Abstract
Background
Neonatal adiposity has many determinants and may be a risk factor for future obesity. Epigenetic regulation of metabolically important genes are a potential contributor.
Objective
To determine whether methylation changes in the LEP gene in cord blood DNA are impacted by the maternal environment or affect neonatal adiposity measures.
Methods
A cross-sectional study of 114 full-term neonates born to healthy mothers with normal glucose tolerance was performed. Cord blood was assayed for leptin and genomewide DNA methylation profiles via the Illumina 450K platform. Neonatal body composition was measured by air displacement plethysmography. Multivariate linear regression models and semi-partial correlation coefficients were used to analyze associations. False discovery rate (FDR) was estimated to account for multiple comparisons.
Results
Maternal pre-pregnancy BMI was associated with decreased methylation at 5 CpG sites near the LEP transcription start site in an adjusted model (FDR<0.022 for each site). The association between maternal BMI and cord blood leptin approached significance (r=0.18, p=0.054). Cord blood leptin was positively correlated with neonatal adiposity measures including birth weight (r=0.45, p < 0.001), fat mass (r=0.47, p < 0.001), and percent body fat (r=0.44, p < 0.001).
Conclusions
Maternal pre-pregnancy BMI is strongly associated with decreased cord blood LEP gene methylation and may mediate the well-known association between maternal pre-pregnancy BMI and neonatal adiposity.
Keywords: Leptin, Epigenetics, Neonatal Adiposity, Neonatal Body Composition, Obesity
Introduction
Childhood obesity is a major public health problem, with prevalence estimates of 8% among infants and toddlers and 16.9% among children aged 2–19 (1). Adiposity at birth is a direct measure of body fat (as opposed to birth weight which comprises both fat and fat-free mass) and may be an important predictor of later childhood obesity. Understanding the early life contributors to and predictors of neonatal body composition is essential for delineating risk factors and mechanisms leading to excess neonatal adiposity, and may inform earlier implementation of obesity prevention efforts.
Epigenetics, or changes to DNA that modify gene expression without altering gene sequence, may represent one mechanism that contributes to the development of an adverse metabolic phenotype for infants and children. DNA methylation is one type of epigenetic change in which increased methylation in a gene promoter region is associated with decreased gene expression. Prenatal exposures, maternal factors and the in-utero environment may alter susceptibility of neonatal genes to aberrant methylation and subsequently change their regulation, expression and potentially the child’s metabolic risk.
Leptin, a hormone encoded by the LEP gene, is produced by adipose tissue. It regulates satiety and energy metabolism, and can be a surrogate marker of fat tissue quantity in humans (2). Cord blood leptin levels have been associated with birth weight and neonatal adiposity, however it is not well determined whether leptin is involved in fat development and deposition or serves solely as an adiposity marker (3,4).
This study focused on determining whether methylation patterns within the LEP gene in cord blood DNA were impacted by known risk factors for neonatal adiposity in a cohort of healthy maternal-neonatal pairs. We hypothesized that the maternal milieu influences methylation patterns within the LEP gene in a way that may translate to adverse neonatal metabolic outcomes or altered gene expression.
Subjects and Methods
Subjects
For this study we utilized a subset of a larger cohort of 168 healthy maternal-neonatal pairs on whom cord blood DNA was available. Participants were recruited from 2011–2014 at a large academic medical center. Women carrying a singleton pregnancy with normal glucose tolerance (as evidenced by normal results on a 2-hour 75g oral glucose tolerance test performed between 24 and 28 weeks gestation) were eligible for the study (5). Women were excluded if they had a history of greater than 3 term pregnancies or smoking during pregnancy, as these factors are associated with excess and restricted growth, respectively (6). All newborns were full-term and were excluded if they required intensive care, were too ill to undergo body composition measurements within the first 24–72 hours of life, or if they had congenital anomalies, as some of these are independently associated with abnormal fetal growth. Cord blood was collected after birth by labor and delivery staff, processed within 30 minutes, and stored at −70°C until laboratory assays were performed. 115 maternal neonatal pairs were initially included in the study; 1 pair was excluded due to poor DNA quality. Of the 114 neonates included in the final analysis, 111 had cord blood leptin levels and 105 had body composition data available. This study was approved by the Northwestern University Institutional Review Board and each mother provided written informed consent for herself and her neonate at the time of study enrollment.
Laboratory Measurements
Samples for leptin were batched and measured in duplicate with a radioimmunoassay kit (Millipore Corp, Billerica, MA). The inter- and intra-assay coefficients were 3.7–5.9% and 3.0–4.0%, respectively. DNA was purified from neonatal cord blood using an Autopure LS Automated DNA Purification System with Autopure reagents (Autogen, Inc., Holliston, MA). The purified DNA samples were stored under −20°C. Methylation levels were measured using the Infinium HumanMethylation 450K Beadchip array (Illumina, Inc. CA, USA), which targets ~486,000 CpG sites, in 114 samples that passed DNA quality testing. Samples were randomly plated on each chip with regard to neonatal sex. A 500-ng DNA sample was used to perform bisulfite conversion followed by methylation profiling as per the Illumina 450K protocol. BeadChips were scanned with an Illumina iScan and analyzed using Illumina GenomeStudio software. All experiments were conducted following manufacturer protocols in the Genomics Core Facility at the Center for Genetic Medicine at Northwestern University.
Neonatal Anthropometrics
Infant length, weight, and body composition were measured by one of two trained clinicians. Length was obtained using a hard surface measuring board, recorded to the nearest 0.1cm, and the results of duplicate measurements averaged to produce the final research measure. Weight and body composition were measured utilizing an air displacement plethysmography system (PeaPod, Cosmed, Rome, Italy) as follows: first, the machine was calibrated prior to use according to manufacturer guidelines. Second, the infant was undressed and placed on the calibrated PeaPod scale and weight recorded to the nearest 0.0001 kg. Next, the infant was placed inside the PeaPod volume chamber for 2 minutes to determine body volume. Density was calculated after which age- and sex-specific fat-free mass density values were used to determine absolute fat-free mass and fat mass. Finally, percent body fat was calculated from these values (7). In accordance with prior studies, body composition measurements were obtained between 24–72 hours of life to mitigate the effects of any possible fluid shifts or weight loss common in the immediate post-birth period (8).
Bioinformatics and Statistical Analysis
Raw Illumina IDAT data were imported using the R package methylumi (9). One sample containing more than 5% of CpG probes with a detection p-value greater than 0.01, as well as 191 probes that were not detectable in more than 5% of samples were removed. Probes in any of the following categories were also removed: 65 built-in SNP probes, 3,091 non-CpG probes, 36,535 probes containing proximal SNPs, and 11,648 probes on sex chromosomes. Signal intensities of the filtered probes were corrected for background noise estimated using normal-exponential convolution with out-of-band probes and then corrected for color bias between the Cy3 and Cy5 channels available using methylumi (10). Methylated and unmethylated intensities were quantile-normalized and corresponding β values were calculated (i.e., the proportion of methylated probe intensity out of total intensity) using the function nasen in the R package watermelon (11). We estimated the proportion of blood cell types using the function estimateCellCounts in the R package minfi (12). ComBat was implemented to account for potential batch effects and effects of cell type heterogeneity on methylation profiles using the function ComBat in R package sva (13). Principle component analysis indicated that the ComBat method was successful in eliminating batch effects.
The focus of this study was to analyze the relationship between the methylation pattern of the LEP gene and neonatal and maternal metabolic measures. We examined a total of 17 CpG sites from the LEP gene, located within the regions 1500 and 200 base pairs upstream of transcription start site (TSS1500 and TSS200, respectively), the 5’ and 3’ untranslated regions (5’UTR and 3’UTR), the first exon (1stExon), and the gene body (Body).
We used linear regression models to identify associations between differentially methylated CpGs in the LEP gene across levels of continuous maternal BMI, and to examine associations between log10-transformed cord blood leptin levels and maternal BMI as well as neonatal outcomes including birth weight, fat mass, and percent body fat. Reported parameter estimates were re-scaled to reflect change in methylation (standardized units) per kg/m2 increase in maternal BMI, percent changes of leptin per unit increase in maternal exposure, or as mean change in neonatal outcome per each 10% increase in leptin, respectively. Unadjusted and multivariate models adjusting for maternal age at delivery, maternal race, gestational age in days, and infant sex were conducted. Unadjusted correlation coefficients and semi-partial correlation (r) between exposure and outcome after controlling the outcome for covariates were calculated using R package ppcor (14). To account for multiple comparisons, we used the Benjamini-Hochberg procedure to calculate the False Discovery Rate (FDR) (15). Data preprocessing and statistical analyses were conducted using R 3.2.3.
Results
Maternal and neonatal characteristics of the cohort are displayed in Table 1. Of note, 61.4% of mothers had a normal pre-pregnancy BMI while 35.1% were overweight or obese. The majority of infants had a birth weight that was appropriate for gestational age.
Table 1.
Maternal and Neonatal Characteristics
| Demographics (n=114) | |
|---|---|
| Maternal Race | 61% White |
| 39% Non-White | |
| Maternal Characteristics (n=114) | Mean ± SD |
| Age at Delivery | 32 ± 3.8 years |
| Pre-Pregnancy BMI | 25.0 ± 6.1 kg/m2 |
| Underweight (BMI <18.5 kg/m2) | 3.5% |
| Normal Weight (BMI 18.5–24.9 kg/m2) | 61.4% |
| Overweight (BMI 25–29.9 kg/m2) | 13.2% |
| Obese (BMI ≥30 kg/m2) | 21.9% |
| OGTT Fasting Glucose | 76 ± 5.3 mg/dl |
| 4.2 ± 0.3 mmol/L) | |
| OGTT 1-hour Glucose | 117 ± 25.9 mg/dl |
| (6.5 ± 1.4 mmol/L) | |
| OGTT 2-hour Glucose | 100 ± 19.2 mg/dl |
| (5.6 ± 1.1 mmol/L) | |
| Neonatal Characteristics (n=114, unless noted) | Mean ± SD |
| Sex | 53% Male |
| 47% Female | |
| Gestational Age | 39.5 ± 1 week |
| Birth Weight | 3502 ± 515 g |
| Length | 51.0 ± 2.3 cm |
| Fat Free Mass (n=105) | 2947 ± 374 g |
| Fat Mass (n=105) | 377 ± 169 g |
| % Body Fat (n=105) | 11 ± 3.8 % |
| Cord Blood Leptin (n=111) | 10.7 ± 8.8 ng/ml |
| (10.7 ± 8.8 mcg/L) | |
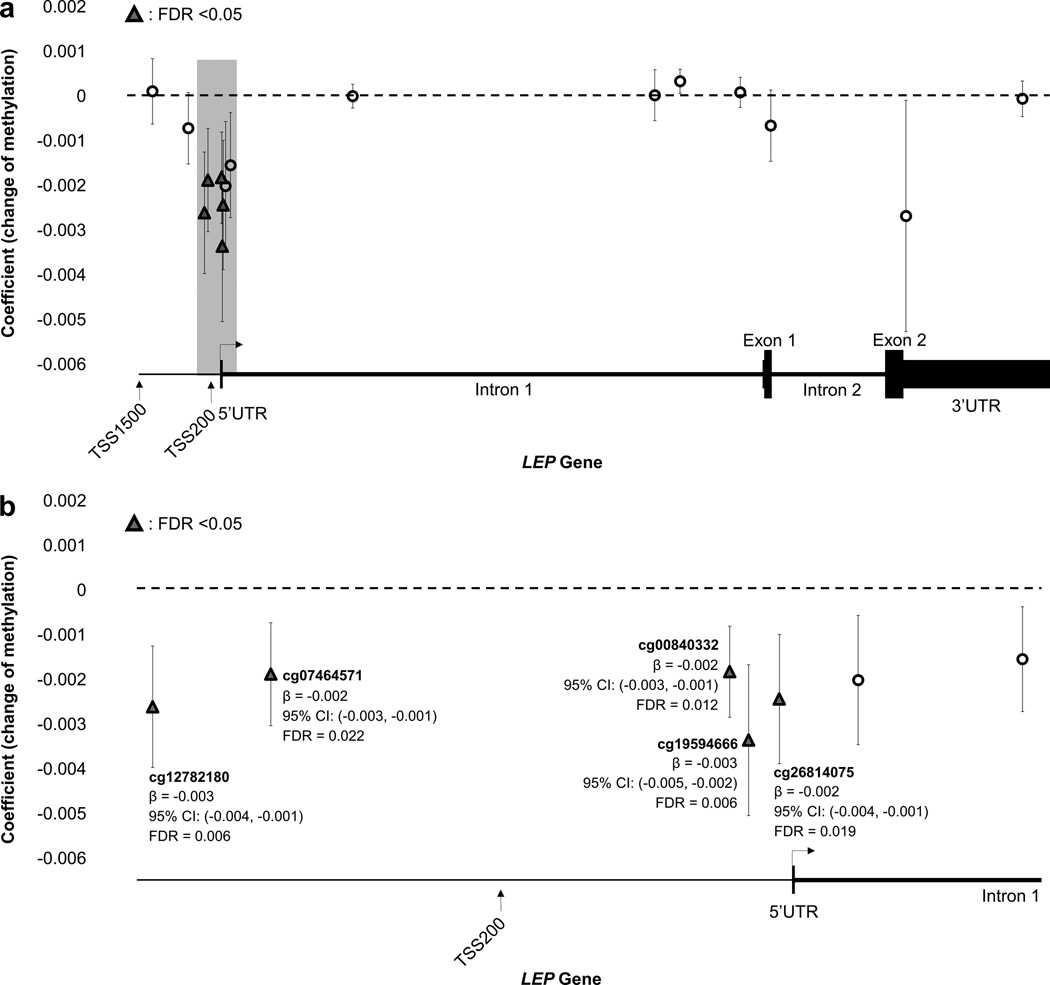
Figure 1 displays the association between increasing maternal pre-pregnancy BMI and decreased methylation at five CpG sites in the LEP gene at FDR <0.05. Each kg/m2 increase in maternal BMI was associated with a mean decrease in methylation of 0.003 standardized units for cg19594666 (95% CI: −0.005, −0.002) and cg00840332 (95% CI: −0.004, −0.001) and a mean decrease in methylation of 0.002 standardized units for cg26714075 (95% CI: −0.003, −0.001), cg12782180 (95% CI: −0.004, −0.001), and cg07464571 (95% CI: −0.003, −0.001). Of these five CpG sites, three are located within 200 base pairs and two within 1500 base pairs of the LEP transcription start site.
Figure 1. Differentially Methylated CpG Sites on the LEP gene.
The triangles and open circles in this schematic represent all CpG sites examined in the analysis; triangles represent CpG sites with decreased methylation that were significantly associated (FDR < 0.05) with maternal BMI in the model adjusted for maternal age at delivery, maternal race, gestational age in days, and infant sex. Parameter estimates and 95% confidence intervals are noted at each significant CpG site. Figure 1a depicts the entire LEP gene while Figure 1b represents a closer look at the portion of the LEP gene promoter where the significant CpG sites are located.
Unadjusted and adjusted associations between cord blood leptin, maternal BMI, and neonatal adiposity measures are displayed in Table 2. Cord blood leptin was positively correlated with measures of neonatal adiposity including birth weight (r=0.45, p<0.001), fat mass (r=0.47, p< 0.001), and percent body fat (r=0.44, p<0.001) when adjusted for maternal age at delivery, maternal race, gestational age, and neonatal sex. The association between increased maternal BMI and cord blood leptin levels (r=0.18, p=0.054) approached significance in the unadjusted model; however, maternal BMI was not significantly associated with birth weight (r=0.17, p=0.067), fat mass (r=0.13, p=0.175), or percent body fat (r=0.06, p=0.502) in adjusted or unadjusted models.
Table 2.
Associations Between Cord Blood Leptin Levels, Maternal BMI, and Neonatal Adiposity
| Univariate | Multivariate* | |||||
|---|---|---|---|---|---|---|
| β value (95%CI) | r | p** | β value (95% CI) | r *** | p** | |
| Exposure | Percent change in leptin per unit increase in maternal BMI | |||||
| Maternal BMI | 2.2 (0.0 – 4.4) |
0.18 | 0.054 | 1.8 (−0.5 – 4.1) |
0.14 | 0.129 |
| Outcomes | Change in outcome per 10% increase in leptin | |||||
| Birth Weight (kg) | 0.024 (0.012 – 0.036) |
0.35 | <0.001 | 0.027 (0.017 – 0.037) |
0.45 | <0.001 |
| Fat Mass (kg) | 0.011 (0.007 – 0.014) |
0.48 | <0.001 | 0.010 (0.007 – 0.014) |
0.47 | <0.001 |
| % Body Fat | 0.235 (0.150 – 0.320) |
0.47 | <0.001 | 0.218 (0.135 – 0.300) |
0.44 | <0.001 |
Analyses were adjusted for maternal age at delivery, maternal race, gestational age in days, and neonatal sex
p values were calculated from linear regression models
Semi-partial correlation between exposure and outcome after controlling the latter for the adjusting covariates
Discussion
Our results suggest that increased maternal pre-pregnancy BMI is associated with decreased methylation near the transcription start site of the neonatal LEP gene in cord blood. The implication of decreased LEP gene methylation near the transcription start site is possible increased neonatal LEP gene expression. We also identified positive associations between cord blood leptin levels and direct measures of neonatal adiposity (birth weight, fat mass, and percent body fat) and a marginally significant association between maternal BMI and cord blood leptin levels, suggesting potential mediation by LEP gene expression. We did not uncover a direct relationship between LEP gene methylation and neonatal adiposity measures or cord blood leptin levels. A full understanding of LEP transcription initiation and regulation has not yet been established though some specific enhancer regions and transcription factors have been identified (16). Further molecular study is necessary to explore how the hypomethylated CpG sites reported in this study affect LEP gene expression. This significant association between increased maternal BMI and decreased neonatal LEP methylation is likely related to the well-described relationship between maternal BMI and maternal leptin (17). Leptin is a known marker of adipose tissue (2). Analogous to the association of insulin resistance in pregnancy with increased neonatal body fat and Freinkel’s theory of “fuel mediated teratogenesis”, it is possible that rising maternal BMI and associated higher maternal leptin levels directly impact placental transfer of lipid substrates. In turn, this leads to greater fetal growth, fat development, and subsequently may indirectly leads to decreased fetal LEP gene methylation, increased LEP gene expression, and increased fetal adipocyte leptin production (18–20). An adjunctive mechanism arises from animal models that have suggested that maternal obesity at conception enhances fetal adipogenesis that may translate in humans as higher fetal fat tissue and leptin production with rising pre-pregnancy BMI (21). There is conflicting literature on the correlation between maternal and neonatal leptin; yet in our study and in others there is a consistent relationship between neonatal adiposity and neonatal leptin levels (17,22). Leptin produced by the placenta primarily enters the maternal circulation (95%) in comparison to the fetal circulation (5%) suggesting that neonatal leptin levels truly reflect production by neonatal fat tissue (22).
Our results did not demonstrate a direct association between decreased LEP methylation and increased neonatal cord blood leptin levels or adiposity measures, as reported by Allard and colleagues (23). Allard’s cohort included women with gestational diabetes and analyses demonstrated that a higher maternal glycemia risk score was associated with decreased LEP methylation and higher cord blood leptin levels. In contrast, all mothers in our cohort had clearly normal glucose tolerance, enabling us to remove the effect of fetal hyperinsulinism as a known contributor to neonatal adiposity and perhaps limiting our ability to observe a more robust correlation between decreased LEP methylation and higher neonatal body fat and cord blood leptin levels as Allard described (24,25). We did demonstrate strong associations between cord blood leptin levels and measures of neonatal adiposity, suggestive of a LEP gene methylation-mediated phenomenon. Our small sample size, lack of gene expression data, and possible additional unknown factors that impact LEP expression, may have prevented us from observing a clear relationship.
Unlike Allard’s study and in line with our results, many studies have not reported a direct correlation between LEP methylation and serum leptin level. Additionally, LEP methylation has not consistently been related to LEP gene expression, suggesting that there may be co-existing genetic, epigenetic, and transcriptional factors affecting LEP methylation, LEP gene expression, and measurable changes in serum leptin levels that are not fully understood. In a sheep model, lack of observable elevations in plasma leptin levels despite increased adipose tissue leptin mRNA expression in infant sheep born to overnourished mothers suggests that other factors are at play (26). Two SNPs in the LEP promoter, rs7799039 and rs2167270, have been associated with serum leptin level, increased BMI, and waist circumference in adults (27). The presence of rs2167270 has also been associated with greater cord blood LEP methylation (28). A recent genome-wide meta-analysis of 23 studies revealed five SNPs that robustly associated with circulating leptin levels in the LEP gene, as well as in or near other genes including SLC32A1, GCKR, CCNL1 and FTO, suggesting that leptin regulation is affected by additional genes (29). In vitro and in vivo studies observed that transcription factor activator protein–2β played a crucial role in regulating promoter activity of LEP in adipocytes (30). Further detailed study in a larger cohort using integrative analyses is necessary to examine how these mechanisms work together to affect LEP gene expression.
Few existing studies have evaluated the epigenetics of the LEP gene in neonates and its relationship to maternal body composition. The results between studies are variable, perhaps because the populations used in our study and existing studies are characteristically different and thus not directly comparable. In a healthy cohort of infants from the Rhode Island Child Health Study, mean cord blood LEP promoter methylation was lower in infants born to obese mothers and higher in SGA infants compared to AGA and LGA. These findings are similar to our correlation between maternal pre-pregnancy BMI and lower cord blood LEP methylation, though we studied specific CpG sites rather than mean methylation across sites (28). Our population was comprised largely of AGA infants while Lesseur’s study primarily included SGA and LGA infants, suggesting methylation changes are evident across the spectrum of infants, not just at the extremes of birth weight. A Dutch cohort comparing 113 preterm infants born SGA vs. average for gestational age (AGA) revealed no difference in methylation patterns at the LEP gene between the two groups (31). However, not all mothers and infants were healthy, therefore other factors may have impacted methylation levels and body size. Garcia-Cardona et al reported that obesity in adolescents was associated with decreased LEP methylation, suggesting that this epigenetic alteration is important even beyond the neonatal period (32).
Pre-pregnancy maternal BMI is well known to be associated with birth weight and measures of neonatal adiposity and may also influence BMI in the toddler years (33–35). In our cohort, the relationship between maternal BMI and birth weight trended towards significance but no relationship was identified with neonatal fat mass, percent body fat, or cord blood leptin; this may have been due to our small sample size.
Strengths of our study include the use of a cohort of healthy mothers with normal glucose tolerance allowing us to remove the known confounder of fetal hyperinsulinism on fetal growth and adiposity (5,24). Another strength was the absence of smoking, a factor known to be associated with abnormal fetal growth. Furthermore, we were able to establish relationships between 1) a maternal exposure (pre-pregnancy BMI) and changes in LEP gene methylation and 2) leptin levels and neonatal adiposity measures, suggesting a plausible physiological mechanism. Weaknesses of our study include its cross-sectional design, only allowing us to report associations rather than causality and small sample size, which may have resulted in false negatives such as the lack of an association between maternal BMI and neonatal adiposity. We also lacked long-term follow up and the ability to assess adiposity or methylation in later infancy or early childhood.
This study demonstrates a strong association between maternal pre-pregnancy BMI and decreased methylation near the LEP gene transcription start site in cord blood, potentially enhancing neonatal LEP gene expression. Higher leptin levels may induce a state of leptin resistance earlier in life, altering metabolic regulation of satiety and energy metabolism and increasing a child’s obesity risk. Research surrounding LEP gene methylation and body composition in neonates and children is an emerging area of clinical investigation. Further study is needed in larger, longitudinal cohorts (perhaps with sibling controls), accounting for known risk factors for obesity and with the inclusion of gene expression studies to determine the clinical impact that the LEP gene has on obesity throughout the life course.
What is already known about this subject
Maternal pre-pregnancy BMI is associated with neonatal adiposity.
Leptin is a surrogate marker of fat tissue quantity in neonates, children, and adults.
What this study adds
Maternal pre-pregnancy BMI may alter epigenetic regulation of the neonatal LEP gene, subsequently impacting neonatal adiposity.
Acknowledgments
RK and JJ conceived the study objectives and design and worked on data interpretation. YZ and ZZ analyzed data. WZ provided support in data analysis. LH provided support in study design, data analysis, and data interpretation. RK wrote the manuscript and all authors edited and had final approval of the submitted manuscript. The authors would like to thank Brian Joyce for his feedback during the preparation of this manuscript. This study was supported by the National Institutes of Child Health and Human Development Grant K12 HD055884 and a Genentech Growth Disorders Grant G-29954.
Footnotes
Conflicts of Interest: The authors of this paper have no conflicts of interest or disclosures.
References
- 1.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of childhood and adult obesity in the United States, 2011–2012. JAMA. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Considine RV, Sinha MK, Heiman ML, et al. Serum immunoreactive-leptin concentrations in normal-weight and obese humans. N Engl J Med. 1996;334:292–295. doi: 10.1056/NEJM199602013340503. [DOI] [PubMed] [Google Scholar]
- 3.Josefson JL, Zeiss DM, Rademaker AW, Metzger BE. Maternal leptin predicts adiposity of the neonate. Horm Res Paediatr. 2014;81:13–19. doi: 10.1159/000355387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papadopoulou FG, Mamopoulos AM, Triantos A, et al. Leptin levels in maternal and cord serum: relationship with fetal development and placental weight. J Matern Fetal Med. 2000;9:298–302. doi: 10.1002/1520-6661(200009/10)9:5<298::AID-MFM9>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 5.Metzger BE, Gabbe SG, Persson B, et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care. 2010;33:676–682. doi: 10.2337/dc09-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh CA, Mahony RT, Foley ME, Daly L, O'Herlihy C. Recurrence of fetal macrosomia in non-diabetic pregnancies. J Obstet Gynaecol. 2007;27:374–378. doi: 10.1080/01443610701327545. [DOI] [PubMed] [Google Scholar]
- 7.Ma G, Yao M, Liu Y, et al. Validation of a new pediatric air-displacement plethysmograph for assessing body composition in infants. Am J Clin Nutr. 2004;79:653–660. doi: 10.1093/ajcn/79.4.653. [DOI] [PubMed] [Google Scholar]
- 8.Hull HR, Thornton JC, Ji Y, et al. Higher infant body fat with excessive gestational weight gain in overweight women. Am J Obstet Gynecol. 2011;205:211, e-1–e-7. doi: 10.1016/j.ajog.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis S, Du P, Bilke S, Triche TJ, Jr, Bootwalla M. methylumi: Handle Illumina methylation data. R package version 2.17.0. 2015 [Google Scholar]
- 10.Triche TJ, Jr, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schalkwyk LC, Pidsley R, Wong CC, et al. wateRmelon: Illumina 450 methylation array normalization and metrics. R package version 101. 2013 [Google Scholar]
- 12.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S. ppcor: An R Package for a Fast Calculation to Semi-partial Correlation Coefficients. Communications for statistical applications and methods. 2015;22:665–674. doi: 10.5351/CSAM.2015.22.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 16.Wrann CD, Rosen ED. New insights into adipocyte-specific leptin gene expression. Adipocyte. 2012;1:168–172. doi: 10.4161/adip.20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geary M, Pringle PJ, Persaud M, et al. Leptin concentrations in maternal serum and cord blood: relationship to maternal anthropometry and fetal growth. Br J Obstet Gynaecol. 1999;106:1054–1060. doi: 10.1111/j.1471-0528.1999.tb08113.x. [DOI] [PubMed] [Google Scholar]
- 18.Hauguel-de Mouzon S, Lepercq J, Catalano P. The known and unknown of leptin in pregnancy. Am J Obstet Gynecol. 2006;194:1537–1545. doi: 10.1016/j.ajog.2005.06.064. [DOI] [PubMed] [Google Scholar]
- 19.Crume TL, Shapiro AL, Brinton JT, et al. Maternal fuels and metabolic measures during pregnancy and neonatal body composition: the healthy start study. J Clin Endocrinol Metab. 2015;100:1672–1680. doi: 10.1210/jc.2014-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freinkel N. Banting Lecture 1980. Of pregnancy and progeny. Diabetes. 1980;29:1023–1035. doi: 10.2337/diab.29.12.1023. [DOI] [PubMed] [Google Scholar]
- 21.Borengasser SJ, Zhong Y, Kang P, et al. Maternal obesity enhances white adipose tissue differentiation and alters genome-scale DNA methylation in male rat offspring. Endocrinology. 2013;154:4113–4125. doi: 10.1210/en.2012-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lepercq J, Challier JC, Guerre-Millo M, Cauzac M, Vidal H, Hauguel-de Mouzon S. Prenatal leptin production: evidence that fetal adipose tissue produces leptin. J Clin Endocrinol Metab. 2001;86:2409–2413. doi: 10.1210/jcem.86.6.7529. [DOI] [PubMed] [Google Scholar]
- 23.Allard C, Desgagne V, Patenaude J, et al. Mendelian randomization supports causality between maternal hyperglycemia and epigenetic regulation of leptin gene in newborns. Epigenetics. 2015;10:342–351. doi: 10.1080/15592294.2015.1029700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Catalano PM, Presley L, Minium J, Hauguel-de Mouzon S. Fetuses of obese mothers develop insulin resistance in utero. Diabetes Care. 2009;32:1076–1080. doi: 10.2337/dc08-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyperglycemia and Adverse Pregnancy Outcome (HAPO) Study: associations with neonatal anthropometrics. Diabetes. 2009;58:453–459. doi: 10.2337/db08-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muhlhausler BS, Duffield JA, McMillen IC. Increased maternal nutrition stimulates peroxisome proliferator activated receptor-gamma, adiponectin, and leptin messenger ribonucleic acid expression in adipose tissue before birth. Endocrinology. 2007;148:878–885. doi: 10.1210/en.2006-1115. [DOI] [PubMed] [Google Scholar]
- 27.Marcello MA, Calixto AR, de Almeida JF, Martins MB. Polymorphism in LEP and LEPR May Modify Leptin Levels and Represent Risk Factors for Thyroid Cancer. 2015;2015:173218. doi: 10.1155/2015/173218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lesseur C, Armstrong DA, Paquette AG, Koestler DC, Padbury JF, Marsit CJ. Tissue-specific Leptin promoter DNA methylation is associated with maternal and infant perinatal factors. Mol Cell Endocrinol. 2013;381:160–167. doi: 10.1016/j.mce.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilpelainen TO, Carli JF, Skowronski AA, et al. Genome-wide meta-analysis uncovers novel loci influencing circulating leptin levels. Nature communications. 2016;7:10494. doi: 10.1038/ncomms10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuke T, Yoshizaki T, Kondo M, et al. Transcription factor AP-2beta inhibits expression and secretion of leptin, an insulin-sensitizing hormone, in 3T3-L1 adipocytes. Int J Obes (Lond) 2010;34:670–678. doi: 10.1038/ijo.2009.295. [DOI] [PubMed] [Google Scholar]
- 31.Tobi EW, Heijmans BT, Kremer D, et al. DNA methylation of IGF2, GNASAS, INSIGF and LEP and being born small for gestational age. Epigenetics. 2011;6:171–176. doi: 10.4161/epi.6.2.13516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Cardona MC, Huang F, Garcia-Vivas JM, et al. DNA methylation of leptin and adiponectin promoters in children is reduced by the combined presence of obesity and insulin resistance. Int J Obes (Lond) 2014;38:1457–1465. doi: 10.1038/ijo.2014.30. [DOI] [PubMed] [Google Scholar]
- 33.Hyperglycaemia and Adverse Pregnancy Outcome (HAPO) Study: associations with maternal body mass index. BJOG. 2010;117:575–584. doi: 10.1111/j.1471-0528.2009.02486.x. [DOI] [PubMed] [Google Scholar]
- 34.Starling AP, Brinton JT, Glueck DH, et al. Associations of maternal BMI and gestational weight gain with neonatal adiposity in the Healthy Start study. Am J Clin Nutr. 2015;101:302–309. doi: 10.3945/ajcn.114.094946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linabery AM, Nahhas RW, Johnson W, et al. Stronger influence of maternal than paternal obesity on infant and early childhood body mass index: the Fels Longitudinal Study. Pediatr Obes. 2013;8:159–169. doi: 10.1111/j.2047-6310.2012.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]