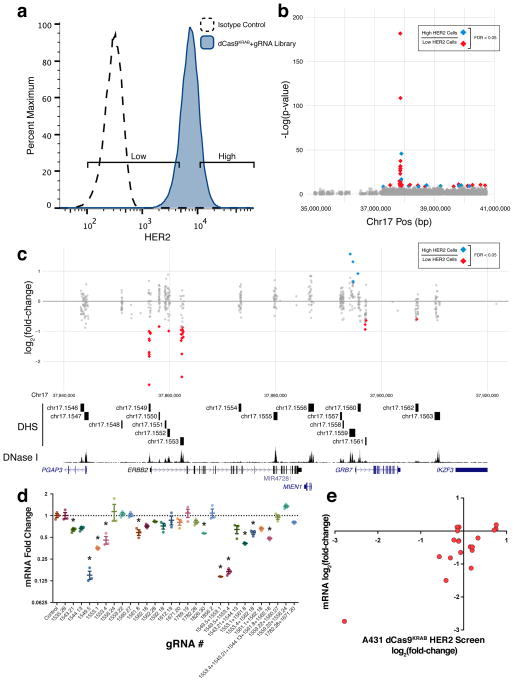
Figure 2.
A dCas9KRAB loss-of-function screen in A431 cells identifies regulatory elements of HER2. (a) Flow cytometry for HER2 expression in A431 cells expressing dCas9KRAB and a gRNA library targeted to all DHSs in 4 Mb surrounding HER2 detected in the HER2-overexpressing SKRB3 breast cancer cell line. (b) Manhattan plot showing regulatory elements affecting HER2 expression. (c) A detailed view of the region around HER2. When comparing high and low HER2-expressing cell populations, gRNAs are enriched in the promoter and an intronic DHS of HER2, as well as surrounding DHSs including the promoter of GRB7, an adapter protein that associates with tyrosine kinases. (d) HER2 mRNA fold-change in response to the most enriched gRNAs from (b) relative to treatment with a control gRNA. * indicates samples are significantly different from control as determined by one-way analysis of variance followed by Dunnett’s Test; adjusted P < 0.05. (n = 3 biological replicates, mean ± SEM). (e) HER2 mRNA log2 fold-change in response to gRNA treatment versus log2 fold-change of the abundance of the corresponding gRNA in the HER2 dCas9KRAB screen (Spearman correlation ρ = 0.5175).
Epigenetic Activation Reveals Gene Regulatory Elements
One application of epigenome editing is the targeted activation of regulatory elements in their natural chromosomal position. This provides the unique opportunity to identify regulatory elements that induce the expression of a gene not normally expressed in a particular cell type. To extend CERES to gain-of-function screens for regulatory elements sufficient to activate target gene expression, we used the dCas9p300 activator30. Human HEK293T cells, which express low levels of HER2, were transduced with dCas9p300 lentivirus and selected with antibiotic to obtain a polyclonal cell line stably expressing the transgene. The HEK293T-dCas9p300 cells were then transduced with the same gRNA library used in the A431-dCas9KRAB screen targeting the 4 Mb region around HER2. At two days after transduction, antibiotic selection was performed for seven days to select cells containing the gRNA vector. Cells were then sorted for high and low HER2 expression (Fig. 3a). We observed a profile that largely mirrored the dCas9KRAB screen, with effects in the opposite direction, including enrichment of individual gRNAs (Figs. 3b and 3c) and DHSs (Supp. Fig. 6), using each gRNA as a replicate, in the promoter region and first intron of HER2. We individually validated the most enriched gRNAs in HEK293T-dCas9p300 cells and detected a high degree correlation of both mRNA fold-change (Spearman ρ = 0.7818) and protein abundance (ρ = 0.8545) with the log2(fold-change) of gRNA abundance in the screen (Figs. 3d, 3e and Supplementary Figs. 7a, b). To confirm that gene regulation at these DHSs was associated with the intended epigenetic editing by dCas9KRAB and dCas9p300, we performed ChIP-qPCR to validate enrichment of H3K9me3 and H3K27ac, respectively (Fig. 4).
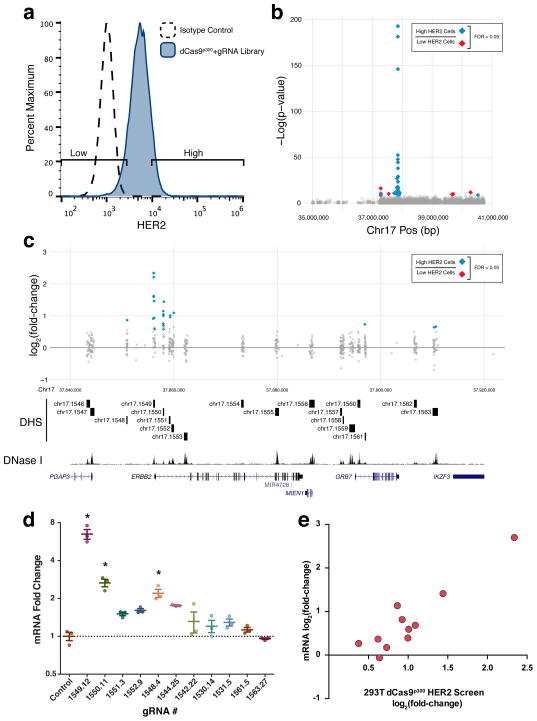
Figure 3.
A dCas9p300 gain-of-function screen in HEK293T cells identifies regulatory elements of HER2. (a) Flow cytometry for HER2 expression in HEK293T cells expressing dCas9p300 and a gRNA library targeted to all the DHSs in a 4 Mb region surrounding HER2 found in the SKBR3 HER2-overexpressing breast cancer cell line. (b) Manhattan plot showing the results of a screen for regulatory elements affecting HER2 expression. (c) A detailed view of the region around HER2. When comparing high and low HER2-expressing cell populations, gRNAs are enriched in the promoter and three intronic DHSs of HER2 as well as several nearby DHSs. (d) HER2 mRNA fold-change in response to the most enriched gRNAs from (b) relative to treatment with a control gRNA. * indicates samples are significantly different from control as determined by one-way analysis of variance followed by Dunnett’s Test; adjusted P < 0.05 (n = 3 biological replicates, mean ± SEM). (e) HER2 mRNA log2 fold-change in response to gRNA treatment versus log2 fold-change of the abundance of the corresponding gRNA in the HER2 HEK293T dCas9p300 screen (Spearman correlation ρ =0.9429).
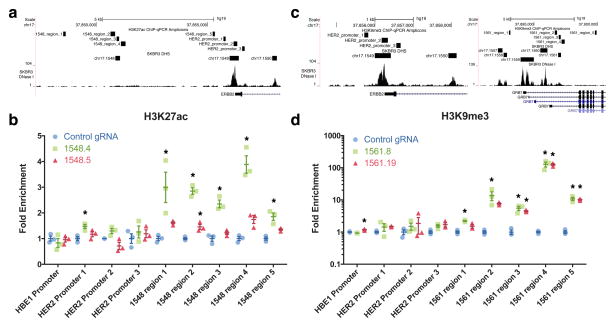
Figure 4.
dCas9p300 and dCas9KRAB remodel epigenetic marks near novel regulatory elements identified from screens. (a) Genomic tracks displaying the qPCR amplicon regions for the H3K27ac ChIP-qPCR assay, DNase-seq signal, and DHS sites for SKBR3 cells near HER2. (b) dCas9p300 targeted to a DHS with a gRNA enriched in the screen (1548.4) in HEK293T cells leads to H3K27ac enrichment near the DHS, in contrast to gRNA 1548.5 targeting the same DHS but was not enriched in the screen. (c) Genomic tracks displaying the qPCR amplicon regions for the H3K9me3 ChIP-qPCR assay, DNase-seq signal, and DHS sites for SKBR3 cells near HER2 and GRB7. (d) dCas9KRAB targeted to a DHS with a gRNA enriched in the screen (1561.8) in A431 cells deposits H3K9me3 near the DHS. A gRNA that was not significantly enriched in the screen (1561.19) also deposits similar amounts of H3K9me3 near the DHS. * indicates samples are significantly different from control as determined by one-way analysis of variance followed by Dunnett’s Test; adjusted P < 0.05 (n = 3 biological replicates, mean ± SEM). All fold enrichments are relative to transduction of a control gRNA and normalized to a region of the ACTB locus.
Screen Results Depend on Cell Type and Direction of Perturbation
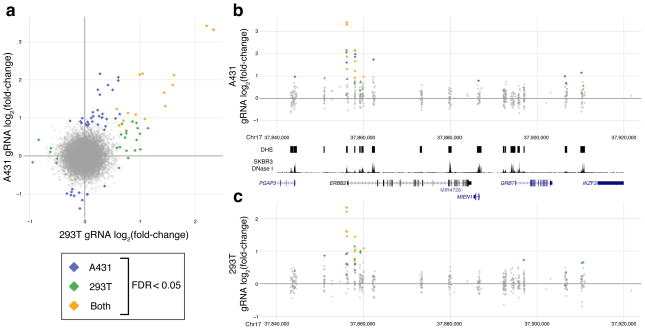
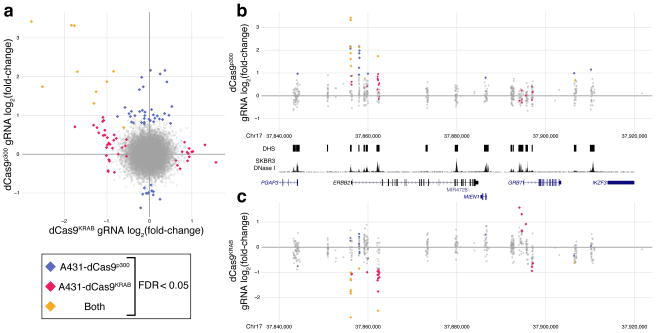
The strongest intronic DHS (DHS_1553) identified in the dCas9KRAB repressor screen in A431 cells was not identified to modulate HER2 expression in the dCas9p300 gain-of-function screen in HEK293T cells, which we further validated with individual gRNAs (Supplementary Figs. 7c, d). However, when we performed the same gain-of-function screen in A431 cells (Supplementary Figs. 8–9), this intronic DHS was selected, indicating that although many of the regulatory elements we found were shared between cell types, some may be cell type-specific. (Fig. 5). This finding indicates that certain cell types have complex and possibly redundant patterns of regulatory element usage, and that CERES can be used to determine cell type-specific enhancer activity. Additionally, the DHSs near the GRB7 promoter that were identified in the dCas9KRAB screen were not enriched in the A431 dCas9p300 screen (Fig. 6). This indicates that active regulatory elements may not be identified in gain-of-function screens and repressed elements may not be identified in loss-of-function screens. This also underscores the need for the combination of repression and activation screens to provide a comprehensive description of gene regulation. Future iterations of the CERES technology, including combinatorial screens that combine activation and repression or target multiple regulatory elements simultaneously, may help to inform these patterns.
Figure 5.
Comparison of HER2 activation screens between different cell types. (a) Log2 fold-change gRNA abundance in the dCas9p300 screen in HEK293T cells versus A431 cells. (b and c) A detailed view of the region around HER2 for the dCas9p300 screens in (b) A431 cells or (c) HEK293T cells. Several of the same DHS contained enriched gRNAs in both screens, including the promoter region, but others, such as an intronic DHS highly enriched in the dCas9KRAB screen in A431 cells, were only enriched in a single screen and may be cell-type specific enhancers.
Figure 6.
Comparisons between HER2 activation and repression screens. (a) Log2 fold-change of the gRNA abundance in the dCas9KRAB screen versus the dCas9p300 screen in A431 cells. (b and c) A detailed view of the region around HER2 for the (b) dCas9p300 or (c) dCas9KRAB screens in A431 cells. Several DHSs contain enriched gRNAs in both screens, including the promoter and an HER2-intronic DHSs. However, DHSs in the GRB7 promoter region are only enriched in the dCas9KRAB screen.
Discussion
New technologies are necessary to address the unique challenges of annotating the function of regulatory elements, in contrast to well-established methods for screening protein-coding genes. Current models of gene regulation often involve multiple regulatory elements that individually may have modest effects on promoter activity but collectively synergize to generate robust gene expression programs in response to complex cell environments and signals. In those models, the perturbations of individual elements in screens such as CERES are likely to have subtle effects, particularly compared to gene knockout screens or screens targeting gene promoters. Indeed, few of the gRNAs in our screens drive more than 2-fold change in gene expression (Fig. 2c, 3c). Nevertheless, targeted validation experiments confirm that these are in fact modulators of target gene expression (Supp. Figs. 5, 7, 9). Furthermore, many of the other gRNAs in those DHS have a consistent direction of effect (e.g. Fig. 2c, DHS 1561; Fig. 3c, DHS 1563; and Supp. Fig. 10). Those findings are consistent with individual regulatory elements having a weak effect on target gene expression, and show the CERES protocol can reproducibly detect those weak effects. Nonetheless, it is possible that our current protocol may miss some of the weak effects of regulatory elements. That limitation may be addressed in the future by improvements to epigenome editing tools and by increasing sample size beyond four biological replicates as used here.
Fulco et al. 49 recently reported a similar loss-of-function screen for regulatory elements using the dCas9KRAB repressor. That study used all gRNAs tiled along a particular genomic region, whereas we focused on putative regulatory elements, such as those defined by DNase-seq. Targeting DHSs allowed us to focus on a much larger genomic window per gRNA, which may be useful to find long range interactions or indirect effects of regulatory elements on a particular gene or phenotype. Although Fulco et al. observed strong correlation of functional regulatory elements with DNase-seq signal, other studies using Cas9-based saturation mutagenesis of non-coding regions found some hits in their screen that were not characterized by DNase I hypersensitivity18, 20. In the context of the gain-of-function dCas9p300 screen, we compared our DHS-focused approach to a saturation screen that included all possible unique gRNAs in a 60 kb window around the HER2 TSS and all possible gRNAs in all DHSs that included any hits in the original screen (Supp. Figs. 11–12). The results were largely overlapping with some differences, indicating that it is valuable to choose gRNA distribution that best suits a particular screen.
To better understand the differences of Cas9-based mutagenesis screens and epigenome editing–based screens, we used this same saturation library with an analogous Cas9 vector and compared the results with the original DHS-focused dCas9KRAB screen (Supp. Figs. 13–15). The hits in the Cas9 screen were predominantly in the coding exons (Supp. Fig. 13) and even the known regulatory elements were missed in this screen (Supp. Fig. 14), supporting the advantages of an epigenome editing approach.
In summary, we show that CERES offers a highly scalable epigenome editing-based platform that enables parallel loss- and gain-of-function methods for screening the function of regulatory regions of the genome in their native genomic context. CERES can identify both proximal and distal regulatory elements that influence gene expression, some of which are located large distances from their target genes. Using CERES, we found all the known regulatory elements of the well-characterized β-globin locus, and also known and—to our knowledge— previously unknown elements controlling levels of the oncogene HER2. In addition to identifying enhancers, CERES also identifies regulatory elements that may act through direct competition with the promoter region or through downstream secondary mechanisms. The ability to switch between screens that utilize either epigenomic repression by dCas9KRAB or activation by dCas9p300 allows further mechanistic understanding of regulatory element activity and reveals unexpected dependencies between genes. In addition, these results further provide evidence that dCas9p300 is sufficient to activate gene expression from distal regulatory elements. Following large-scale screens to identify regulatory element function by CERES, saturation mutagenesis screens with genome editing could be performed on targeted regulatory elements to identify the essential transcription factor binding motifs that facilitate enhancer activity16–20, 50. Although this study used chromatin accessibility DNase-seq maps to define the regulatory element landscape, the gRNA library could be designed based on any input, including locations based on ChIP-Seq, Gro-Seq, or SNPs identified in genome-wide association studies or eQTL analyses. The gRNA library may target all putative regulatory elements or target elements with differential epigenetic signatures between cell types or treatments.
Additionally, CERES could be used to screen for regulatory elements that impact cellular phenotype, including drug resistance or cell growth. Although we targeted gRNA libraries to putative regulatory elements within a region of the genome, scaling up to regulatory elements across the entire genome is feasible. Our study shows that epigenome editing-based screening is a powerful tool for dissecting the regulatory networks that coordinate gene expression. Ultimately, we envision these methods will inform how altered regulation of gene expression contributes to disease, drug response, and regeneration. By illuminating the function of the non-coding genome, we also expect these technologies will provide a new class of drug targets for diverse indications.
Supplementary Methods
Plasmids
The lentiviral dCas9-KRAB plasmid was generated by cloning in a Hygromycin resistance gene driven by the PGK promoter after dCas9-KRAB using Gibson assembly (NEB, E2611L). The lentiviral gRNA expression plasmid was cloned by combining a U6-gRNA cassette with an EGFP-P2A-Bsr or DsRed-P2A-Bsr cassette into a lentiviral expression backbone using Gibson assembly. The donor plasmid was cloned by inserting homology arms (surrounding the HBE1 stop codon), amplified by PCR from genomic DNA of K562 cells, flanking a P2A-mCherry sequence with a LoxP-puromycin resistance cassette using Gibson assembly. Individual gRNAs were ordered as oligonucleotides (IDT-DNA), phosphorylated, hybridized, and cloned via into the EGFP gRNA plasmid for the HBE1 screen or DsRed gRNA plasmid for the HER2 screens using BsmBI sites.
The lentiviral dCas9-p300 construct was generated by PCR-amplification of Cas9 from lentiCRISPRv251 (Addgene #52961) with primers generating D10A and H840A mutations in overlapping fragments. The p300 core effector domain was PCR-amplified from Addgene #6135730, with a C-terminal FLAG epitope included. These fragments were cloned into a lentiCRISPRv2 backbone lacking the U6/sgRNA cassette using Gibson assembly (NEB, E2611L).
Cell Culture
K562, HEK293T, and A431 cells were obtained from the American Tissue Collection Center (ATCC) via the Duke University Cancer Center Facilities. K562 cells were maintained in RPMI 1640 media supplemented with 10% FBS and 1% penicillin-streptomycin. HEK293T and A431 cells were maintained in DMEM High Glucose supplemented with 10% FBS and 1% penicillin-streptomycin. All cell lines were cultured at 37 °C and 5% CO2.
The K562-dCas9KRAB HBE1 reporter cell line was generated by electroporation of 2 × 106 cells with a donor plasmid (6.67 μg), a HBE1 stop codon-targeting gRNA plasmid (6.67 μg), and a Cas9 plasmid (6.67 μg), as described previously32. Transfected cells were selected with puromycin (Sigma, P8833) at a concentration of 1 μg/mL, beginning 2 days after electroporation, for 7 days. To remove the puromycin selection cassette, cells were transfected with a CMV-CRE plasmid (20 μg) and cultured for 10 days without antibiotic selection. Next, the cells were transduced with a lentivirus containing dCas9KRAB in media containing 4 μg/mL polybrene (Santa Cruz, sc-134220). Cells were selected with Hygromycin B (200 μg/mL) for 7 days. Cells were then clonally isolated via serial dilutions to obtain a single clone harboring the knocked-in reporter without the puromycin cassette and dCas9KRAB.
The A431-dCas9p300 and HEK293T-dCas9p300 cell lines were derived by transducing either A431 or HEK293T cells with a lentivirus containing dCas9p300-P2A-puromycin and, two days after transduction, selected with puromycin for 7 days.
DNase-Seq
DNase-seq was performed on SKBR3 cell line as previously described52, with the addition of phosphorylation modification to linker 1b to increase ligation efficiency.
gRNA Library Design and Cloning
DNase I hypersensitive sites (DHS) for the K562 cell line were downloaded from www.encodeproject.org while SKBR3 DHS regions were obtained as described earlier and used to extract genomic sequences as input for gRNA identification. For each cell line’s set of DHSs, we used the gt-scan algorithm to identify gRNA protospacers within each DHS region and identify possible alignments to other regions of the genome53. The result is a database containing all possible gRNAs targeting all DHSs in the given cell line and each gRNA’s possible off target locations. For both libraries, gRNAs were selected based on minimizing off target alignments. For the HBE1 library, 10,739 gRNAs were selected, targeting 281 DHSs surrounding HBE1, limited to a maximum of 50 gRNAs per DHS. For the HER2 library, 12,189 gRNAs were selected, targeting 433 DHSs surrounding HER2, limited to a maximum of 30 gRNAs per DHS. For each library, controls were designed by searching for gRNAs in non-DHS regions. For the HBE1 library, 1,733 gRNAs were selected, and for the HER2 library, 283 gRNAs were selected. For the saturation library targeting a 60 kilobase region surrounding the HER2 promoter and DHSs containing at least one significantly enriched gRNA identified in the dCas9KRAB and dCas9p300 screens, all gRNAs within those regions were selected except those that had a perfect alignment to another area of the genome. All libraries were synthesized by Custom Array and the oligo pools were cloned into the lentiviral gRNA expression plasmid using Gibson assembly as described by Shalem et al., with minor modifications13.
Lentivirus Production and Titration
For the gRNA libraries and dCas9KRAB, lentivirus was produced by transfecting 5×106 HEK293T cells with the lentiviral gRNA expression plasmid pool or dCas9KRAB plasmid (20 μg), psPAX2 (Addgene, 12260, 15 μg), and pMD2.G (Addgene, 12259, 6 μg) using calcium phosphate precipitation54. After 14-20 hours, the transfection media was exchanged with fresh media. Media containing produced lentivirus was collected 24 and 48 hours later. The lentivirus was concentrated 20X the initial media volume using Lenti-X concentrator (Clontech, 631232), following manufacturer’s instructions.
The titer of the lentivirus containing either the HBE1 or HER2 pool of gRNAs was determined by transducing 4×105 cells with varying volumes of lentivirus and 4 days later, measuring the level of GFP or DsRed using the MACSQuant VYB flow cytometer (Miltenyi Biotec) to determine the percent transduction.
For dCas9p300, 15×106 HEK293T cells were transfected with dCas9p300 plasmid (120 μg), psPAX2 (90 μg), and pMD2.G (36 μg) using calcium phosphate precipitation. After 14–20 hours, the transfection media was exchanged with fresh media, and 24 hours later, the media was harvested. The lentivirus was pooled using ultracentrifugation (Beckman) at 24,000 rpm for 2 hours at 4 °C. The lentiviral pellet was resuspended in 1X PBS overnight to achieve a concentration of 400x.
To produce lentivirus for individual gRNA validations, 2×105 cells were transfected with gRNA plasmid (200 ng), psPAX2 (600 ng), and pMD2.G (200 ng) using Lipofectamine 2000 following the manufacturer’s instructions. After 14 to 20 hours, transfection media was exchanged with fresh media. Media containing produced lentivirus was harvested 24 and 48 hours later, centrifuged for 10 minutes at 800xG, and directly used to transduce cells.
Lentiviral gRNA Screening
For all screens, 6.236×106 cells were transduced during seeding in a 15 cm dish in 20 mL of media supplemented with 4 μg/mL of polybrene across 4 replicates, except the A431-dCas9p300 HER2 screen targeting DHSs, which was performed across 3 replicates. Cells were transduced at an MOI of 0.2 to try and achieve 1 gRNA per cell and 100-fold coverage of each gRNA library. After 2 days, cells were treated with Blasticidin S (ThermoFisher, A1113903) at a concentration of 20 μg/mL. For the dCas9KRAB screen targeting DHSs of the HBE1 locus, cells were grown for an additional 12 days, for the dCas9KRAB screen targeting DHSs of the HER2 locus in A431 cells, an additional 9 (Supplementary Figure 16) or 17 days, and for the dCas9p300 screen targeting DHSs or saturating a 60 kilobase region of the HER2 locus in HEK293T or A431 cells, an additional 7 days. For the Cas9 screen targeting a 60 kilobase region surrounding the HER2 promoter and all DHSs containing enriched gRNAs in the dCas9KRAB screen with all possible gRNAs in A431s, cells were grown an additional 9 days. Cells were passaged to ensure adequate fold coverage of the gRNA library to maintain representation. After culturing, 5×107 K562 cells were harvested, washed once with PBS, and resuspended in DMEM without phenol red and supplemented with 100 units of DNase I (NEB, M0303S) for sorting. For HEK293T or A431 cells, 5×107 cells were harvested using 0.25% Trypsin-EDTA (ThermoFisher, 25200056) and resuspended in 5% goat serum in PBS to block for 30 minutes at 4 °C. Next, cells were incubated in 125 μg of HER2 primary antibody (Monoclonal Mouse IgG2B Clone 191924, R&D Systems) in 5 mL of 5% goat serum in PBS for 30 minutes at 4 °C. Cells were then washed once in PBS and resuspended in secondary antibody (Goat anti-Mouse IgG2b Secondary Antibody, Alexa Fluor 488 conjugate, ThermoFisher A-21141) diluted 1:500 in 5 mL of 5% goat serum in PBS and incubated for 30 minutes at 4 °C. Finally, cells were washed once with PBS and resuspended in DMEM without phenol red supplemented with 100 units of DNase I for sorting.
An aliquot of 1.3×106 cells was taken before sorting for a bulk unsorted sample for each screen. The highest and lowest 10% of cells were sorted based on mCherry signal for the dCas9KRAB screen of HBE1 in K562 cells, or Alexa Fluor 488 signal for all HER2 screens. 1.3×106 cells were sorted for each group. Cell sorting was performed using a MoFlo Astrios (Beckman Coulter) or SH800 (Sony Biotechnology). After sorting, cells were harvested for genomic DNA using the DNeasy Blood and Tissue Kit (Qiagen, 69506).
Genomic DNA Sequencing
To amplify the gRNA libraries from each sample, 8.3 μg of gDNA was used as template across 8 100 μL PCR reactions using Q5 hot start polymerase (NEB, M0493L). Amplification was carried out following the manufacturer’s instructions using 25 cycles at an annealing temperature of 60 °C using the following primers:
Fwd 5′-AATGATACGGCGACCACCGAGATCTACACAATTTCTTGGGTAGTTTGCAGTT
Rev 5′-CAAGCAGAAGACGGCATACGAGAT (6 bp index sequence) GACTCGGTGCCACTTTTTCAA
Amplified libraries were purified using Agencourt AMPure XP beads (Beckman Coulter, A63881) using double size selection of 0.65X and then to 1X the original volume. Each sample was quantified after purification using the Qubit dsDNA High Sensitivity Assay Kit (ThermoFisher, Q32854). Samples were pooled and sequenced on a MiSeq (Illumina) with 21bp paired-end sequencing using the following custom read and index primers:
Read1 5′-GATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCG
Index 5′-GCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTC
Read2 5′-GTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC
Data Processing and Enrichment Analysis
FASTQ files were aligned to custom indexes (generated from the bowtie2-build function) using Bowtie255 with the options -p 32 --end-to-end --very-sensitive -3 1 -I 0 -X 200. Counts for each gRNA were extracted and used for further analysis. All enrichment analysis was performed using R. For DHS level analysis, gRNAs for each DHS were grouped together and a linear regression model (normalized_gRNA_count = β1*(sorted_bin)+ β2*(replicate)) was used to detect differences between the high and low conditions using the Holm method for multiple hypothesis correction. For individual gRNA enrichment analysis, the DESeq256 package was used to compare between high and low, unsorted and low, or unsorted and high conditions for each screen.
Individual gRNA Validations
The protospacers from the top enriched gRNAs found in each screen (listed in supplementary table 3) were order as oligonucleotides from IDT and cloned into a lentiviral gRNA expression vector as described earlier. The same modified cell lines used in the corresponding screen were used for the individual gRNA validations. The cells were transduced with individual gRNAs and after 2 days were selected with Blasticidin S (20 μg/mL). Samples with combinations of gRNAs were transduced by delivering equal volume viral supernatant of each gRNA. Cells were selected for 12 days for the HBE1 dCas9KRAB screen hits, 9 days for HER2 dCas9KRAB screen hits, and 7 days for the HER2 dCas9p300 screen hits.
For all screen validations, mRNA expression was done in triplicate. Total mRNA was harvested from cells using the Qiagen RNeasy Plus Mini kit (Qiagen, 74136). cDNA was generated using the SuperScript VILO cDNA Synthesis Kit (ThermoFisher, 11754250). qRT-PCR was performed using the Perfecta SYBR Green FastMix (Quanta Biosciences, 95072-012) with the FX96 Real-Time PCR Detection System (Bio-Rad) with the primers listed in supplementary table 4. The results are expressed as fold-increase mRNA expression of the gene of interest normalized to GAPDH expression by the ΔΔCt method.
For flow cytometry analysis of the HBE1 dCas9KRAB screen validations, cells were harvested, washed once in PBS, and resuspended in PBS. For the HER2 dCas9KRAB and dCas9p300 screen validations, cells were harvested, washed once with PBS, then resuspended in 5% goat serum in PBS and blocked for 30 minutes at 4 C. HER2 primary antibody (Monoclonal Mouse IgG2B Clone 191924, R&D Systems) was then added and allowed to incubated for 30 minutes at 4 C. Cells were then washed once in 5% goat serum in PBS. Secondary antibody (Goat anti-Mouse IgG2b Secondary Antibody, Alexa Fluor 488 conjugate, ThermoFisher A-21141) was then added and cells were allowed to incubate at 4 C for 30 minutes. Cells were then washed once in PBS. All cells were analyzed using the MACSQuant VYB flow cytometer (Miltenyi Biotec).
ChIP qPCR
The same cell lines used in the screens described earlier were used for ChIP-qPCR experiments: H3K27ac samples used HEK293T cells expressing dCas9p300 and H3K9me3 samples used A431 cells expressing dCas9KRAB. Cells were transduced with lentivirus containing gRNAs and allowed to grow for 9 days for H3K27ac samples or 11 days for H3K9me3 samples. Cells were fixed in 1% formaldehyde for 10 minutes at room temperature. The reaction was quenched with 0.125 M glycine, and the cells were lysed using Farnham lysis buffer with a protease inhibitor cocktail (Roche). Nuclei were collected by centrifugation at 2,000 rpm for 5 minutes at 4°C and lysed in RIPA buffer with a protease inhibitor cocktail (Roche). The chromatin was sonicated using a Bioruptor Sonicator (Diagenode, model XL) and immunoprecipitated using the following antibodies: anti-H3K27ac (abcam, ab4729) and anti-H3K4me3 (abcam, ab8580). The formaldehyde crosslinks were reversed by heating overnight at 65°C, and genomic DNA fragments were purified using a spin column. qPCR was performed using SYBR Green Fastmix (Quanta BioSciences) with the CFX96 Real-Time PCR Detection System (Bio-Rad). 1 ng of genomic DNA was used in each qPCR reaction. The data are presented as fold change gDNA relative to negative control and normalized to a region of the ACTB locus.
Data Availability
DNase-seq and gRNA library sequencing data is deposited in the NCBI’s Gene Expression Omnibus and is accessible through GEO Series accession number GSEXXXXX.
Supplementary Material
Acknowledgments
This work was supported by the Thorek Memorial Foundation, US National Institutes of Health (NIH) grants to G.E.C., T.E.R., and C.A.G. (R01DA036865, R41GM119914, and U01HG007900), a NIH Director’s New Innovator Award (DP2OD008586) and National Science Foundation (NSF) Faculty Early Career Development (CAREER) Award (CBET-1151035) to C.A.G., and NIH grant P30AR066527. T.S.K. and J.B.B. were supported by a NIH Biotechnology Training Grant (T32GM008555).
Footnotes
AUTHOR CONTRIBUTIONS
T.S.K, G.E.C., T.E.R., and C.A.G. designed experiments. T.S.K., J.B.B., A.S., L.S., and M.C. performed the experiments. I.B.H. provided critical reagents. T.S.K, G.E.C., T.E.R., and C.A.G. analyzed the data. T.S.K. and C.A.G. wrote the manuscript with contributions by all authors.
CONFLICT OF INTEREST
TSK, JBB, IBH, GEC, TER, and CAG are named inventors on patent applications related to genome engineering. TSK, GEC, TER, and CAG are founders of Element Genomics.
References
- 1.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hindorff LA, Junkins HA, Mehta JP, Manolio TA. OPG: Catalog Published Genome-Wide Assoc Studies. 2009. [Google Scholar]
- 3.Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA. The NIH Roadmap Epigenomics Mapping Consortium. Nature Biotechnology. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A. Genome-Wide Quantitative Enhancer Activity Maps Identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 6.Vockley CM, Guo C, Majoros WH, Nodzenski M, Scholtens DM, Hayes MG, Lowe WL, Reddy TE. Massively parallel quantification of the regulatory effects of noncoding genetic variation in a human cohort. Genome Research. 2015;25:1206–1214. doi: 10.1101/gr.190090.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vockley CM, D’Ippolito AM, McDowell IC, Majoros WH, Safi A, Song L, Crawford GE, Reddy TE. Direct GR Binding Sites Potentiate Clusters of TF Binding across the Human Genome. Cell. 2016;166:1269–1281. e1219. doi: 10.1016/j.cell.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 9.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature Biotechnology. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 13.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nature Biotechnology. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 16.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, Luc S, Kurita R, Nakamura Y, Fujiwara Y, Maeda T, Yuan GC, Zhang F, Orkin SH, Bauer DE. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han R, Myacheva K, Zwart W, Elkon R, Agami R. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nature Biotechnology. 2016;34:192–198. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopal N, Srinivasan S, Kooshesh K, Guo Y, Edwards MD, Banerjee B, Syed T, Emons BJM, Gifford DK, Sherwood RI. High-throughput mapping of regulatory DNA. Nature Biotechnology. 2016;34:167–174. doi: 10.1038/nbt.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diao Y, Li B, Meng Z, Jung I, Lee AY, Dixon J, Maliskova L, Guan K-l, Shen Y, Ren B. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Research. 2016;26:397–405. doi: 10.1101/gr.197152.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanjana NE, Wright J, Zheng K, Shalem O, Fontanillas P, Joung J, Cheng C, Regev A, Zhang F. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545–1549. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vierstra J, Reik A, Chang KH, Stehling-Sun S, Zhou Y, Hinkley SJ, Paschon DE, Zhang L, Psatha N, Bendana YR, O’Neil CM, Song AH, Mich AK, Liu PQ, Lee G, Bauer DE, Holmes MC, Orkin SH, Papayannopoulou T, Stamatoyannopoulos G, Rebar EJ, Gregory PD, Urnov FD, Stamatoyannopoulos JA. Functional footprinting of regulatory DNA. Nat Methods. 2015;12:927–930. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Housden BE, Muhar M, Gemberling M, Gersbach CA, Stainier DY, Seydoux G, Mohr SE, Zuber J, Perrimon N. Loss-of-function genetic tools for animal models: cross-species and cross-platform differences. Nat Rev Genet. 2017;18:24–40. doi: 10.1038/nrg.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nature Methods. 2016;13:127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi Lei S, Larson Matthew H, Gilbert Luke A, Doudna Jennifer A, Weissman Jonathan S, Arkin Adam P, Lim Wendell A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert Luke A, Larson Matthew H, Morsut L, Liu Z, Brar Gloria A, Torres Sandra E, Stern-Ginossar N, Brandman O, Whitehead Evan H, Doudna Jennifer A, Lim Wendell A, Weissman Jonathan S, Qi Lei S. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao X, Yang J, Tsang JCH, Ooi J, Wu D, Liu P. Reprogramming to Pluripotency Using Designer TALE Transcription Factors Targeting Enhancers. Stem Cell Reports. 2013;1:183–197. doi: 10.1016/j.stemcr.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao X, Tsang JCH, Gaba F, Wu D, Lu L, Liu P. Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic Acids Research. 2014;42:e155–e155. doi: 10.1093/nar/gku836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert Luke A, Horlbeck Max A, Adamson B, Villalta Jacqueline E, Chen Y, Whitehead Evan H, Guimaraes C, Panning B, Ploegh Hidde L, Bassik Michael C, Qi Lei S, Kampmann M, Weissman Jonathan S. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nature Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nature Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu X, Tao Y, Gao X, Zhang L, Li X, Zou W, Ruan K, Wang F, Xu G-l, Hu R. A CRISPR-based approach for targeted DNA demethylation. Cell Discovery. 2016;2:16009. doi: 10.1038/celldisc.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choudhury SR, Cui Y, Lubecka K, Stefanska B, Irudayaraj J, Choudhury SR, Cui Y, Lubecka K, Stefanska B, Irudayaraj J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget. 2016;7:46545–46556. doi: 10.18632/oncotarget.10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, Klasić M, Zoldoš V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Research. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167:233–247. e217. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-Resolution Mapping and Characterization of Open Chromatin across the Genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, Weaver M, Shafer A, Lee K, Neri F, Humbert R, Singer MA, Richmond TA, Dorschner MO, McArthur M, Hawrylycz M, Green RD, Navas PA, Noble WS, Stamatoyannopoulos JA. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nature Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 40.Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian β-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 41.Baron MH. Developmental Regulation of the Vertebrate Globin Multigene Family. Gene Expression. 1996;6:129–137. [PMC free article] [PubMed] [Google Scholar]
- 42.Dean A, Ley TJ, Humphries RK, Fordis M, Schechter AN. Inducible transcription of five globin genes in K562 human leukemia cells. Proceedings of the National Academy of Sciences. 1983;80:5515–5519. doi: 10.1073/pnas.80.18.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Gräf S, Huss M, Keefe D, Liu Z, London D, McDaniell RM, Shibata Y, Showers KA, Simon JM, Vales T, Wang T, Winter D, Zhang Z, Clarke ND, Birney E, Iyer VR, Crawford GE, Lieb JD, Furey TS. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Research. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vu T, Claret FX. Trastuzumab: Updated Mechanisms of Action and Resistance in Breast Cancer. Frontiers in Oncology. 2012;2 doi: 10.3389/fonc.2012.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER2. New England Journal of Medicine. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 46.Gajria D, Chandarlapaty S. HER2-amplified breast cancer: mechanisms of trastuzumab resistance and novel targeted therapies. Expert Review of Anticancer Therapy. 2011;11:263–275. doi: 10.1586/era.10.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurtado A, Holmes KA, Geistlinger TR, Hutcheson IR, Nicholson RI, Brown M, Jiang J, Howat WJ, Ali S, Carroll JS. Regulation of ERBB2 by oestrogen receptor–PAX2 determines response to tamoxifen. Nature. 2008;456:663–666. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu PY, Li TK, Ding ST, Lai IR, Shen TL. EGF-induced Grb7 Recruits and Promotes Ras Activity Essential for the Tumorigenicity of Sk-Br3 Breast Cancer Cells. The Journal of Biological Chemistry. 2010;285:29279–29285. doi: 10.1074/jbc.C110.114124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, Engreitz JM. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:769–773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Brien A, Bailey TL. GT-Scan: identifying unique genomic targets. Bioinformatics. 2014;30:2673–2675. doi: 10.1093/bioinformatics/btu354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salmon P, Trono D. Production and titration of lentiviral vectors. Curr Protoc Neurosci. 2006;Chapter 4(Unit 4):21. doi: 10.1002/0471142301.ns0421s37. [DOI] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNase-seq and gRNA library sequencing data is deposited in the NCBI’s Gene Expression Omnibus and is accessible through GEO Series accession number GSEXXXXX.