Abstract
Expectations of results from genome sequencing by end users are influenced by perceptions of uncertainty. This study aimed to assess uncertainties about sequencing by developing, evaluating, and implementing a novel scale. The Perceptions of Uncertainties in Genome Sequencing (PUGS) scale comprised ten items to assess uncertainties within three domains: clinical, affective, and evaluative. Participants (n=535) from the ClinSeq® NIH sequencing study completed a baseline survey that included the PUGS; responses (mean=3.4/5, SD=0.58) suggested modest perceptions of certainty. A confirmatory factor analysis identified factor loadings that led to elimination of two items. A revised eight-item PUGS scale was used to test correlations with perceived ambiguity (r=−0.303, p<0.001), attitudinal ambivalence (r=−0.111, p=0.011), and ambiguity aversion (r=−0.093, p=0.033). Results support nomological validity. A correlation with the MICRA uncertainty subscale was found among 175 cohort participants who had received results (r=−0.335, p<0.001). Convergent and discriminant validity were also satisfied in a second sample of 208 parents from the HudsonAlpha CSER Project who completed the PUGS (mean=3.4/5, SD=0.72), and configural invariance was supported across the two datasets. As such, the PUGS is a promising scale for evaluating perceived uncertainties in genome sequencing, which can inform interventions to help patients form realistic expectations of these uncertainties.
Keywords: Genome sequencing, Perceptions of uncertainty, Practical uncertainties, PUGS scale
Introduction
Uncertainty abounds in medicine and affects how patients regard information about their health. Although genomics is not exceptional in this respect, the scope and degree of uncertainty in genome sequencing are not yet well defined. A recent conceptual taxonomy categorizes medical uncertainties according to multiple discrete dimensions: source, issue and locus.1 The second dimension, issue, represents the manifestations or substantive objects of uncertainty, which may be scientific (data-centered), practical (system-centered), or personal (patient-centered) in nature. Practical uncertainties encompass issues related to access and utilization of health care services, while personal uncertainties encompass uncertainties pertaining to psychosocial and existential issues important to patients, including the personal meaning and implications of illness or health information. Any of these uncertainties may influence patients’ expectations of, and responses to, genomic information.1,2
In genome sequencing, practical uncertainties may be related to a variety of specific issues including competence of health care providers and the behaviors or actions that need to be undertaken in response to test results. A recent overview of priorities for research in genome sequencing highlights the importance of assessing and understanding how patients’ perceptions of these and personal uncertainties affect the use of genome sequencing.3 Accomplishing this task requires reliable and valid measures; however, such measures are currently lacking. Past efforts to measure patients’ perceptions of uncertainties in health care have focused on uncertainties related to living with chronic illness and are not directly applicable to the types of uncertainties inherent in genomic information.4,5 One widely used scale, the Multidimensional Impact of Cancer Risk Assessment (MICRA) scale, includes an uncertainty subscale that is broad, focusing on perceptions of general uncertainties in genetic test results and patients’ future plans. The uncertainty subscale yields results with minimal practical application and limited specificity, in that it does not discriminate among the various genetic testing-related uncertainties that patients may perceive.6
The objective of the current study was to address this research gap. Building on practical findings elicited from exploratory research with participants receiving genome and exome sequencing, we developed and conducted initial psychometric evaluation of a new measure, the Perceptions of Uncertainties in Genome Sequencing (PUGS) scale. Our goal was to produce a reliable, valid, and parsimonious measure that would enable researchers to quantify perceptions of important uncertainties among end-users of genome sequencing, and to ultimately determine how best to support them when contemplating or undergoing sequencing. Data from the PUGS scale could help to identify areas of perceived uncertainty, pre and post-results where perceptions of certainty may exceed their interpretation.
To achieve our objective, we studied adult participants from an NIH research cohort who consented to undergo sequencing and receive results. All but urgent medically actionable findings were returned as a matter of personal choice. We assessed other psychological outcomes as a means of evaluating the psychometric properties of the PUGS scale in conjunction with perceptions of uncertainty, including perceptions of ambiguity (a specific source of uncertainty consisting of the reliability, credibility, or adequacy of risk information)7 and attitudinal ambivalence (simultaneous positive and negative appraisals of risk information).8 We hypothesized that higher perceptions of practical uncertainty would be associated with higher perceptions of ambiguity and greater attitudinal ambivalence. We also assessed individual differences in propensity towards ambiguity aversion, a phenomenon characterized by pessimistic appraisals of ambiguous risks and choice options, and avoidance of decision-making.7,9 We had insufficient evidence to hypothesize that participants with higher individual-level ambiguity aversion would have lower perceptions of uncertainty. However, one possibility is that those who are more averse to ambiguity may be motivated to mitigate the threat of uncertainty by minimizing it, thus reporting lower perceptions of uncertainty. In this cohort we also used the PUGS to assess perceptions of uncertainty arising from return of genomic test results one month after receipt for convergent validity testing using the MICRA uncertainty subscale.6 Data from a cohort of parents and children with developmental disabilities was used to extend a confirmatory factor analysis with a more diverse sample.
Methods
ClinSeq® is an NIH cohort study that initially focused on coronary artery disease (CAD) as a model for studying the genetic architecture of common disease.10 The cardiovascular health of participants within a spectrum of CAD risk was evaluated. To ensure diversity in CAD risk, we stratified participants evenly across “bins” using a Framingham risk score (bin 1 contained those at <5% 10-year-risk for the development of coronary artery disease; bin 2 contained those at 5–10% risk; bin 3 contained those at >10% risk; and bin 4 contained those with known CAD). Self-referred participants learned of the study through local notices and word of mouth. Stratification was accomplished by declining participants who met criteria for bins that were complete. Eighty percent of participants were unaffected with CAD.10
The PUGS scale development was based on the findings of prior ClinSeq® qualitative studies that used open-ended survey questions and focus groups to explore perceptions of uncertainties in genome sequencing among adult participants.2,11 Participants discussed uncertainty about how to act on the information, how they might feel about the information upon receipt, and whether they could trust the results. Although scale development was not initially our objective, the descriptions of practical uncertainties generated from these efforts inspired development of items ascertaining perceptions of uncertainties distinguishable into three domains related to genome sequencing: clinical uncertainty, affective uncertainty, and evaluative uncertainty. Candidate scale items were evaluated for clarity, meaning, and format of presentation. With clinical utility in mind, our specific aim was to develop a scale to assess perceptions of uncertainty most likely to affect responses to receipt of results.
The scale was initially piloted with a convenience sample of four NIH post-baccalaureate research fellows, and revised following content assessment interviews. Responses contributed to assessment of the face validity of the scale and the decision to use higher scores to represent greater certainty, rather than uncertainty. Additional piloting of the PUGS was conducted with two volunteers from the ClinSeq® cohort,12 and minor wording revisions were made to individual items. The first version of the scale included ten items.
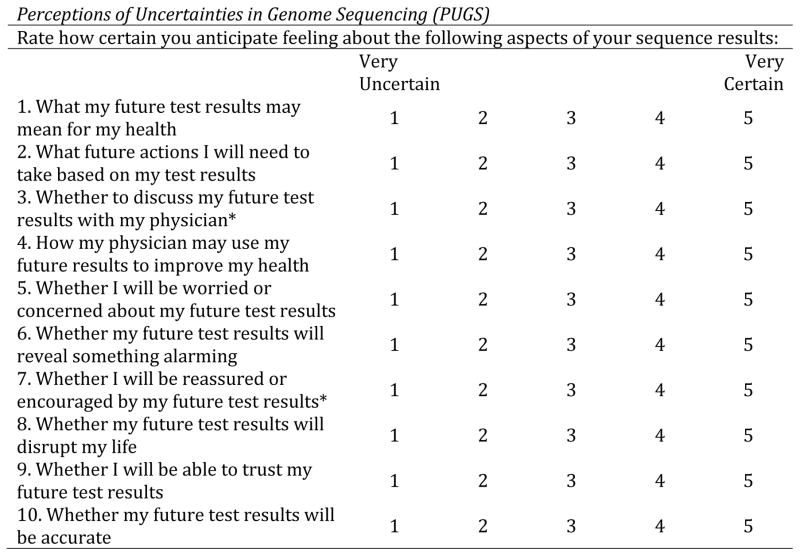
Prior to receipt of results or counseling, ClinSeq® participants completed baseline surveys that requested they rate how certain they anticipated being from 1 (very uncertain) to 5 (very certain) about each item. Examples of items in each domain include: “What my future test results may mean for my health” (clinical), “Whether I will be worried or concerned about my future test results” (affective), and “Whether I will be able to trust my future test results” (evaluative). Questions 1–4 were developed to capture clinical uncertainty, questions 5–8 for affective uncertainty and questions 9 and 10 for evaluative uncertainty about the trustworthiness of sequence information (see Figure 2).
Figure 2.
The PUGS scale
* Items removed from the PUGS scale based on results from the CFA
The first version of the PUGS scale was included in a baseline ClinSeq® social and behavioral survey composed of multiple scales that took participants about 45 minutes to complete.10 The survey also included scales that address components of uncertainty including perceptions of ambiguity, medical ambiguity aversion, and ambivalence toward learning results from sequencing. ClinSeq® participants who chose to receive carrier results completed a one-month follow-up survey to assess cognitive, affective, and behavioral responses to receipt of those results. Goals included assessing differences between anticipated and realized uncertainties. The follow-up survey included assessment of the impact of receiving results including a slightly modified version of the PUGS (Figure 2).
The following scales were used to assess these states:
Perceptions of Ambiguity
Perceived ambiguity regarding the lack of clarity in sequence information was assessed using items developed based on prior research1,7 that were scored on a five-point scale from 1 (Strongly disagree) to 5 (Strongly agree), and were summed and averaged for an overall score. The α coefficient was 0.74.
Ambivalence toward learning results
Ambivalence was assessed using two items,8 “I have mixed feelings about whether to receive this type of sequencing result” and “I am torn about whether to learn this type of sequencing result” across three different types of results (medically actionable results, carrier results, and variants of unknown significance). Responses to the three types of results were not significantly different. As such, an average across the six items was calculated to represent overall attitudinal ambivalence toward receipt of sequence results. The α coefficient was 0.89.
Medical Ambiguity Aversion
The six-item Ambiguity Aversion in Medicine scale13 was used to assess individual-level propensity towards aversion to ambiguity about medical tests or treatments. An example item is: “I don't think my test results will give clear answers about my future health.” Items were scored on a scale from 1 (Strongly disagree) to 5 (Strongly agree). Two items are reversed scored then all six are summed and averaged for an overall score. The α coefficient was 0.80.
Perceptions of Uncertainty following receipt of results
Uncertainty of test results was assessed using a nine-item subscale of the Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire, which was initially developed to measure cancer-specific test distress.6 The wording was modified from a cancer risk result to assess perceptions of uncertainty about a genetic test result. Respondents were asked to rate the degree to which they had experienced each item in the last two weeks using the responses: 0 (Never), 1 (Rarely), 3 (Sometimes), and 5 (Often). An example item was “being uncertain about what your result means about your future health.” All items were summed and averaged for an overall score. The α coefficient was 0.74.
Nomological validity was assessed by testing hypothesized relationships of the degree of uncertainty perceptions to related variables: perceived ambiguity, ambiguity aversion and attitudinal ambivalence. Ambiguity is one source of uncertainty1 and hypothesized to be correlated with perceptions of uncertainty about genome sequencing. Perceived uncertainty was expected to be higher among individuals with greater attitudinal ambivalence, providing a “known-groups” test of validity. Ambiguity aversion, on the other hand, was hypothesized to be associated with lower perceptions of uncertainty about genome sequencing.
Data from parent pairs participating in the HudsonAlpha CSER Project14 were used to further assess the validity and reliability of the ten-item version of the PUGS. The CSER project is a translational study designed to examine the effectiveness and clinical utility of genome sequencing to identify genetic causes for intellectual disability, developmental delay, and related phenotypes, and involves the sequencing of mother-father-child trios from Alabama. The PUGS was administered to both mothers and fathers as a part of a questionnaire involving assessments of a number of constructs including genomic knowledge, health literacy, numeracy, and perceived severity of the child’s illness. The questionnaire was administered from 2012 to 2015, at the time of enrollment in the study.
Statistical Analysis
Exploratory factor analysis (EFA) and confirmatory factor analysis (CFA) were used to determine whether scale items mapped onto their intended domains and the degree of variance accounted for by each factor among the ClinSeq® participants. In order to more effectively accommodate the categorical nature of the response scales, polychoric correlations and a robust weighted least squares (MLSMV) estimation was utilized in MPLUS to perform the CFA. A hierarchical EFA and CFA model was fit in MPLUS to accommodate for the hierarchical nature of the CSER data with families at the highest level and mothers and fathers within families as the second level of the hierarchy.
The National Human Genome Research Institute Institutional Review Board approved the ClinSeq® study. The HudsonAlpha CSER project was approved by Western IRB and the University of Alabama at Birmingham IRB.
Results
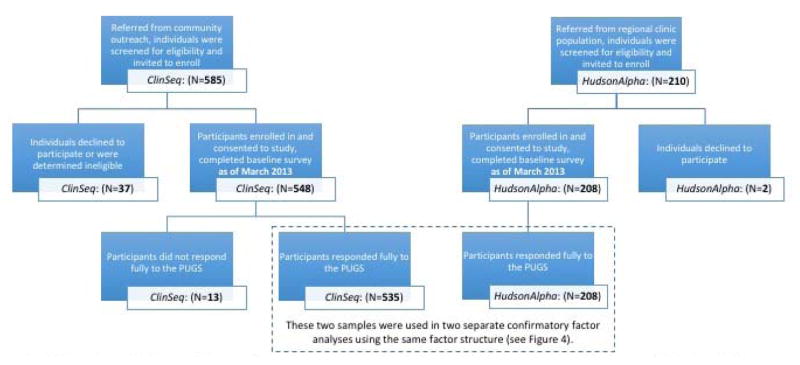
Five hundred forty-eight participants from the ClinSeq® cohort had completed a baseline survey as of the time of analysis (March 2013), thirteen of whom had not responded to each item on the PUGS. These non-responders did not differ from the 535 responders comprising our sample on any demographic. Responders to the PUGS (35.3% from ClinSeq®bin 1, 34.2% from bin 2, 11.2% from bin 3, and 19.3% from bin 4) were on average 61 years of age (SD=5.5), and most were married (76.2%). In addition, 104 parent pairs (dependent data on 208 individuals) from the HudsonAlpha CSER Project completed a respective baseline survey; 97% of participants responded to each item on the PUGS. Most (88.5%) of these individuals had one affected child. The degree of formal education differs between these two samples as members of the ClinSeq® cohort were more likely to have a post-graduate education (63.1% vs. 12.5%; X2=143.04, p<0.001). The flow of participants in both samples is shown in Figure 1, and demographic characteristics from both samples are presented in Table 1.
Figure 1.
A flowchart of the participants from the two samples included in the analyses. ClinSeq and HudsonAlpha. While recruitment for both studies continued, the totals reflect all enrollees with baseline data at the time of analysis.
Table 1.
Demographic characteristics of both samples used for the PUGS CFA.
| Characteristic | Response Option | ClinSeq® (N=535) | HudsonAlpha (N=208) |
|---|---|---|---|
|
| |||
| n (%) | n (%) | ||
| Gender | Male | 297 (55.5) | 104 (50.0) |
| Female | 238 (44.5) | 104 (50.0) | |
|
| |||
| Race | White | 496 (92.7) | 182 (89.2) |
| Other | 39 (7.3) | 35 (17.2) | |
|
| |||
| Ethnicity | Hispanic or Latino | 7 (1.3) | 8 (3.8) |
| Not Hispanic or Latino | 526 (98.7) | 199 (95.7) | |
|
| |||
| Highest Level of Education | High school or less | 19 (3.6) | 60 (28.8) |
| Technical or professional school | 7 (1.3) | 5 (2.4) | |
| Associate’s degree | – | 23 (11.1) | |
| Some college | 31 (5.8) | 45 (21.6) | |
| Bachelor’s degree | 135 (25.2) | 49 (23.6) | |
| Post-graduate degree | 328 (61.3) | 26 (12.5) | |
|
| |||
| Annual Household Income | Less than $25,000 | 7 (1.3) | – |
| $25,000 $49,999 | 21 (3.9) | – | |
| $50,000 $74,999 | 27 (5.0) | – | |
| $75,000 $99,999 | 57 (10.7) | – | |
| More than $100,000 | 397 (74.2) | – | |
Exploratory Factor Analysis on ClinSeq® data
An EFA was performed on the items of the PUGS scale. The measures of sampling adequacy were satisfied for all items (>0.62), the overall KMO=0.77 and Bartlett’s test was significant indicating that an EFA could be supported by the data. Three factors, accounting for 61% of the variation in the 10 items, had eigenvalues greater than 1.0. A varimax rotation (using 0.5 as a cutoff) indicated that the first four items loaded on the first (clinical) factor, the second four items loaded on the second (affective) factor and the last two items loaded on the third (evaluative) factor. There were no serious cross loadings; however, the communality for the “Discuss Physician” item was low at 0.3. The communalities for Worried and Reassured were 0.47 whereas all others had values greater than 0.50. Based on these results, an initial hypothesized model, consisting of the three factors (representing clinical, affective and evaluative uncertainty) along with the items associated with each factor in the EFA, was utilized in a CFA.
Confirmatory Factor Analysis on ClinSeq® data
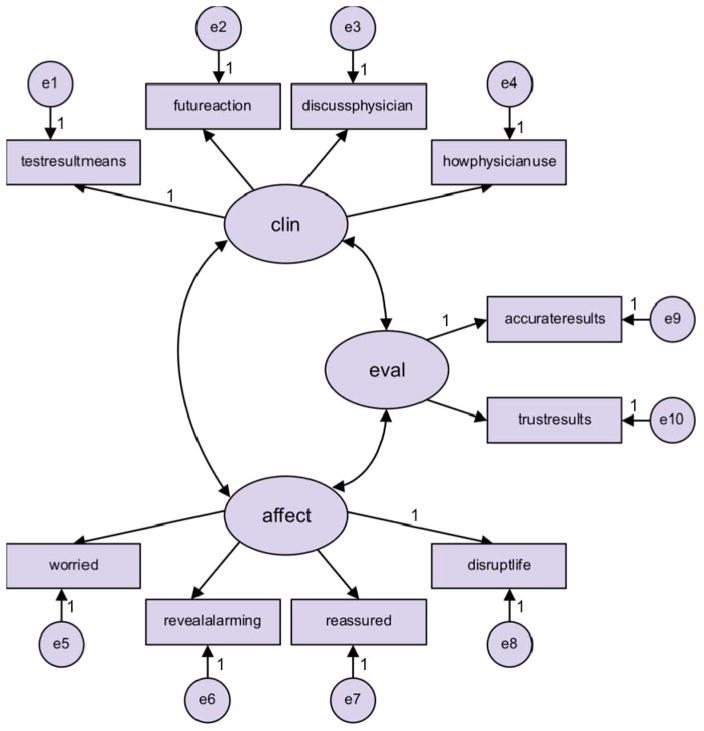
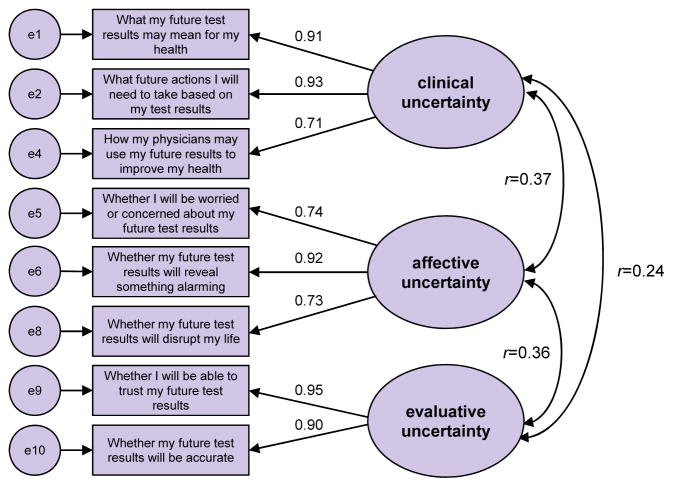
An initial CFA was performed on the hypothesized factors and associated items derived from the EFA but allowing for correlation between the factors as shown in Figure 3. This analysis indicated poor fit statistics and cross loading associated with the item “Reassured”, in the affective uncertainty factor, and the clinical uncertainty factor (highly significant modification index). Eliminating “Reassured” from the model and performing a CFA analysis on the resulting model again yielded poor fit statistics and indicated that “Discuss Physician” in the clinical uncertainty factor cross loaded with the evaluative uncertainty factor (highly significant modification index). Eliminating the “Discuss Physician” item from the scale resulted in adequate CFA model fit (ChiSq/df = 3.8, CFI = 0.99 and RMSEA = 0.08) with all communalities greater than 0.51, all loadings positive and significant and all factor correlations positive and significant. Moreover, the factor reliabilities were all greater than 0.84 and the average variance extracted by each factor was greater than 0.64 implying convergent validity. Discriminant validity was also satisfied. This final model can be found in Figure 4.
Figure 3.
The measurement model for the confirmatory factor analysis.
Figure 4.
The CFA model with standardized factor loadings for the items and correlations between the factors.
Confirmatory Factor Analysis on HudsonAlpha data
An exploratory analysis of the HudsonAlpha CSER Project data indicated that the responses of mothers and fathers exhibited a similar skewed distribution pattern as observed in the baseline ClinSeq® sample. A MANOVA for the differences in response between mothers and fathers in the same family indicated a significant gender effect with the item “whether to discuss my future test results with my physician” (p=0.027; this item was not included in subsequent analysis to coincide with the baseline ClinSeq® analysis) and the item “whether I can trust my future test results”(p=0.010) indicated a significant difference in the average responses of mothers and fathers. No other items indicated a significant difference between the responses of mothers and fathers, both of which were included in the analysis.
A two-level EFA and CFA model for an eight-item scale was used to accommodate the family dependency inherent in the sampling design of this data. Intra-class correlations were all greater than 0.150 (the largest was 0.357) except for one item (0.055). The two-level EFA model indicated a three-factor solution at the individual level and a one-factor solution at the family level. The three-factor solution at the individual level was consistent with the baseline ClinSeq®model. A two-level CFA model was fit using the EFA results as an initial hypothesis. The resulting CFA model, when fit using the same algorithm that was utilized with the baseline ClinSeq® data, resulted in unstable parameter estimates for the loadings due to a combination of the two-level model, the categorical indicators, the limited sample size and the skewed response distributions. As a result, a Bayesian two-level CFA model was fit in an attempt to overcome the estimation difficulties associated with the more traditional estimation algorithm. The Bayesian model assumes the parameters are random and calculates the posterior distribution of the parameters given the observed data. Confidence intervals for each parameter can then be used to assess the parameter significance. Using MPlus and the MCMC algorithm, after 300,000 iterations it demonstrated convergence. The results of the Bayesian two-level CFA model indicated no significant difference between the observed and replicated model Chi-Square values, and that all the factor loadings were significantly different from zero at the individual level of the model. The Bayesian estimates for the factor loadings at the family level were all insignificant indicating that the higher level of the model was not required. Based on this result, a one-level non-Bayesian model was also fit (ChiSq/df=2.66, CFI=0.99, RMSEA=0.09) yielding positive significant raw and standardized factor loadings as well as positive significant factor correlations. The reliability of all the constructs was greater than 0.85 and the average variance extracted was greater than 0.66 indicating that the model satisfied convergent validity. The constructs also satisfy discriminant validity. These results indicate that the model for the HudsonAlpha CSER Project data demonstrates configural invariance with the model developed for the baseline ClinSeq®data.
The CFA results were used to modify the proposed PUGS scale by eliminating the items “Reassured” and “Discuss Physician.” (denoted with an asterisk in Figure 2). To score the PUGS, the items in each domain were summed and averaged for the three sub-scales. Summing and averaging all items resulted in an overall perceived uncertainty score with high internal consistency (α=0.83).
At the one-month follow up the eight-item PUGS was further modified to reflect that results had been returned. This version of the PUGS can be found in Figure 2.
Perceptions of anticipated uncertainties
Baseline responses to the eight-item PUGS among the ClinSeq® cohort were positively skewed with an overall mean score of 3.4 out of 5 (SD=0.58). Higher scores convey greater certainty (i.e., lower uncertainty), and therefore this suggests that the respondents anticipated being somewhat more certain than uncertain. Within the HudsonAlpha cohort, baseline responses to the eight-item PUGS were also positively skewed with a mean of 3.4 out of 5 (SD=0.72).
Perceptions of genome sequencing information in the ClinSeq® cohort
Participants perceived a relatively low degree of ambiguity about sequencing results with a mean of 2.2 out of 5 and SD of 0.60. Their attitudinal ambivalence toward learning sequence results was similarly low with a mean of 1.7 out of 5 and SD of 0.74. As hypothesized, level of perceived uncertainty was positively correlated with both variables. That is, those with the lowest perceived uncertainty (i.e., highest perceived certainty) had the lowest perceived ambiguity regarding sequence information (r=−0.303, p<0.001) and the lowest ambivalence regarding learning it (r=−0.111, p=0.011). Negative correlations reflect the PUGS scoring: higher scores indicate greater certainty, and lower scores greater uncertainty.
Ambiguity aversion was normally distributed with a mean of 2.5 out of 5 and a SD of 0.65. Perceptions of uncertainty were negatively correlated with ambiguity aversion as we anticipated. Those who were more ambiguity-averse had lower perceptions of uncertainty (i.e., higher perceptions of certainty; r=−0.093, p=0.033).
Perceptions of post-result uncertainties in the ClinSeq® cohort
At the one-month follow-up, 175 ClinSeq® participants’ PUGS responses were skewed toward certainty with a mean of 4.1 out of 5 and SD of 0.77. The ClinSeq® participants similarly reported experiencing relatively little uncertainty with a mean on the MICRA uncertainty subscale of 0.4 out of 5 and SD of 0.49. We anticipated a strong association between the PUGS and MICRA scores as they measure similar constructs. Indeed, perceived uncertainty as measured by the PUGS following receipt of results was positively correlated with the MICRA uncertainty subscale (r=−0.335, p<0.001). Negative correlations reflect the PUGS scoring: higher scores indicate greater certainty, and lower scores greater uncertainty.
Discussion
Due to the broad anticipated scope of uncertainties in genome sequencing and the increasing and novel nature of clinical genome sequencing, objective assessment of perceptions of uncertainty is critically important. The PUGS scale provides a new means of accomplishing this task. Ascertaining three primary domains of uncertainties judged to be important by respondents anticipating receipt of sequencing results, the PUGS scale shows promising evidence of reliability and validity.
We conclude that the PUGS scale is a useful tool for assessing patients’ perceptions of uncertainties regarding the clinical, affective, and evaluative implications of genome sequencing results. The reliability (factor analysis and Cronbach’s alphas) and validity (convergent and known-groups) data support our conclusion that the PUGS scale provides a respectable measure of perceived uncertainties related to genome sequencing. The high correlation between data from the PUGS and the MICRA uncertainty subscale administered simultaneously at one month following receipt of carrier results suggests the scale has promise as an assessment of post-result perceptions of uncertainty. Additional use of the scale will provide data for further psychometric, and concurrent and discriminant validity testing.
The PUGS was designed to capture the dynamic state of perceived uncertainties associated with specific genome sequencing results and in various medical contexts including, but not limited to, results that are diagnostic or secondary and unexpected. The CFA indicates that the same factor structure was appropriate across these distinct applications for the PUGS in two samples that differ substantially in intention (to return results generally to adults and to identify causes of developmental conditions in children) and in formal education of respondents. And while it remains for future investigation to determine its validity in the context of other types of results and the generalizability of the factor weights, the data presented here suggest its utility is unlikely to be contingent upon testing indication or socioeconomic status.
Comparing anticipated perceptions of uncertainties to post hoc perceptions once patients have received results will indicate how well anticipated uncertainties predict perceptions of uncertainties following receipt of results. The PUGS will be useful in longitudinal studies that assess change in perceptions over time and in studies aimed at assessing the effectiveness of interventions to help patients achieve realistic perceptions of uncertainties.
Limitations
The ClinSeq® cohort is highly educated, and primarily of European origin.10 Yet the HudsonAlpha CSER Project data come from a population lower in formal education, suggesting that the PUGS may be used to similar effect with more diverse populations. We are administering the PUGS to other diverse populations undergoing sequencing, which will facilitate further assessment of its psychometric properties. It should be noted that the Hudson Alpha CFA may have been impacted by the relatively small sample size given the estimation requirements of the polychoric correlations and other model parameters. Consequently, despite the fact that the RMSEA of 0.09 was slightly higher than 0.08 (a generally accepted standard for CFA analysis), the final CFA model reported for thse data was deemed acceptable given that the other model fit measures met acceptable standards and that there were no model deficiencies observed in the model diagnostics.
While we believe generating candidate items for development of this scale directly from the end users is a strength in representing their perceptions rather than those determined by clinical experts, we also acknowledge the limitations of relying on the input of one genome sequencing cohort with high formal education. In particular, it remains for future investigation in novel settings to determine the extent of generalizability; mixed-methods studies could serve to ensure that relevant uncertainties are maximally captured. To date the personal dimension of the taxonomy of medical uncertainties–that is, psychosocial and existential factors–remains incompletely delineated and should be addressed in future efforts to measure uncertainties in genome sequencing.
Conclusions
Our earlier data suggested that expectations for benefit among early adopters of genome sequencing were high and may have been unrealistic.2,11 It behooves us as practitioners to help our patients appreciate the practical uncertainties that may accompany the receipt of variants. We hypothesize that a more realistic pretest understanding of the practical uncertainties of genomic testing will allow patients to better calibrate their expectations and respond to their test results. Genetic counseling that helps patients understand the specific issues of uncertainty in genome testing can be an important resource for both untested patients at the time of consent, and tested patients during the disclosure of results. Evidence from studies using the PUGS scale may help counselors explore clients’ areas of greatest uncertainty, and suggest targeted resources to help them better manage these uncertainties following receipt of results. Given the results described here, the PUGS scale exhibits promising properties of a valid and reliable scale for assessing perceived uncertainties about results from genome sequencing.
Acknowledgments
The authors thank Drs. Gillian Hooker, Jennifer Taber, and Holly Peay for their contributions to development of the PUGS scale and comments on an earlier version of the manuscript. We also thank Cris Price for statistical consultation and comments on an earlier version of this paper. The Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health funded this study through grant HG200317-12.
Footnotes
Conflict of Interest Statements:
Barbara Biesecker, Kyle Brothers, Paul Han, Bill Klein, Katie Lewis, Kendall Umstead and Samuel Woolford have no conflicts to declare. Leslie Biesecker receives royalties from Genentech Corp., is an unpaid advisor to Illumina Corp. and receives honoraria from Wiley-Blackwell Inc.
References
- 1.Han PK, Klein WM, Arora NK. Varieties of uncertainty in health care: A conceptual taxonomy. Med Decis Making. 2011;31:828–838. doi: 10.1177/0272989X11393976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biesecker BB, Klein WM, Lewis K, et al. How do research participants perceive “uncertainty” in genome sequencing? Genet Med. 2014;16:977–980. doi: 10.1038/gim.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray SW, Martins Y, Feuerman LZ, et al. Social and behavioral research in genomic sequencing: Approaches from the Clinical Sequencing Exploratory Research Consortium Outcomes and Measures Working Group. Genet Med. 2014;16:727–735. doi: 10.1038/gim.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishel MH. Reconceptualization of the uncertainty in illness theory. Image J Nurs Sch. 1990;22:256–262. doi: 10.1111/j.1547-5069.1990.tb00225.x. [DOI] [PubMed] [Google Scholar]
- 5.Mishel MH, Padilla G, Grant M, et al. Uncertainty in illness theory: A replication of the mediating effects of mastery and coping. Nurs Res. 1991;40:236–240. [PubMed] [Google Scholar]
- 6.Cella D1, Hughes C, Peterman A, Chang CH, Peshkin BN, Schwartz MD, Wenzel L, Lemke A, Marcus AC, Lerman C. A brief assessment of concerns associated with genetic testing for cancer: the Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire. Health Psychol. 2002;21:564–572. [PubMed] [Google Scholar]
- 7.Ellsberg D. Risk, ambiguity, and the Savage axioms. Q J Econ. 1961;75:643–669. [Google Scholar]
- 8.Conner M, Sparks P. Ambivalence and attitudes. Eur Rev Soc Psychol. 2002;12:37–70. [Google Scholar]
- 9.Camerer CF, Weber M. Recent Developments in Modelling Preferences: Uncertainty and Ambiguity. J Risk Uncertain. 1992;5:325–370. [Google Scholar]
- 10.Lewis K, Klein WM, Han PK, et al. Characterizing the ClinSeq cohort. PlosOne. 2015;10(7):e0132690. doi: 10.1371/journal.pone.0132690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Facio FM, Brooks S, Loewenstein J, et al. Motivators for participation in a whole- genome sequencing study: Implications for translational genomics research. Eur J Hum Genet. 2011;19:1213–1217. doi: 10.1038/ejhg.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biesecker LG, Mullikin JC, Facio FM, et al. The ClinSeq Project: Piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19:1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han PK, Reeve BB, Moser RP, et al. Aversion to ambiguity regarding medical tests and treatments: Measurement, prevalence, and relationship to sociodemographic factors. J Health Commun. 2009;14:556–572. doi: 10.1080/10810730903089630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brothers KB, East KM, Kelley WV, et al. Eliciting Preferences on Secondary Findings: The Preferences Instrument for Genomic Secondary Results (PIGSR) Genet Med. doi: 10.1038/gim.2016.110. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]