Abstract
We report a case of premature chromatid separation/mosaic variegated aneuploidy syndrome identified by microcephaly on fetal ultrasound and confirmed by cytogenetic analysis of amniotic fluid. Initial mutational analysis of the entire coding region of the BUB1B gene failed to identify any causative mutations. However, further analysis revealed a known compound heterozygous mutation in the upstream region of this gene and a novel Alu insertion mutation in the intron.
Premature chromatid separation (PCS) is a phenomenon observed as separate and splayed chromatids with discernible centromeres and involves most chromosomes during metaphase. PCS is a genetic trait that is dominantly inherited and can be observed in metaphase chromosomes in a standard cytogenetic examination.1 Since PCS is innocuous and is observed in the normal healthy population, it is occasionally identified as an incidental finding during screenings for other purposes.
The homozygosity of PCS manifests another clinical entity, PCS/MVA syndrome (OMIM #176430, #257300), an autosomal recessive disorder that is characterized by an increased frequency of PCS in >50% metaphase cells and mosaic aneuploidies.1,2 PCS/MVA also manifests a variety of mosaic aneuploidies, especially trisomies, double trisomies and monosomies. Affected patients have clinical characteristics such as growth retardation, microcephaly, cataracts, Dandy–Walker complex, uncontrollable clonic seizures, polycystic kidneys, and a high risk of Wilms tumor and rhabdomyosarcoma.1
PCS is caused by mutations in the BUB1B (budding uninhibited by benzimidazoles 1 homolog beta) gene (encoding BubR1) that is the core component of the mitotic spindle checkpoint for mitotic fidelity and genome stability.2 PCS/MVA syndrome is mostly caused by compound heterozygous mutations in the BUB1B gene. Both parents are often the healthy carrier of a heterozygous PCS trait as they harbor the monoallelic BUB1B mutation.
In this study, we report a case of PCS/MVA syndrome in which the initial mutational analysis of the entire coding region of the BUB1B gene failed to identify any causative mutations. However, further analysis revealed a compound heterozygous known mutation in the upstream region and a novel Alu insertion mutation in the intron of BUB1B.
A 24-year-old G1P1 pregnant female in a nonconsanguineous Japanese couple was referred to our facility because of fetal abnormalities. In the 24th week of gestation, her fetus was found to have a growth restriction with extreme microcephaly (−5.0 s.d.). Screening for an intrauterine infection was negative and no family history was recorded. We performed standard cytogenetic test of the amniotic fluid and analyzed 50 metaphases. The results were 46, XY, inv(9)(p11q13)[11]/47, XY, inv(9)(p11q13), +7[12]/48, XY, inv(9)(p11q13), +3, +17[6]/47, XY, inv(9)(p11q13), +5[4]/48, XY, inv(9)(p11q13), +2, +7[4]/49, XY, inv(9)(p11q13), +3, +17, +18[4]/47, XY, inv(9)(p11q13), +16[2]/others [7]. Besides the well-known but innocuous inv(9)(p11q13) polymorphism, a mosaicism of aneuploidy was observed. A baby boy was born via caesarian section at 38 weeks of gestation. The Apgar score was 8 points for 1 min and 9 points for 5 min after birth. The body weight was 1,934 g (−3.2 s.d.) and the head circumference was 26.7 cm (−4.5 s.d.). He also had congenital cataracts, a prominent nasal bridge, a low-set ear, micrognathia and ambiguous genitalia.
Since PCS/MVA syndrome was suspected, we evaluated the PCS frequencies in blood samples from the infant and his parents. Cord blood or peripheral blood lymphocytes were cultured in RPMI1640 in the presence of PHA-M, arrested with 0.1 μg/ml colcemid for 2 h and treated with 0.075 M KCl at 32 °C for 20 min. Chromosomes were spread onto glass slides and stained with Giemsa solution. Hundreds of metaphases were used to evaluate the frequency of cells with PCS. Whereas the metaphases from the baby showed a 27.2% frequency of PCS, those from his father and mother showed 11.9% and 7.3%, respectively.
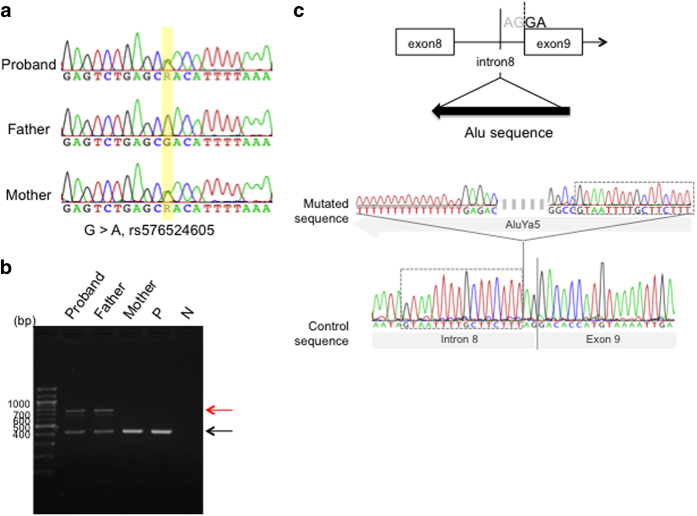
After receiving approval from the Ethics Review Board for Human Genome Studies at Fujita Health University and written informed consent from the patients, we obtained cord blood or peripheral blood lymphocyte samples for genomic analyses. We screened all 23 coding exons of the BUB1B gene baby and parents by means of PCR direct sequencing. However, we failed to detect any mutations within the coding region. Recently, a G>A mutation located 44 kb upstream of BUB1B (rs576524605) was identified among Japanese PCS cases with only one mutation detected in the coding region.3 We therefore examined the upstream region of BUB1B using the Sanger method. The expected G>A mutation was identified both in the baby and the unaffected mother (Figure 1a).
Figure 1.
Genomic sequence of the BUB1B gene. (a) Sequence of the 44 kb upstream region of the BUB1B gene. A heterozygous G>A mutation (rs576524605) was detected in the current study patient and mother, but not in the father. (b) Agarose gel electrophoresis of PCR products that were amplified with primers for intron 8 and exon 9 of BUB1B. A ~400 bp normal band is indicated with a black arrow and a ~700 bp aberrant band with a red arrow. (c) Sequence analysis of BUB1B PCR products from the baby and parents. The upper panel is a schematic illustration of the region around the boundary between intron 8 and exon 9. Arrowheads indicate the PCR primers used. An Alu element was found to be inserted just before the AG consensus dinucleotide at the splicing acceptor in the proband and father. The lower panel shows the sequence around the inserted element. Target site duplications are indicated by dashed lined boxes.
When we further surveyed the coding mutation in exon 9 of BUB1B, we noticed an extra larger sized PCR product in both the baby and unaffected farther (Figure 1b). As shown in Figure 1c, sequencing of this larger amplicon revealed a novel insertion mutation, albeit with a 16 nt target site duplication (5′- GTAATTTTGCTTCTTT-3′). The inserted element was an AluYa5 orientated inversely and located at the polypyrimidine tract near the 3′ splice site of intron 8. This insertion was not detected in 150 control samples from a normal Japanese population (data not shown).
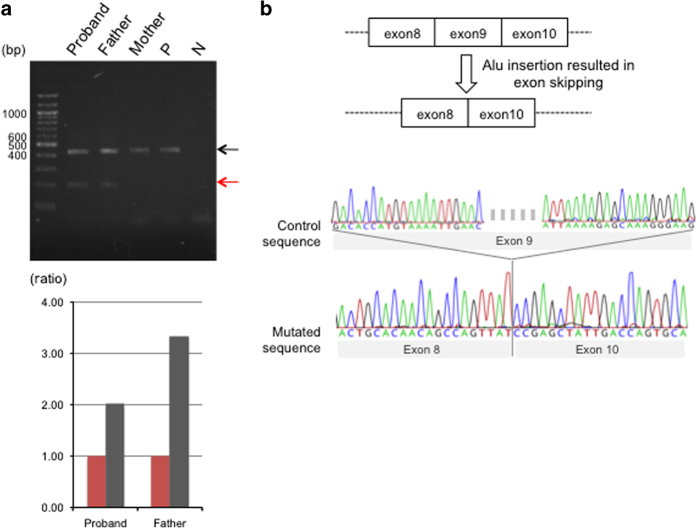
The novel Alu insertion disrupts the upstream region of the 3′ splice site (‘AG’) in the BUB1B gene, suggesting a possible influence on normal splicing. We performed reverse transcription PCR using primers specific for flanking exons 8 and 10. In addition to the product derived via normal splicing, we detected an additional shorter product in both the infant and his father (Figure 2a). Direct sequencing of these reverse transcription PCR products revealed that the smaller product did not include exon 9 of BUB1B, indicating that exon skipping had occurred due to the insertion mutation (Figure 2b). This exon skipping would result in a frameshift that might produce a truncated nonfunctional protein. Further, this analysis also allowed us to separately quantify the transcript level from normal splicing and aberrant splicing using ImageJ software (Bethesda, MD, USA). In the baby, the ratio of the product from normal splicing to aberrant splicing was reduced compared to the father. This suggested that the BUB1B transcript level from the maternal allele was decreased due to the upstream mutation (Figure 2a).
Figure 2.
RNA analysis of the BUB1B gene in the proband and parents. (a) Agarose gel electrophoresis of RT-PCR products of exons 8–10. Larger products (black arrows) were found to be derived from the normal allele and smaller products (red arrows) from the allele with the Alu insertion. The lower panel shows the quantification of these amplified products. Vertical bar indicates the ratio of the product from normal splicing to aberrant splicing (internal control). Red bars indicate the smaller PCR product levels, whereas black bars indicate the relative amounts of the larger amplicons. (b) Sequence analysis of the RT-PCR products. The upper panel displays a predictive structure of the mutant transcript. Arrowheads indicate the position of the PCR primers. The lower panel shows the sequence around the exon–exon boundaries of BUB1B. RT-PCR, reverse transcription PCR.
We have identified a novel Alu insertion mutation in the BUB1B gene in a patient with PCS/MVA syndrome. The insertion of a mobile element is a common pathway for the generation of new mutations. The mutation in our current case resulted in aberrant splicing, possibly leading to the production of a nonfunctional protein. The other allele was found to be a known hypomorphic allele due to a mutation in the upstream region of the BUB1B gene. In mice, complete loss of the BUB1B causes embryonic lethality, whereas decreased expression results in a phenotype similar to PCS/MVA syndrome.4,5 Our current patient might have survived the early embryonic stage due to a small level of BUB1B transcript and then developed PCS/MVA phenotype in the uterus.
PCS/MVA syndrome is a well-known disorder associated with a very high incidence of malignant rhabdomyosarcoma, Wilms tumor and leukemia in childhood.6 It might be important therefore for PCS/MVA patients to avoid antimitotic drugs such as vincristine or taxol that inhibit polymerization of microtubules since this might cause secondary chromosomal abnormalities. Early genetic diagnosis might enable us to select an appropriate treatment for such childhood tumors. Further, this genetic information might give the couple an opportunity to undergo prenatal or preimplantation genetic diagnosis.
Acknowledgments
We thank Narumi Kamiya for technical assistance. This study was supported by a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (15H04710 and 24390085) and that from Ministry of Health, Welfare and Labor (16ek0109067h0003).
Footnotes
The authors declare no conflict of interest.
References
- Tadashi K, Tohru K, Tohru T, Hideo M, Osamu M, Yasuhiko T et al. Mosaic variegated aneuploidy with multiple congenital abnormalities: homozygosity for total premature chromatid separation trait. Am J Med Genet 1998; 78: 245–249. [DOI] [PubMed] [Google Scholar]
- Sandra H, Kim C, Sarah R, Alberto P, Helen F, David F et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet 2004; 36: 1159–1161. [DOI] [PubMed] [Google Scholar]
- Hiroshi O, Tatsuo M, Akinori K, Kosuke H, Tetsushi S, Yoshiki K et al. TALEN-mediated single-base-pair editing identification of an intergenic mutation upstream of BUB1B as causative of PCS (MVA) syndrome. Proc Natl Acad Sci USA 2014; 111: 1461–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Tongyi L, Yuqiang F, Suqing X, Xuan H, Radma M et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood 2004; 103: 1278–1285. [DOI] [PubMed] [Google Scholar]
- Darren JB, Karthik BJ, J Douglas C, Michael T, Subhash J, Alena K et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet 2004; 36: 744–749. [DOI] [PubMed] [Google Scholar]
- Sébastien J, Michelle B, Jean MR, Françoise M, Albert D. High risk of malignancy in mosaic variegated aneuploidy syndrome. Am J Med Genet 2002; 109: 17–21. [DOI] [PubMed] [Google Scholar]
Data Citations
- Kurahashi Hiroki.HGV Database. 2017. 10.6084/m9.figshare.hgv.1364. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Kurahashi Hiroki.HGV Database. 2017. 10.6084/m9.figshare.hgv.1364. [DOI]