Abstract
Background
The genetics of depression has been explored in genome-wide association studies that focused on major depressive disorder or depressive symptoms with mostly negative findings. A broad depression phenotype including both phenotypes has not been tested previously using a genome-wide association approach. We aimed to identify genetic polymorphisms significantly associated with a broad phenotype from depressive symptoms to major depressive disorder.
Methods
We analysed two prior studies of 70,017 participants of European ancestry from general and clinical populations in the discovery stage. We performed a replication meta-analysis of 28,328 participants. SNP-based heritability and genetic correlations were calculated using LD score regression. Discovery and replication analyses were performed using a P-value based meta-analysis. Lifetime major depressive disorder and depressive symptom scores were used as the outcome measures.
Results
The SNP-based heritability of major depressive disorder was 0.21 (SE=0.02), the SNP-based heritability of depressive symptoms was 0.04 (SE=0.01), and their genetic correlation was 1.001 (SE=0.2). We found one genome-wide significant locus related to the broad depression phenotype (rs9825823, chromosome 3: 61,082,153, P=8.2×10−9) located in an intron of the FHIT gene. We replicated this SNP in independent samples (P= 0.02) and the overall meta analysis of the discovery and replication cohorts (1.0×10−9).
Conclusions
This large study identified a new locus for depression. Our results support a continuum between depressive symptoms and major depressive disorder. A phenotypically more inclusive approach may help achieve the large sample sizes needed to detect susceptibility loci for depression.
Keywords: Depressive symptoms, major depressive disorder, CHARGE consortium, Psychiatric Genomics Consortium, genome-wide association study, FHIT gene
Introduction
The etiology of depression – a worldwide leading cause of disability (1) – is not well understood. As indicated by family, twin and adoption studies, genetic factors mediate part of vulnerability to major depressive disorder (MDD) with a modest heritability of around 40% (2). However, we understand little of the specific genetic architecture of MDD. Multiple genome-wide association studies (GWAS) for MDD have been published (3–10). The largest MDD GWAS was the mega-analysis by the MDD Working Group of the Psychiatric Genomics Consortium (PGC). In that study, over 9,000 MDD cases and 9,500 controls were analyzed but no association with MDD reached genome wide significance (7). Recently, CONVERGE (China, Oxford and VCU Experimental Research on Genetic Epidemiology) consortium identified two genome-wide significant associations in 5,303 Chinese women with severe and recurrent MDD (near the SIRT1 gene, P=2.53×10−10 and in an intron of the LHPP gene, P=6.45×10−12) (11). A GWAS of depressive symptoms (heritability 23%–29%) (12, 13) in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium in approximately 50,000 people from the general population found no genome-wide significant associations (14). Due to the relatively small sample sizes, the previous GWAS of depressive disorders and depressive symptoms were arguably underpowered to detect small genetic effects (15, 16).
Depression can be conceptualized along a continuum of severity from subthreshold or minor depression to MDD of varying severity (e.g., mild, moderate, and severe) (17). Using a continuum approach may augment statistical power as sample size can be increased substantially and patients who fall into the ‘grey area’ can be assessed. Several lines of evidence support a depression continuum. In longitudinal studies, there is an increased risk of MDD in patients with minor depression and subthreshold depression (18, 19). Statistical studies of disorder classification (taxometric) suggested that severity of depression is continuously distributed and there is no discontinuity in the latent structure of depression (19, 20). Family studies report that relatives of probands with milder forms of depression have greater risk of MDD compared to relatives of probands without any mood disorders (21–24). A higher number of depressive symptoms is related to greater disability, worse quality of life, and a higher mortality risk (18, 25–29). MDD and continuous measures of depression are highly correlated and severity of depressive symptoms along the continuum is linear (30, 31).
The goal of the current study was to combine the results of the largest GWAS using categorical lifetime MDD and continuous measures of depression to identify genetic variants underlying the entire depression continuum.
Methods and Materials
Study Design and Samples
This study was a collaboration between investigators on the PGC MDD and CHARGE genome-wide association meta-analyses (GWAMA). In the discovery phase, we aggregated two GWAMA published in 2013 (7, 14). Basic descriptive features and phenotype definitions of the contributing samples are provided in Table S1. The mega-analysis of MDD consisted of nine studies of 9,240 cases meeting international criteria for lifetime MDD and 9,519 healthy controls. The CHARGE meta-analysis of depressive symptoms included 22 cohorts and comprised 51,258 persons. Each cohort contributing to the GWAMA of the PGC and CHARGE were distinct. In the replication analyses, 16 case-control studies with DSM-IV MDD (6,718 cases and 13,453 controls) were included along with 8,157 subjects from the general population with assessment of depressive symptoms. All subjects were of European ancestry. Institutional review boards approved all studies and all participants provided written informed consent.
Phenotype Characteristics
In the PGC GWAMA, MDD was established with structured clinical interviews (e.g., Clinical Interview Schedule-Revised, Diagnostic Interview for Genetic Studies, and the Structured Clinical Interview for DSM-IV). All clinical evaluations were made by experienced clinicians/interviewers. Most cases were ascertained from clinical sources. Controls were screened in most of the studies to require the absence of MDD and recruited from the general population. Full details about the PGC samples can be found in the previous publication (7). In the CHARGE GWAMA, depressive symptoms were assessed with validated questionnaires. Measures include the Center for Epidemiological Studies-Depression (CES-D) scale, Geriatric Depression Scale (GDS), Patient Health Questionnaire-9, and the Beck Depression Inventory-II (BDI-II) mostly assessing depressive symptoms in previous weeks rather than lifetime MDD (14). Persons with schizophrenia, bipolar disorder, and dementia were excluded. Persons aged 40 and older and with genotype data and depressive symptom score were included.
The 16 MDD case-control replication samples were part of an expanded but unpublished PGC MDD analysis. MDD was diagnosed with interviews. In the depressive symptom replication cohort, the Health and Retirement Study, the 8-item CESD was applied. Respondents were excluded if they were less than 40 years of age or evidence of cognitive impairment.
Genotyping and Imputation
In the PGC samples, (Table S1), individual genotypes were assembled, processed through a central quality control pipeline and imputed using the CEU and TSI HapMap3 reference panels. Quality control procedures were extensive (7). In the CHARGE cohorts, genotype quality control and imputation were conducted in each study separately. The imputation reference was the HapMap2 Central EUrope (CEU) panel (14). In the MDD replication cohorts (Table S3), imputation was performed using IMPUTE2 / SHAPEIT (chunk size of 3 Mb and default parameters). The imputation reference set consisted of 2,186 phased haplotypes from the 1000 Genomes Project. In the Health and Retirement Study, imputation was performed using the HapMap2 CEU reference panel.
Statistical Analyses
LD score regression was used to compute the SNP-based heritability and the genetic correlation using the 1000 Genomes CEU reference panel (32).
In the PGC GWAMA, a logistic regression analysis was used to test the association between MDD and imputed SNP dosages under an additive model and adjusting for study indicators and five principal components (7). In the CHARGE GWAMA, a linear regression analysis was applied to test the association of depressive symptom score on imputed SNP dosages in the contributing studies adjusting for age and sex. Analyses were adjusted for principal components for most but not all cohorts in the CHARGE GWAMA. A P value based meta-analysis was applied in the CHARGE GWAMA (14). Effect size estimates were based on a dichotomous outcome in the PGC and on a continuous outcome in the CHARGE GWAMA. To combine these effect estimates, a P value based meta-analysis weighted by sample size with METAL was used. This method allows different weights for each study and takes into account the direction of effect at each SNP (33). To specify the direction of the effect, the PGC used the logistic regression coefficient beta and the CHARGE used z-scores. Weights were based on the number of the MDD cases in the PGC study (n=9,240) and the number of individuals in the CHARGE with clinically significant depressive symptoms (n=5,976) using population specific cutoff scores of the questionnaires were considered for weighting. To test whether the results are affected by different sample size weightings, equal weights per study, or no weight as suggested by Stouffer (34), we carried out a series of sensitivity analyses.
We selected the genome wide significant SNPs in two loci from the discovery stage for replication. After analyzing these data, we performed a P value based meta-analysis combining all replication samples. Further, we analyzed the results of the discovery and all replication samples weighting for number of cases.
Results
In the discovery stage, we performed a GWAMA in 70,017 participants of European ancestry by combining the PGC MDD (7) and CHARGE GWAMA (14). We applied an LD score regression to the summary statistics from each study to compute the SNP-based heritabilities and the genetic correlation. As reported previously (35), the SNP-based liability scale heritability of MDD was 0.2 (standard error 0.02) for 20% of prevalence. The lambda was 1.1 and the regression intercept was 1.0 (standard error 0.01). The SNP-based heritability of depressive symptoms was 0.04 (standard error 0.01). The lambda was 1.1 and the regression intercept was 1.0 (standard error 0.01). The SNP-based heritability of the broad depression phenotype was 0.3 (standard error 0.04). MDD and depressive symptoms showed significant co-heritability (1.001, standard error 0.2, Z-score= 4.6, P=4.6×10−6). This result supports the contention of a continuum between depressive symptoms and MDD. However, the genetic correlation should be interpreted carefully as LD regression is quite sensitive to environmental confounding and like twin studies often lacks precision. Also, different evaluation methods of the depression phenotypes might cause different genetic correlation estimates that cannot easily be compared.
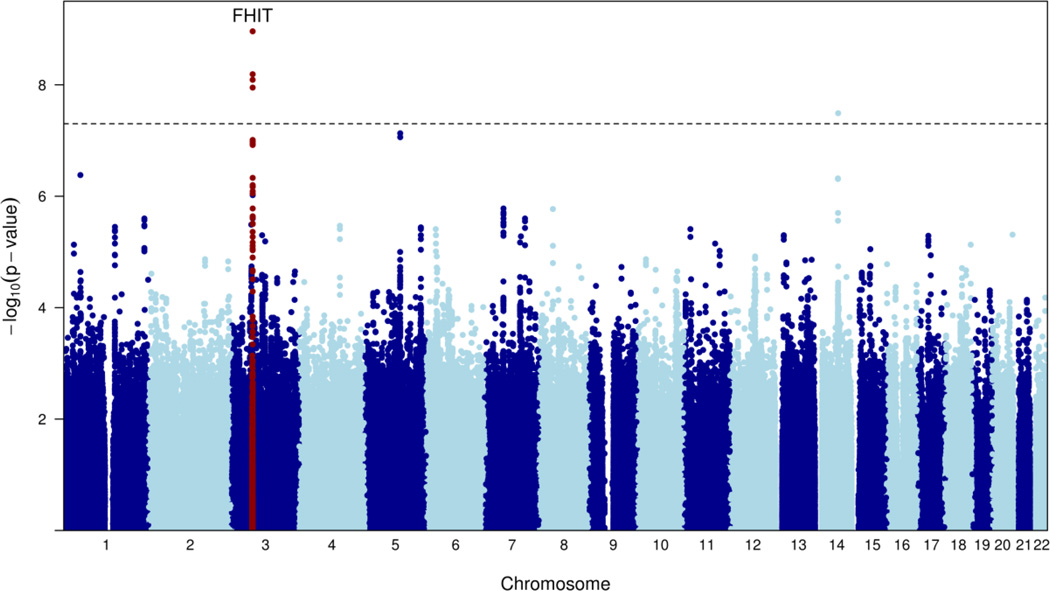
We conducted a meta-analysis of the PGC MDD and the CHARGE depressive symptoms GWAMA using a weighted, P-value based meta-analysis. The results are summarized in Figure 1 and Figures S1–S3. The combined meta-analysis was conducted for 918,921 SNPs. Two loci were genome-wide significant: a SNP in an intron of FHIT (rs9825823, chr3: 61,082,153, P =8.2×10−9) and a SNP in an intron of PLEK2 (rs9323497, chr14: 67,873,128, P=3.3×10−8) (Table 1). All SNPs with a P value of association <5×10−5 are presented in Table S2. Using different weights or Stouffer’s unweighted method had only slight effects on the results (data not shown). Figures S4 and S5 shows forest plots for two SNPs shown in Table 1.
Figure 1. Manhattan Plot.
X-axis represents the chromosomal position for each SNP, and y-axis the −log10 P value for association with depression.
Table 1.
Meta-analysis results of the PGC MDD GWAMA and the CHARGE depressive symptoms GWAMA(discovery)
| Combined meta-analysis* (Ntotal=70,017) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | BP | Closest Gene |
Location | Allele | MAF | Direction |
z- score |
P |
| rs9825823 | 3 | 61,082,153 | FHIT | intron | T/C | 0.46 | ++ | 5.8 | 8.2×10−9 |
| rs9323497 | 14 | 67,873,128 | PLEK2 | intron | T/C | 0.05 | ++ | 5.5 | 3.3×10−8 |
Chr: chromosome, BP: base pair, MAF: minor allele frequency (GRCh37/hg19). Allele: Minor/Major on the + strand. Direction of effect: CHARGE (N=51,258; continuous outcome analysis), PGC (N=18,759; of which 9,240 were MDD cases).
This analysis was weighted for number of participants with clinically significant depressive symptoms in the CHARGE study (n=5,976) and number of cases in the PGC study (n=9,240).
Table 2 presents the replication analyses and the meta-analysis of discovery and replication results. One of the genome-wide significant variants within the FHIT gene (rs9825823) was associated with depression continuum in the replication cohorts (z-score=2.4, P=0.02). The result of the final metaanalysis of discovery and replication samples also indicated a positive replication as indexed by a lower p-value (z-score=6.1, P=1.0×10−9). This SNP had a positive association with depressive symptoms in the CHARGE study (P=5.5 × 10−4) and a similar pattern was observed in the PGC study (P=4.1 × 10−6). The SNP in an intron of PLEK2 (rs9323497) was not related to depression continuum significantly (z-score=0.2, P=0.9).
Table 2.
Replication analyses and final meta-analysis of discovery and replication stages
| PGC replication (N=20,171) |
HRS replicatio n (N= 8,157) |
Overall replication (N=28,328) |
Meta-analysis of discovery & two replication samples (N= 98,345) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr:BP | Allel e |
Direc t.a |
P | Direc t. |
P | Direct .b |
P | MA F |
Direc t.c |
z- scor e |
P |
| rs98258 23 |
3:61,082,1 53 |
T/C | +++−− ++−+− ++−− ++ |
0.0 4 |
+ | 0.2 | ++ | 0.0 2 |
0.4 6 |
+++ | 6.10 | 1.0×1 0−9 |
| rs93234 97 |
14:67,873, 128 |
T/C | +−+−−− − ++++− −−++ |
0.8 | − | 0.8 | +− | 0.9 | 0.0 5 |
−++ | 4.61 | 4.0×1 0−6 |
Chr: Chromosome, BP: base pair, MAF: minor allele frequency (GRCh37/hg19), Direct=Direction. Allele: Minor/Major on the + strand.
Order of the studies: The Cognitive Function and Mood Study (CoFaMS), The PsyCoLaus Study, MDD2000-Edinburgh, GENPOD/NewMeds, Depression Genes Networks, GenRED2, Harvard i2b2, Janssen, The Marx Planck Institute of Psychiatry (MPIP) Munich Antidepressant Response Signature (MARS) Study OMNIex, QIMR COEX, Radiant Irish, Radiant US Cases, Radiant Denmark Cases, Roche, SHIP Trend, TwinGene.
Order of the studies: PGC- replication Health and Retirement Study.
Order of the studies: PGC-replication, Health and Retirement Study, combined meta-analysis of discovery samples.
We performed an additional replication analysis of our two genome-wide significant SNPs using the publicly available data of the recently published GWAMA of depressive disorders in a sample of Chinese women (the CONVERGE study) (11). In CONVERGE, rs9825823 (odds ratio=1.01, P=0.12) and rs9323497 (odds ratio=0.97, P=0.0002, with a different direction of association than in our discovery sample) were not related to depression at the genome-wide significance level although the latter reached nominal significance. However, in the joint meta-analysis of the HRS, PGC MDD and the CONVERGE studies, we found that the association between the rs9825823 and the depression continuum (z-score=2.85, P=0.004) was slightly stronger than our initial replication analysis. When these replication and discovery samples were combined, the association with our top hit also became stronger (analyses without the CONVERGE data: z-score= 6.1, P = 1×10−9; with the CONVERGE data z-score= 6.2, P = 6.8×10−10). Results of additional replication analyses are given in the Table S4.
Discussion
We report the results of a combined GWAMA of depression continuum including MDD (18,759 cases and controls) and depressive symptoms (51,258 participants). In the discovery stage, we found genome-wide significant associations in the FHIT and PLEK2 genes. One SNP in the intron of the FHIT gene showed a significant association in the combined analysis of discovery and replication samples of MDD and depressive symptoms samples, and exceeded a genome-wide significance threshold.
The significant locus (rs9825823, chr3: 61,082,153) maps to the intronic region of the fragile histidine triad (FHIT) gene, a tumor suppressor protein implicated in several cancers (36). FHIT is expressed in multiple brain regions (amygdala, anterior cingulate cortex, caudate nucleus, prefrontal cortex, hippocampus, and hypothalamus, http://www.gtexportal.org/home/gene/FHIT, accessed 10.07.2016). It plays an important role in oxidative stress and level of DNA damage (37), biological processes implicated in MDD (38, 39). FHIT is a circadian clock modifier gene (40) and has been related to daytime sleepiness (41), which may be salient to the etiology of depression.
In a GWAS of recurrent, early-onset MDD, three SNPs located in the FHIT gene were among the strongest associations in the overall and sex-stratified analyses (8) although none was genome-wide significant. Genetic variants located in FHIT have been reported in genetic studies of anxiety,(42) autism (43), mental stress (44), comorbid depressive syndromes and alcohol dependence (45), citalopram-induced side effects (46) and in a latent class analysis of MDD symptoms (7), but none met genome-wide significance.
Several methodological aspects should be discussed. First, we evaluated depression continuum by combining cases from clinical populations diagnosed with MDD and participants from the general population who had been assessed for depressive symptoms. Such an inclusive approach may increase heterogeneity of the phenotype especially because lifetime MDD was evaluated whereas depressive symptoms indicate past weeks only. If anything, such approach would cause an underestimation of the effects as less information on depressive symptoms were obtained. However, the advantages of a large sample can outweigh the disadvantages of a less precisely defined phenotype. This has been observed in the GWAS of educational attainment which was successfully used as a proxy for intelligence (47). Our additional replication analysis showed that increasing the sample size yielded a stronger association of the top hit with depression continuum. It is complex to calculate statistical power of the current analysis as quantitative and qualitative measures were combined. In the current study, a genetic association with the depression continuum may reflect an effect on broad depressive phenotypes but could also be accounted for by an association with low levels of general well-being (12–18% heritability) that co-occur with depressive symptoms (48). Second, we used a P-value based meta-analysis, as effect estimates could not be directly evaluated in a straightforward manner. Third, the heterogeneity of the imputation methods used in the PGC and CHARGE discovery samples might reduce the statistical power. However, different imputation references did not change the results in the published PGC MDD study (7).
In conclusion, in this large GWAMA of a broad depression phenotype, we detected a locus associated with depression in clinical and general population samples. Our results suggest the importance a broader depression phenotype to identify genetic variants underlying depression. Large samples with different depression phenotypes may also help disentangle the genetic background of different forms of depression.
Supplementary Material
Acknowledgments
The authors acknowledge the essential role of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium in development and support of this manuscript. CHARGE members include the Netherland’s Rotterdam Study (RS), the NHLBI’s Framingham Heart Study (FHS), Cardiovascular Health Study (CHS), the NHLBI’s Atherosclerosis Risk in Communities (ARIC) Study, and the NIA’s Iceland Age, Gene/Environment Susceptibility (AGES) Study. Core funding for the Psychiatric Genomics Consortium is from the US National Institute of Mental Health (U01MH094421). The PGC was supported by NIMH R01 MH094421 and NIMH R01 MH094421. HRS is supported by the National Institute on Aging (NIA U01AG009740). The genotyping was funded separately by the National Institute on Aging (RC2 AG036495, RC4 AG039029). Our genotyping was conducted by the NIH Center for Inherited Disease Research (CIDR) at Johns Hopkins University. Genotyping quality control and final preparation of the data were performed by the Genetics Coordinating Center at the University of Washington. The CoLaus|PsyCoLaus study was and is supported by research grants from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, and the Swiss National Science Foundation (grants 3200B0–105993, 3200B0-118308, 33CSCO-122661, 33CS30-139468 and 33CS30-148401). The University of Edinburgh is a charitable body, registered in Scotland, with registration number SC005336. DJM is an NRS Research Fellow, supported by the CSO. AMM is supported by The Wellcome Trust (104036/Z/14/Z), The Dr Mortimer and Theresa Sackler Foundation and by a SFC Senior Clinical Fellowship (SCD/12). GenPod was funded by the Medical Research Council (MRC, UK G0200243) and we acknowledge the other coinvestigators, Cowen, Nutt, Peters and Sharp. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113, 085475 and 090355. NEWMEDS: The research leading to these results has received support from the Innovative Medicine Initiative Joint Undertaking (IMI-JU) under grant agreement no. 115008 of which resources are composed of European Union and the European Federation of Pharmaceutical Industries and Associations (EFPIA) in-kind contribution and financial contribution from the European Union's Seventh Framework Programme (FP7/2007-2013). Funding for the NESDA and NTR studies were obtained from the Netherlands Organization for Scientific Research (NWO), Center for Medical Systems Biology (CSMB, NWO Genomics), NBIC/BioAssist/RK(2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI –NL, 184.021.007). VU University’s Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA); the European Science Foundation (ESF, EU/QLRT-2001-01254), the European Community's Seventh Framework Program (FP7/2007-2013), ENGAGE (HEALTH-F4-2007-201413); the European Science Council (ERC Advanced, 230374), Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute, Sioux Falls, South Dakota (USA) and the National Institutes of Health (NIH, R01D0042157-01A, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health. Computing was supported by BiG Grid, the Dutch e-Science Grid, which is financially supported by NWO. The RADIANT studies present independent research part funded by a joint grant from the Medical Research Council, UK and GlaxoSmithKline (G0701420), and by financial support from the National Institute for Health Research (NIHR) Biomedical Research Centre for Mental Health at the South London and Maudsley NHS Foundation Trust and King's College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. The GENDEP project was funded by the European Commission Framework 6 grant, EC Contract Ref.: LSHB-CT-2003-503428. Lundbeck provided nortriptyline and escitalopram for the GENDEP study. GlaxoSmithKline and the UK NIHR of the Department of Health contributed to the funding of the sample collection at the Institute of Psychiatry, London. GENDEP was funded by the European Commission Framework 6 grant, EC Contract Ref.: LSHB-CT-2003-503428. Lundbeck provided nortriptyline and escitalopram for the GENDEP study. GlaxoSmithKline and the UK National Institute for Health Research of the Department of Health contributed to the funding of the sample collection at the Institute of Psychiatry, London. The collection of samples and genotyping of the Danish controls was supported by grants from The Danish Strategic Research Council, The Stanley Research Foundation and H. Lundbeck A/S and we thank David M Hougaard, Section of Neonatal Screening and Hormones, Statens Serum Institute, Copenhagen, Denmark, Preben Bo Mortensen, National Centre for Register-based Research, Aarhus University, Denmark and The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH, Denmark. SHIP-TREND is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania, and the network ‘Greifswald Approach to Individualized Medicine (GANI_MED) funded by the Federal Ministry of Education and Research (grant 03IS2061A). Genomewide genotyping in SHIP-TREND-0 was supported by the Federal Ministry of Education and Research (grant no. 03ZIK012). This work was also funded by the German Research Foundation (DFG: GR 1912/5-1). The Cognitive Function and Mood Study (CoFaMS) is supported by the Faculty of Health Science, University of Adelaide, Adelaide, Australia. We are grateful to the Janssen study volunteers for participating in the clinical studies, participating clinical investigators and the staff for enabling patient recruitment and blood sample collection. We thank the staff in Janssen Neuroscience for sample banking, DNA extraction, quality control, plating, and clinical data anonymizaiton, and the staff at Illumina for genotyping of the Janssen DNA samples.
The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
ND had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. ND (Department of Epidemiology, Erasmus Medical Center) and SW (Department of Genetics, University of North Carolina at Chapell Hill) were conducted and are responsible for the data analysis.
Dr. Sullivan is a consultant for Pfizer, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting galley proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: ND had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. ND (Department of Epidemiology, Erasmus Medical Center) and SW (Department of Genetics, University of North Carolina at Chapell Hill) were conducted and are responsible for the data analysis.
Conflict of Interest: The other authors report no conflicts.
References
- 1.Murray CJ, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 3.Kohli MA, Lucae S, Saemann PG, Schmidt MV, Demirkan A, Hek K, et al. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron. 2011;70:252–265. doi: 10.1016/j.neuron.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K, et al. Genome-wide association study of major recurrent depression in the U.K. population. Am J Psychiatry. 2010;167:949–957. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- 5.Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ, et al. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry. 2010;15:589–601. doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- 6.Rietschel M, Mattheisen M, Frank J, Treutlein J, Degenhardt F, Breuer R, et al. Genome-wide association-, replication-, and neuroimaging study implicates HOMER1 in the etiology of major depression. Biol Psychiatry. 2010;68:578–585. doi: 10.1016/j.biopsych.2010.05.038. [DOI] [PubMed] [Google Scholar]
- 7.Major Depressive Disorder Working Group of the Psychiatric GC. Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2011;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shyn SI, Shi J, Kraft JB, Potash JB, Knowles JA, Weissman MM, et al. Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol Psychiatry. 2011;16:202–215. doi: 10.1038/mp.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, et al. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.consortium C. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansson M, Gatz M, Berg S, Johansson B, Malmberg B, McClearn GE, et al. Gender differences in heritability of depressive symptoms in the elderly. Psychol Med. 2004;34:471–479. doi: 10.1017/s0033291703001375. [DOI] [PubMed] [Google Scholar]
- 13.Johnson W, McGue M, Gaist D, Vaupel JW, Christensen K. Frequency and heritability of depression symptomatology in the second half of life: evidence from Danish twins over 45. Psychol Med. 2002;32:1175–1185. doi: 10.1017/s0033291702006207. [DOI] [PubMed] [Google Scholar]
- 14.Hek K, Demirkan A, Lahti J, Terracciano A, Teumer A, Cornelis MC, et al. A genome-wide association study of depressive symptoms. Biol Psychiatry. 2013;73:667–678. doi: 10.1016/j.biopsych.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry. 2012;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levinson DF, Mostafavi S, Milaneschi Y, Rivera M, Ripke S, Wray NR, et al. Genetic Studies of Major Depressive Disorder: Why Are There No Genome-wide Association Study Findings and What Can We Do About It? Biol Psychiatry. 2014;76:510–512. doi: 10.1016/j.biopsych.2014.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobo DM, Agius M. The mental illness spectrum. Psychiatria Danubina. 2012;24(Suppl 1):S157–S160. [PubMed] [Google Scholar]
- 18.Cuijpers P, de Graaf R, van Dorsselaer S. Minor depression: risk profiles, functional disability, health care use and risk of developing major depression. J Affect Disord. 2004;79:71–79. doi: 10.1016/S0165-0327(02)00348-8. [DOI] [PubMed] [Google Scholar]
- 19.Cuijpers P, Smit F. Subthreshold depression as a risk indicator for major depressive disorder: a systematic review of prospective studies. Acta psychiatrica Scandinavica. 2004;109:325–331. doi: 10.1111/j.1600-0447.2004.00301.x. [DOI] [PubMed] [Google Scholar]
- 20.Prisciandaro JJ, Roberts JE. A comparison of the predictive abilities of dimensional and categorical models of unipolar depression in the National Comorbidity Survey. Psychol Med. 2009;39:1087–1096. doi: 10.1017/S0033291708004522. [DOI] [PubMed] [Google Scholar]
- 21.Lewinsohn PM, Klein DN, Durbin EC, Seeley JR, Rohde P. Family study of subthreshold depressive symptoms: risk factor for MDD? J Affect Disord. 2003;77:149–157. doi: 10.1016/s0165-0327(02)00106-4. [DOI] [PubMed] [Google Scholar]
- 22.Remick RA, Sadovnick AD, Lam RW, Zis AP, Yee IM. Major depression, minor depression, and double depression: are they distinct clinical entities? American journal of medical genetics. 1996;67:347–353. doi: 10.1002/(SICI)1096-8628(19960726)67:4<347::AID-AJMG6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 23.Kessler RC, Zhao S, Blazer DG, Swartz M. Prevalence, correlates, and course of minor depression and major depression in the National Comorbidity Survey. J Affect Disord. 1997;45:19–30. doi: 10.1016/s0165-0327(97)00056-6. [DOI] [PubMed] [Google Scholar]
- 24.Chen LS, Eaton WW, Gallo JJ, Nestadt G, Crum RM. Empirical examination of current depression categories in a population-based study: symptoms, course, and risk factors. Am J Psychiatry. 2000;157:573–580. doi: 10.1176/appi.ajp.157.4.573. [DOI] [PubMed] [Google Scholar]
- 25.Cuijpers P, Vogelzangs N, Twisk J, Kleiboer A, Li J, Penninx BW. Differential mortality rates in major and subthreshold depression: meta-analysis of studies that measured both. The British journal of psychiatry : the journal of mental science. 2013;202:22–27. doi: 10.1192/bjp.bp.112.112169. [DOI] [PubMed] [Google Scholar]
- 26.Ayuso-Mateos JL, Nuevo R, Verdes E, Naidoo N, Chatterji S. From depressive symptoms to depressive disorders: the relevance of thresholds. The British journal of psychiatry : the journal of mental science. 2010;196:365–371. doi: 10.1192/bjp.bp.109.071191. [DOI] [PubMed] [Google Scholar]
- 27.Sakashita C, Slade T, Andrews G. Empirical investigation of two assumptions in the diagnosis of DSM-IV major depressive episode. The Australian and New Zealand journal of psychiatry. 2007;41:17–23. doi: 10.1080/00048670601050440. [DOI] [PubMed] [Google Scholar]
- 28.de Graaf LE, Huibers MJ, Cuijpers P, Arntz A. Minor and major depression in the general population: does dysfunctional thinking play a role? Comprehensive psychiatry. 2010;51:266–274. doi: 10.1016/j.comppsych.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Backenstrass M, Frank A, Joest K, Hingmann S, Mundt C, Kronmuller KT. A comparative study of nonspecific depressive symptoms and minor depression regarding functional impairment and associated characteristics in primary care. Comprehensive psychiatry. 2006;47:35–41. doi: 10.1016/j.comppsych.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Radloff LS. The CES-D Scale: A Self-Report Depression Scale for Research in the General Population. Applied Psychological Measurement. 1977;1:385. [Google Scholar]
- 31.Rakofsky JJ, Schettler PJ, Kinkead BL, Frank E, Judd LL, Kupfer DJ, et al. The prevalence and severity of depressive symptoms along the spectrum of unipolar depressive disorders: a post hoc analysis. The Journal of clinical psychiatry. 2013;74:1084–1091. doi: 10.4088/JCP.12m08194. [DOI] [PubMed] [Google Scholar]
- 32.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stouffer S, Suchman E, DeVinnery L, Star S, Williams R. The American Soldier, volume I: Adjustment during Army Life. Princeton University Press; 1949. p. 56. [Google Scholar]
- 35.Cross-Disorder Group of the Psychiatric Genomics C. Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wali A. FHIT: doubts are clear now. The Scientific World Journal. 2010;10:1142–1151. doi: 10.1100/tsw.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karras JR, Paisie CA, Huebner K. Replicative Stress and the FHIT Gene: Roles in Tumor Suppression, Genome Stability and Prevention of Carcinogenesis. Cancers. 2014;6:1208–1219. doi: 10.3390/cancers6021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen A, Krogh J, Miskowiak K, Bolwig TG, Kessing LV, Fink-Jensen A, et al. Systemic oxidatively generated DNA/RNA damage in clinical depression: associations to symptom severity and response to electroconvulsive therapy. J Affect Disord. 2013;149:355–362. doi: 10.1016/j.jad.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 39.Lopresti AL, Maker GL, Hood SD, Drummond PD. A review of peripheral biomarkers in major depression: the potential of inflammatory and oxidative stress biomarkers. Progress in neuro-psychopharmacology & biological psychiatry. 2014;48:102–111. doi: 10.1016/j.pnpbp.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 40.Byrne EM, Heath AC, Madden PA, Pergadia ML, Hickie IB, Montgomery GW, et al. Testing the role of circadian genes in conferring risk for psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:254–260. doi: 10.1002/ajmg.b.32230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gottlieb DJ, O'Connor GT, Wilk JB. Genome-wide association of sleep and circadian phenotypes. BMC medical genetics. 2007;8(Suppl 1):S9. doi: 10.1186/1471-2350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luciano M, Huffman JE, Arias-Vasquez A, Vinkhuyzen AA, Middeldorp CM, Giegling I, et al. Genome-wide association uncovers shared genetic effects among personality traits and mood states. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:684–695. doi: 10.1002/ajmg.b.32072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsang KM, Croen LA, Torres AR, Kharrazi M, Delorenze GN, Windham GC, et al. A genome-wide survey of transgenerational genetic effects in autism. PLoS One. 2013;8:e76978. doi: 10.1371/journal.pone.0076978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikpay M, Seda O, Tremblay J, Petrovich M, Gaudet D, Kotchen TA, et al. Genetic mapping of habitual substance use, obesity-related traits, responses to mental and physical stress, and heart rate and blood pressure measurements reveals shared genes that are overrepresented in the neural synapse. Hypertens Res. 2012;35:585–591. doi: 10.1038/hr.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edwards AC, Aliev F, Bierut LJ, Bucholz KK, Edenberg H, Hesselbrock V, et al. Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr Genet. 2012;22:31–41. doi: 10.1097/YPG.0b013e32834acd07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adkins DE, Clark SL, Aberg K, Hettema JM, Bukszar J, McClay JL, et al. Genomewide pharmacogenomic study of citalopram-induced side effects in STAR*D. Transl Psychiatry. 2012;2:e129. doi: 10.1038/tp.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rietveld CA, Cesarini D, Benjamin DJ, Koellinger PD, De Neve JE, Tiemeier H, et al. Molecular genetics and subjective well-being. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9692–9697. doi: 10.1073/pnas.1222171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.