Abstract
It is well known that different coral species have different tolerances to thermal or cold stress, which is presumed to be related to the density of Symbiodinium. However, the intrinsic factors between stress-tolerant characteristics and coral-associated bacteria are rarely studied. In this study, 16 massive coral and 9 branching coral colonies from 6 families, 10 genera, and 18 species were collected at the same time and location (Xinyi Reef) in the South China Sea to investigate the bacterial communities. The results of an alpha diversity analysis showed that bacterial diversities associated with massive corals were generally higher than those with branching corals at different taxonomic levels (phylum, class, order, and so on). In addition, hierarchical clustering tree and PCoA analyses showed that coral species were clustered into two large groups according to the similarity of bacterial communities. Group I consisted of massive Goniastrea, Plesiastrea, Leptastrea, Platygyra, Echinopora, Porites, and Leptoria, and group II consisted of branching Acropora and Pocillopora. These findings suggested that both massive corals and branching corals have their own preference for the choice of associated bacteria, which may be involved in observed differences in thermal/cold tolerances. Further analysis found that 55 bacterial phyla, including 43 formally described phyla and 12 candidate phyla, were detected in these coral species. Among them, 52 phyla were recovered from the massive coral group, and 46 phyla were recovered from the branching coral group. Formally described coral pathogens have not been detected in these coral species, suggesting that they are less likely to be threatened by disease in this geographic area. This study highlights a clear relationship between the high complexity of bacterial community associated with coral, skeletal morphology of coral and potentially tolerances to thermal or cold stress.
Keywords: coral-associated bacteria, coral bleaching, thermal or cold stress susceptibility, skeletal morphology, South China Sea
Introduction
Coral-associated microorganisms, including bacteria, fungi, Archaea, dinoflagellates, eukaryotic viruses and phage (Wegley et al., 2007), play significant roles in the biogeochemical cycle, material transformation and maintaining health of coral reef ecosystems (Lesser et al., 2004; Fiore et al., 2010; Mao-Jones et al., 2010; Mahmoud and Kalendar, 2016). Among them, bacteria are the most complex group in terms of species, function, and variability (Wegley et al., 2007; Li et al., 2013; Glasl et al., 2016). Recently, studies have shown that the diversities of bacterial communities associated with corals are extremely high (Li et al., 2013), and significantly affected by the factors including species (Hong et al., 2009), geography (McKew et al., 2012), season (Chen et al., 2011; Li et al., 2014), and more (Bourne et al., 2008; Ceh et al., 2012). For example, the communities of coral-associated bacteria from the Caribbean were significantly more diverse than their Indonesian counterparts and some of the coral host-specific communities (e.g., Clostridiales associated with Acropora spp.) were distinct (McKew et al., 2012). Bacterial communities associated with Acropora tenuis, Tubastrea faulkneri, and Pocillopora damicornis were also analyzed before and after mass spawning on Ningaloo Reef in Western Australia. The results indicated that bacterial diversity increased for all coral species after spawning; some bacterial groups (e.g., Roseobacter, Erythrobacter, and Alteromonadales) that may play an important role in coral reproduction were found to be prominent in these coral species (Ceh et al., 2012). Bacterial communities associated with Porites lutea from Luhuitou fringing reef located in Sanya, southern Hainan Island, northern South China Sea, had a much more dynamic seasonal response and were also significantly different between mucus, tissue and skeleton (Li et al., 2014). In addition, Meron et al. (2011) further showed that ocean acidification would affect the community structure of coral-associated bacteria, and some potential pathogens, such as Vibrionaceae and Alteromonadaceae, would become active with an increasing abundance.
With the development of DNA sequencing technology, an increasing number of bacterial phyla (including some candidate phyla) associated with corals have been identified. Overall, the dominant bacterial phyla were similar among different coral species, of which Proteobacteria, Bacteroidetes, Firmicutes, Cyanobacteria, and Chloroflexi were the most predominant bacterial groups with an abundance of over 90% (Meron et al., 2011; Li et al., 2014). In addition, many candidate phyla (WS3, BRC1, OD1, and so on) were reported in corals for the first time (Lee et al., 2012; Li et al., 2013), and the number of associated bacteria from one coral sample could reach more than 2000 on the species or OUT level (Sunagawa et al., 2010; Li et al., 2014). However, a comparative analysis of bacterial diversities associated with corals is lacking. So the characteristics of coral-associated bacteria in different coral species are poorly understood.
Coral thermal bleaching events occur frequently and have become more and more serious with global warming and frequent El Niño phenomena (Glynn, 1993; Edwards et al., 2001; Berkelmans et al., 2004; McDermott, 2016). However, different coral species have different tolerances to thermal stress. In 1998, large-scale coral colonies had been investigated on reefs fringing inshore islands on the Great Barrier Reef during a large-scale coral bleaching event. A detailed analysis showed that most of the branching corals (BC), such as Acroporidae, Pocilloporidae, and Milleporidae (Isopora, Pocillopora damicornis, Stylophora pistillata, and so on), were highly susceptible, while massive corals (MC) such as Faviidae, Fungiidae, Oculinidae, and so on (e.g., Cyphastrea, Turbinaria, and Galaxea) were relatively unaffected by bleaching stress (Marshall and Baird, 2000). The data on coral bleaching, subsequent mortality and community succession were collected from 1990 to 1998 on artificial and natural reefs in the Maldives (Edwards et al., 2001). Owing to sea surface temperature (SST) anomalies, approximately 98% of branching Pocilloporidae and Acroporidae on artificial structures deployed on a reef flat in 1990 died, whereas the most of massive Poritidae, Agariciidae, and Faviidae survived the bleaching event. The coral communities contained 95% BC and 5% MC before a bleaching event of the artificial reefs in 1994, and the post-bleaching communities consisted of 3% BC and 97% MC. These results were also confirmed in a laboratory simulation experiment. For example, the response of nine species of stony coral (Acropora spp., Pocillopora damicornis, Pavona decussate, P. lutea, and Montipora sp.) from Luhuitou fringing reef to high temperature treatments (ranging from 26 to 32°C) was surveyed (Li et al., 2008). The results showed that BC of Acropora spp. and P. damicornis were most susceptible to bleaching, while the MC P. decussate, P. lutea, and Montipora sp. had better endurance to high temperature (32°C).
In contrast to coral thermal bleaching, cold bleaching will occur when the SST is abnormally low (Saxby et al., 2003; Hoegh-Guldberg and Fine, 2004; Yu et al., 2004; Yu, 2012). In this case, MC also show stronger tolerance to cold stress than BC. In 2003, large-scale branching Acroporidae exposed at low tide bleached and died because of extremely cold temperatures (12°C) at Heron Island, on the southern end of the Great Barrier Reef (Hoegh-Guldberg and Fine, 2004). In laboratory experiments, the tolerance and response of different coral species in low water temperature (ranged from 24 to 14°C) showed similar results: (Li et al., 2009) the branching Acropora spp. died most easily, while massive P. lutea and P. decussate had a significantly higher tolerance of low water temperature.
It is well known that coral-associated bacteria play an important role in the coral holobiont, and show extremely high diversity. Furthermore, different coral species have different tolerances to thermal or cold stress. However, the relationship between coral-associated bacteria and coral susceptibility to thermal or cold stress are currently unclear. Some researchers think that coral bleaching tolerance is related to Symbiodinium density because previous studies had revealed that MC generally had a higher algal density than the BC. In addition, the coral-associated bacterial communities were structured by multiple factors at different scales. Coral bleaching events caused by high or low temperature have occurred frequently in the South China Sea (Yu, 2012), which is a natural laboratory for studying the response of coral reefs to extreme climate events as well as evolutionary characteristics of coral. In this study, a certain amount of scale massive and BC were used to analyze bacterial diversity; the corals were collected at the same time and location (Xinyi Reef of the South China Sea), to control for the influences of geography, season, and some environmental factors. The main purpose is to explore the characteristics of bacterial diversities associated with different coral species, and further explore the relationship between coral-associated bacteria and coral susceptibility to thermal or cold stress.
Materials and Methods
Study Site, Sample Collection, and Species Identification
Our study site is located at Xinyi Reef of Nansha Islands, in the South China Sea. In May 2015, coral samples were collected using a hammer and chisel within 100 m and at a depth of 9–10 m of site A (9°20′6″ N, 115°55′49″ E) at Xinyi Reef (Figure 1). The SST, pH value and salinity were approximately 31.2°C, 8.21 and 33.1, respectively. Three replicated samples (approximately 8 cm × 8 cm) per coral species were collected from the side of the colonies. The distance between two repeated samples is above 10 m. As many different coral species as possible were collected. The collected samples were washed with sterile seawater and then placed in sterile plastic bags. All samples were briefly stored at low temperatures (0–4°C) and then immediately transported back to the laboratory for DNA extraction.
FIGURE 1.
Location map of sampling site A at Xinyi Reef, South China Sea. (A) Map of the whole South China Sea with labeled coral reef area. The Xinyi Reef is marked by a box. (B) The Xinyi Reef is magnified showing sampling site A (9°20′6″ N, 115°55′49″ E). The map was constructed using software ArcGIS (ver. 10.1). The offshore reef area was drawn using remote sensing images (fused by landsat 8 multispectral bands and panchromatic band) with resolution of 15 m.
Coral tissue was removed using airbrush and coral skeleton was immediately ready for each species identification. All coral samples were identified according to their ecological and morphological characteristics.
DNA Extraction, PCR Amplification, and Illumina MiSeq Sequencing
Small pieces of coral sample including tissue, mucus and skeleton (approximately 50 mg), cut with a pair of scissors, were used to extract genomic DNA using TIANamp Marine Animals DNA Kit [Tiangen Biotech (Beijing) Co., Ltd., Beijing, China] according to the manufacturer’s instructions. The V3–V4 region of bacterial 16S rRNA gene was amplified using the bacterial-specific forward primer 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′), where barcode is an eight-base sequence unique to each sample (Mori et al., 2014; Xu et al., 2016). The reaction system and procedure of PCR using a ABI GeneAmp® 9700 thermal cycler are the same as previously described (Sun et al., 2014). Triplicate PCR products were pooled for each sample, and then fragments with size in the range of 421–460 bp were purified and quantified using the AxyPrep DNA gel extraction kit (Axygen Biosciences, Union City, CA, United States) and QuantiFluorTM-ST Fluorescence quantitative system (Promega, United States). Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 250) on an Illumina MiSeq platform according to the standard protocols (Majorbio Bio-Pharm Technology Co. Ltd., Shanghai, China). The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Submission Number: SUB2295385).
Data Analysis
Raw sequences were optimized using the software platform Trimmomatic to exclude the reads with homopolymer inserts and low quality scores (<20) (Bolger et al., 2014). The remaining high-quality reads were used for taxonomic analysis. After removing the chimeric sequences, all reads were identified and classified using the Ribosomal Database Project (RDP) by setting a bootstrap confidence level of 70% to cluster into operational taxonomic units (OTUs) at a 97% identity threshold and obtain the representative sequence for each OTU (Edgar, 2010). Based on the OTUs cluster analysis, a series of statistical analysis indices, including Coverage, ACE estimator, Shannon index, and so on, were calculated for each sample using the software Mothur (version v.1.30.1) (Schloss et al., 2011). Taxonomy was aligned and compared with the SILVA database1 (Release 123) (Quast et al., 2013) using the Qiime platform2. The beta diversity analysis was executed using the principal co-ordinates analysis (PCoA) at the OTU level.
Results
Identification of Coral Species and Skeletal Morphology Analysis
A total of 25 scleractinian coral samples were selected to test for symbiotic bacteria. These coral samples were identified as belonging to 6 families, 10 genera, and 18 species based on their ecological and morphological characteristics (Table 1). They were then divided into two groups according to skeletal morphology: 16 MC and 9 BC. Massive corals included Faviidae, Poritidae, Fungiidae, and Mrulinidae, while BC included Acroporidae and Pocilloporidae.
Table 1.
Coral species identification.
| Index | Family | Genus | Species | |
|---|---|---|---|---|
| Massive coral | M01 | Faviidae | Goniastrea | Goniastrea edwardsi |
| M02 | Faviidae | Plesiastrea | Plesiastrea curta | |
| M03 | Faviidae | Leptastrea | Leptastrea bottae | |
| M04 | Faviidae | Leptastrea | Leptastrea bottae | |
| M05 | Faviidae | Leptastrea | Leptastrea sp. | |
| M06 | Faviidae | Platygyra | Platygyra daedalea | |
| M07 | Faviidae | Platygyra | Platygyra crosslandi | |
| M08 | Faviidae | Platygyra | Platygyra crosslandi | |
| M09 | Faviidae | Echinopora | Echinopora gemmacea | |
| M10 | Poritidae | Porites | Porites rus | |
| M11 | Poritidae | Porites | Porites rus | |
| M12 | Poritidae | Porites | Porites lutea | |
| M13 | Poritidae | Porites | Porites lutea | |
| M14 | Fungiidae | Fungia | Fungia scutaria | |
| M15 | Fungiidae | Fungia | Fungia fungites | |
| M16 | Mrulinidae | Leptoria | Leptoria phrygia | |
| Branching coral | B01 | Acroporidae | Acropora | Acropora brueggemanni |
| B02 | Acroporidae | Acropora | Acropora brueggemanni | |
| B03 | Acroporidae | Acropora | Acropora humilis | |
| B04 | Acroporidae | Acropora | Acropora sp. | |
| B05 | Pocilloporidae | Pocillopora | Pocillopora eydouxi | |
| B06 | Pocilloporidae | Pocillopora | Pocillopora eydouxi | |
| B07 | Pocilloporidae | Pocillopora | Pocillopora eydouxi | |
| B08 | Pocilloporidae | Pocillopora | Pocillopora sp. | |
| B09 | Pocilloporidae | Pocillopora | Pocillopora verrucosa | |
Diversity of Coral-Associated Bacteria
The number of recovered reads was no less than 30,460 for each sample after quality filtering; these were clustered into different 97% OTUs. The length of those sequences ranged from 421 to 460 bp. The Good’s coverage of each sample library was more than 99%. These sequencing results thus represented the true condition of the bacteria in the sample. Other statistical analysis indices including ACE, Chao, and Shannon are listed in Table 2. These indices showed an obvious difference between MC and BC, with community diversity higher in MC than that in the BC. The number of OTUs was also generally higher in MC than branching. Among them, there are six samples with greater than 1000 OTUs including M02, M05, etc. Contrarily, the number of OTUs was lower than 700 from all the BC (Table 2). The Shannon indices are similar in all MC but higher than in BC. This means that the diversity and equitability of bacteria associated with MC are higher than BC. However, they also show that a few MC are relatively low (1.91 in M04, 3.53 in M06, and 3.17 in M16) compared to the other massive samples (5.81 in M02, 5.88 in M09, etc.). The value of the Shannon index in BC ranged from 2.28 to 4.89.
Table 2.
Numbers of sequences and operational taxonomic units (OTUs) (97%) and diversity estimates of bacteria associated with different morphologic corals.
| Index | No. of Seq | OTUs | ACE | Chao | Coverage | Shannon | |
|---|---|---|---|---|---|---|---|
| Massive coral | M01 | 41680 | 766 | 794.87 | 836.04 | 0.998412 | 5.21 |
| M02 | 35444 | 1416 | 1500.66 | 1543.55 | 0.994396 | 5.81 | |
| M03 | 31922 | 624 | 659.02 | 681.78 | 0.997869 | 4.68 | |
| M04 | 39438 | 410 | 422.98 | 430.32 | 0.999059 | 1.91 | |
| M05 | 39070 | 1277 | 1345.71 | 1361.36 | 0.995907 | 5.63 | |
| M06 | 33930 | 729 | 758.92 | 760.36 | 0.997774 | 3.53 | |
| M07 | 36843 | 965 | 1044.91 | 1056.97 | 0.996263 | 5.28 | |
| M08 | 43063 | 639 | 659.84 | 669.75 | 0.998929 | 5.17 | |
| M09 | 42064 | 1262 | 1320.19 | 1360.11 | 0.996746 | 5.88 | |
| M10 | 33750 | 1274 | 1352.08 | 1389.64 | 0.994473 | 5.64 | |
| M11 | 40612 | 568 | 581.87 | 587.50 | 0.999298 | 5.21 | |
| M12 | 41705 | 1186 | 1410.66 | 1443.40 | 0.99196 | 5.71 | |
| M13 | 31995 | 1090 | 1258.90 | 1274.29 | 0.992344 | 5.59 | |
| M14 | 37203 | 544 | 557.94 | 569.38 | 0.999094 | 4.97 | |
| M15 | 39468 | 955 | 1021.05 | 1038.38 | 0.996649 | 5.45 | |
| M16 | 31134 | 492 | 539.49 | 547.86 | 0.997282 | 3.17 | |
| Branching coral | B01 | 30460 | 644 | 682.26 | 693.52 | 0.997745 | 4.81 |
| B02 | 34571 | 643 | 668.94 | 694.04 | 0.998468 | 4.89 | |
| B03 | 35185 | 451 | 466.51 | 476.62 | 0.998879 | 3.85 | |
| B04 | 40201 | 497 | 515.65 | 517.50 | 0.998926 | 3.50 | |
| B05 | 34747 | 430 | 466.22 | 475.64 | 0.997978 | 2.28 | |
| B06 | 31229 | 689 | 709.98 | 717.68 | 0.998239 | 4.58 | |
| B07 | 39195 | 321 | 332.22 | 334.59 | 0.999414 | 3.82 | |
| B08 | 44505 | 498 | 513.35 | 527.06 | 0.999273 | 4.52 | |
| B09 | 35440 | 680 | 884.31 | 870.41 | 0.993343 | 2.77 | |
Bacterial Community Composition
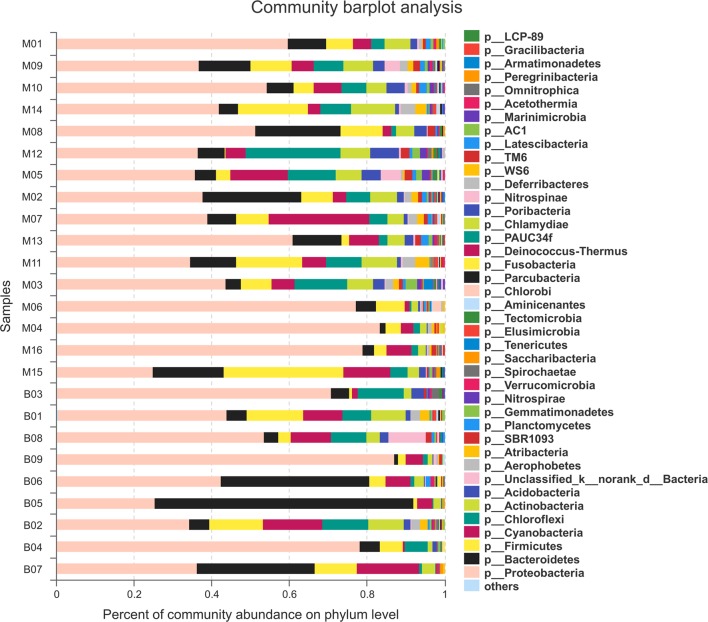
Fifty-five bacterial phyla, including 43 formally described bacterial phyla and 12 candidate phyla, were recovered from the 25 coral samples (Figure 2). Among them, 43 bacterial phyla were present in both MC and BC groups. Nine bacterial phyla (including five candidate phyla) were unique groups to the MC sample libraries and 3 (including 2 candidate phyla) to BC, respectively (Supplementary Figure S1). According to the average value of abundance for each bacterial group, the dominant bacteria were similar to those previously reported, of which Proteobacteria (52.56%), Bacteroidetes (12.83%), Firmicutes (8.66%), Cyanobacteria (6.75%), and Chloroflexi (5.63%) were predominant in the all of sample libraries. The highest abundance of Proteobacteria reached 87.39, 83.78, and 79.81% in B09, M04. and M16 libraries, respectively. However, Bacteroidetes was the most predominant bacterial phylum in B05 (66.64%), B06 (37.98%), and B07 (30.73%) sample libraries relative to the all MC (ranged from 1.71 to 25.71%) and the other BC (1.11–5.24%). Firmicutes was the most predominant in M15 library (31.34%), though low abundance was detected in the other sample libraries (ranged from 0.67 to 17.89%). Additionally, Cyanobacteria and Chloroflexi were the second dominant bacterial phylum in M07 (25.99%) and M12 (24.40%) libraries, respectively.
FIGURE 2.
Bacterial composition profiles. Taxonomic classification of bacterial reads retrieved from different coral species at phylum level using RDP classifier. “others” represent the bacterial phyla with abundances of less than 0.01%.
At the class level, Gammaproteobacteria and Alphaproteobacteria were the most abundant group in most of the coral species libraries (Supplementary Figure S2). For example, the abundance of Gammaproteobacteria in M04, M06, and M16 could reach 74.42, 46.39, and 46.83%, respectively. Alphaproteobacteria were the most predominant bacterial class, and in M01, M13, and B09 could reach 38.98, 39.07, and 41.44%, respectively. On the other hand, it was unique that Cytophagia was the first dominant group in B05 (64.53%) and B06 (28.25%). Although the bacterial compositions exhibited high complexity at other taxonomic levels (order, family, genus, and species) (Supplementary Figures S3–S6), bacterial diversities associated with MC were generally higher than those in BC (Table 3). However, the complexity of few samples was lower, e.g., M04, B07, B05, and B03 (Table 3 and Supplementary Figures S3–S6).
Table 3.
Numbers of bacteria associated with different skeletal morphologic corals in different taxonomic levels.
| Index | Phylum | Class | Order | Family | Genus | Species | Unclassified species | |
|---|---|---|---|---|---|---|---|---|
| Massive coral | M01 | 26 | 61 | 138 | 222 | 335 | 486 | 280 |
| M02 | 40 | 84 | 182 | 292 | 497 | 751 | 665 | |
| M03 | 31 | 60 | 139 | 204 | 317 | 450 | 174 | |
| M04 | 29 | 54 | 109 | 154 | 247 | 318 | 92 | |
| M05 | 32 | 77 | 173 | 277 | 433 | 676 | 601 | |
| M06 | 26 | 57 | 126 | 207 | 338 | 467 | 262 | |
| M07 | 37 | 77 | 163 | 263 | 427 | 601 | 364 | |
| M08 | 20 | 47 | 107 | 173 | 296 | 408 | 231 | |
| M09 | 38 | 80 | 169 | 286 | 494 | 725 | 537 | |
| M10 | 38 | 83 | 183 | 304 | 488 | 721 | 553 | |
| M11 | 29 | 57 | 129 | 202 | 332 | 412 | 156 | |
| M12 | 26 | 62 | 127 | 192 | 290 | 501 | 685 | |
| M13 | 29 | 66 | 133 | 221 | 354 | 538 | 552 | |
| M14 | 28 | 55 | 125 | 201 | 329 | 414 | 130 | |
| M15 | 28 | 59 | 136 | 235 | 437 | 607 | 348 | |
| M16 | 27 | 55 | 118 | 192 | 311 | 387 | 105 | |
| Branching coral | B01 | 31 | 60 | 135 | 217 | 370 | 479 | 165 |
| B02 | 29 | 54 | 135 | 216 | 351 | 463 | 180 | |
| B03 | 16 | 36 | 90 | 145 | 225 | 304 | 147 | |
| B04 | 21 | 41 | 92 | 165 | 287 | 365 | 132 | |
| B05 | 17 | 36 | 90 | 154 | 261 | 324 | 106 | |
| B06 | 28 | 56 | 122 | 201 | 351 | 476 | 213 | |
| B07 | 17 | 39 | 84 | 139 | 211 | 252 | 69 | |
| B08 | 24 | 46 | 104 | 158 | 236 | 317 | 181 | |
| B09 | 29 | 60 | 136 | 218 | 362 | 498 | 182 | |
There were 532 families detected from the 25 corals samples, including 123 families in MC groups and 21 families in BC groups, as well as 388 families that were shared in both MC and BC groups. Rhodobacteraceae, Hahellaceae, Flammeovirgaceae, and Vibrionaceae were dominant bacteria groups with a higher average abundance of 11.40, 7.23, 6.51, and 3.38% in all coral samples, respectively (Supplementary Figure S7). Among them, Rhodobacteraceae, Vibrionaceae, and Alteromonadaceae were potential coral pathogens and had an average abundance of 11.40, 3.38, and <1.00%, respectively. However, confirmed coral pathogens were not detected from all coral species.
Bacterial Composition Associated with Different Coral Species
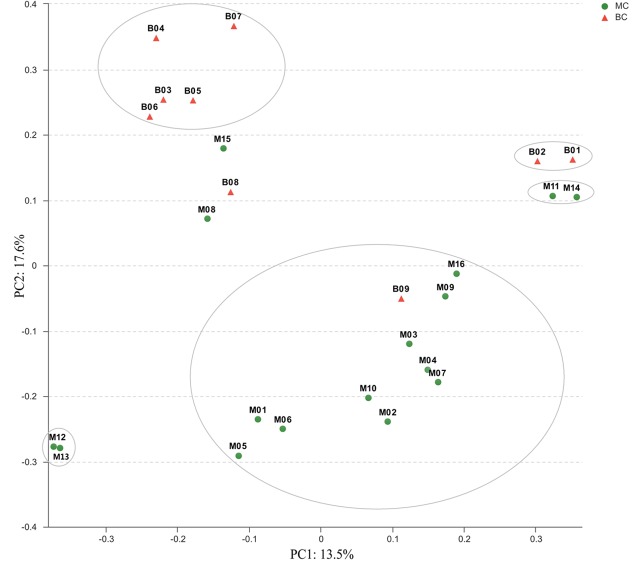
The similarity among the bacterial communities associated with 25 coral samples was evaluated using PCoA and hierarchical clustering analysis on OTU level. The PCoA analysis using the Bray–Curtis metric showed that coral samples with different bacterial communities could roughly cluster into two groups (Figure 3). Group I contained all of the MC species except the M08, M11, M12, M13, M14, and M15 samples. However, M11/M14 and M12/M13 samples were very closely clustered together. Group II mainly contained BC species. Similar results were also confirmed by the Hierarchical clustering trees analysis (Supplementary Figure S8). Clustering and PCoA results indicated that the most significant factor affecting bacterial species composition was skeletal morphology of the coral host.
FIGURE 3.
Principal co-ordinates analysis (PCoA) plot based on the OTU level from 25 coral species. The scatter plot is of principal coordinate 1 (PC1) vs. principal coordinate 2 (PC2). PC1 and PC2 represent the principal factors affecting bacterial composition associated with corals.
Discussion
Coral Species Size and Bacterial Diversity Analysis
Presently, an ever-increasing number of scleractinian corals are being selected for surveys of bacterial diversity associated with coral (Sunagawa et al., 2010; Chen et al., 2011; Ceh et al., 2012; Lee et al., 2012; McKew et al., 2012; Li et al., 2013, 2014). These previously described coral species cover different genera (including Isopora, Pocillopora, Astreopora, Stylophora, Acropora, Porites, Montastraea, Diploria, Gorgonia, Tubastrea, etc.) and were collected from different locations (Caribbean Sea, Red Sea, South China Sea, Great Barrier Reef, Maldives, and so on). These studies indicate that the diversities of coral-associated bacteria were extremely high, and many bacterial phyla (BRC1, OD1, and SR1) were reported in corals for the first time (Li et al., 2013). However, the number of coral species for each study was small (less than 10 species) and the main attention of the bacterial-coral association surveys was focused on species specificity (Hong et al., 2009; Morrow et al., 2012), compartment specificity (Sweet et al., 2011; Li et al., 2014), geographic specificity (McKew et al., 2012), and season specificity (Hong et al., 2009; Chen et al., 2011; Li et al., 2014). In this study, large-scale coral samples involving 18 species, 10 genera, and 6 families were collected to analyze the bacterial communities in the Xinyi Reef of South China Sea (Table 1). These coral species could be clearly divided into two groups according to their skeletal morphology, which were massive and branching (Supplementary Material). Compared with previous studies, more bacterial phyla were detected from single coral libraries in this study. For example, the higher numbers of bacterial phyla associated with massive Plesiastrea curta, Echinopora gemmacea, Porites rus (M10), Platygyra crosslandi (M07), and Leptastrea sp. were 40, 38, 38, 37, and 32, respectively. And the bacterial phyla of other coral species ranged from 16 to 31 (most coral species > 25) (Table 3). While using the same sequencing method, Li et al. (2013) indicated that the bacterial diversity was higher in corals Porites lutea and Galaxea fascicularis from Sanya Bay in South China Sea than those from the other locations. In that study, 23 bacterial phyla were detected, which was higher than in previous studies (bacterial phyla < 20). Altogether, 55 bacterial phyla including 43 formally described bacterial phyla and 12 candidate phyla were recovered from the 25 coral samples in this study. Among them, plenty of bacterial phyla may represent phyla that have not been found in corals previously (Figure 2). Therefore, our study can provide more significant information in coral-associated bacteria based on the advantages of coral species size and bacterial diversities.
The most dominant bacteria were Proteobacteria in most of the coral species, followed by members of the Bacteroidetes, Firmicutes, Cyanobacteria, and Chloroflexi, which was similar to previous studies (Ceh et al., 2012; Lee et al., 2012; Li et al., 2013). The highest abundance of Proteobacteria could reach 87.39, 83.78, and 79.81% in B09, M04, and M16 libraries, respectively. However, Bacteroidetes was the most predominant bacteria in B05 (66.64%), B06 (37.98%), and B07 (30.73%) libraries relative to all the MC (ranged from 1.71 to 25.71%) and the other BC (1.11–5.24%); this comparison was reported for the first time in this study. In addition, Firmicutes was the most predominant in M15 library (31.34%), which was similar to that in the mucus and skeleton of Porites lutea collected in August in the Luhuitou fringing reef of the South China Sea (Li et al., 2014), but Firmicutes was rarely found in other previous studies. Furthermore, 12 candidate phyla, including WS2, RBG-1L, CP-89, and so on, were detected in all coral species. Many novel bacterial species are not annotated.
The Relationship between Bacterial Diversity and Coral Potentially Tolerances to Thermal or Cold Stress
The bacterial diversities associated with corals from MC versus BC groups had significant characteristic differences. The bacterial diversities associated with MC groups were generally higher than those of BC groups at different taxonomic levels (Table 3). For example, the average values of number for bacterial phyla in MC and BC groups were 30 and 24, respectively. In addition, the average OTU level values were 887 and 539, respectively, in MC and BC groups. Although Li et al. (2013) indicated that MC (Porites lutea and Porites lutea) had higher estimated diversities than BC (Acropora millepora), only three coral species were analyzed. Therefore, this study further certified that coral skeletal morphology plays a key role in determining the diversities of bacteria associated with corals by increasing the number of coral species. Meanwhile, the more similar studies from other geographic locations are needed to prove this possible relationship.
This study also showed that the potentially tolerance of scleractinian coral to thermal or cold stress is related to its skeletal morphology. As described in the introduction, MC species have better tolerances to thermal or cold stress than BC species. Combining with the analysis of bacterial diversity in this study, it can be inferred that abundant bacterial species may help corals respond to abnormal changes of temperature. The mechanism is not clear, which may be related to the microbial regulation of coral reef ecosystems. Shinzato et al. (2011) completed the genome sequencing of Acropora digitifera and identified more than 20,000 protein-coding genes. This study not only disclosed the development age of coral, but also found that Acropora might have lost the essential gene of cysteine biosynthesis, thus be metabolically dependent on its symbionts. Such relationship made the coral particularly sensitive to the climate change which endangered its symbiotic. Lin et al. (2015) successfully decoded the genome of Symbiodinium kawagutii and identified protein-coding genes and Symbiodinium-specific gene families. From the genetic evidence, this study strongly demonstrated that S. kawagutii could alter their genetic structure in the history of symbiosis to suit for the environmental changes better. Metagenomic analysis of the microbial community associated with coral also provided some insights into the coral responding to environmental changes (Wegley et al., 2007). Therefore, more related studies in the future will be helpful in unraveling the mechanism of coral susceptibility to thermal or cold stress.
Cluster Analysis Based on the Similarity of Coral-Associated Bacteria
Although the bacterial communities associated with different coral species were very complex, PCoA and hierarchical clustering analysis obviously showed that massive and branching corals could roughly cluster into different groups (Figure 3 and Supplementary Figure S8). Comparing with previous studies, this study improved the number of coral species and classified samples according to their skeletal types, and then implemented clustering analysis based on the similarity of coral-associated bacteria. These results revealed that both MC and BC have their own associated bacteria, which will provide a theoretical basis for future studies of genetic evolution of coral holobionts.
Potential Pathogens in Corals
Over the past 30 years, various coral diseases have caused severe damage to coral reefs worldwide. However, these coral diseases occurred mostly because environmental changes led to colonization and abnormal reproduction of pathogens in coral hosts (Rosenberg and Ben-Haim, 2002; Ben-Haim et al., 2003; Reshef et al., 2006; Rosenberg et al., 2007; Tout et al., 2015). Currently, the confirmed coral pathogens mainly include Vibrio shiloi (Kushmaro et al., 1996), Vibrio coralliilyticus (Ben-Haim et al., 2003), Vibrio carchariae (Ritchie and Smith, 1995), Vibrio alginolyticus (Cervino et al., 2004), Aspergillus sydowii (Geiser et al., 1998), Aurantimonas coralicida (Denner et al., 2003), Thalassomonas loyana (Barash et al., 2005), and Serratia marcescens (Patterson et al., 2002). Fortunately, none of these coral pathogens were detected from 924,854 reads in 25 coral sample libraries in this study. This suggests that these corals are less likely to be threatened by common bacterial diseases in this sea area. Sharon and Rosenberg (2010) indicated that Vibrio species in coral mucus are largely in the viable-but-non-culturable (VBNC) state, and may contribute to the health of corals by preventing infections from pathogens. In our study, the abundance of Vibrionaceae was also found to be high (average abundance of 3.38% in all coral species) (Supplementary Figure S7). This further suggests that Vibrio species are common in coral holobionts.
Conclusion
Our results from this study showed that bacterial diversities associated with MC were generally higher than those associated with BC at different taxonomic levels. Furthermore, many bacterial phyla and unannotated bacterial species were detected in coral sample libraries. Combined with the differences in thermal or cold stress between massive and branching corals, we infer that abundant bacterial species can help corals respond to abnormal changes of temperature. Although the mechanism is not clear, this study provides novel insights into the relationship between the high complexity of bacterial community associated with coral, skeletal morphology of coral and potentially tolerances to thermal or cold stress.
Author Contributions
KY and JL conceived the research, YW, XH, WH, and ZW contributed the materials, JL performed all experiments, ZQ and ZP drawn all pictures, QY and WW identified coral species, JL and KY wrote the manuscript, all authors edited and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the National Basic Research Program of China (2013CB956102), the National Natural Science Foundation of China (91428203 and 41666005), the BaGui Scholars Program Foundation (2014BGXZGX03), and the Natural Science Foundation of Guangdong Province (2014A030310353).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00979/full#supplementary-material
References
- Barash Y., Loya Y., Sulam R., Rosenberg E. (2005). Bacterial Strain BA-3 and a filterable factor cause a white plague-like disease in corals from the Eilat coral reef. Aquat. Microb. Ecol. 40 183–189. 10.3354/ame040183 [DOI] [Google Scholar]
- Ben-Haim Y., Zicherman-Keren M., Rosenberg E. (2003). Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 69 4236–4242. 10.1128/AEM.69.7.4236-4242.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelmans R., De’ath G., Kininmonth S., Skirving W. J. (2004). A comparison of the 1998 and 2002 coral bleaching events on the Great Barrier Reef: spatial correlation, patterns, and predictions. Coral Reefs 23 74–83. 10.1007/s00338-003-0353-y [DOI] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D., Iida Y., Uthicke S., Smith-Keune C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2 350–363. 10.1038/ismej.2007.112 [DOI] [PubMed] [Google Scholar]
- Ceh J., Raina J. B., Soo R. M., Van K. M., Bourne D. G. (2012). Coral-bacterial communities before and after a coral mass spawning event on Ningaloo Reef. PLoS ONE 7:e36920 10.1371/journal.pone.0036920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervino J. M., Hayes R. L., Polson S. W., Polson S. C., Goreau T. J., Martinez R. J., et al. (2004). Relationship of Vibrio species infection and elevated temperatures to yellow blotch/band disease in Caribbean corals. Appl. Environ. Microbiol. 70 6855–6864. 10.1128/AEM.70.11.6855-6864.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-P., Tseng C.-H., Chen C. A., Tang S.-L. (2011). The dynamics of microbial partnerships in the coral Isopora palifera. ISME J. 5 728–740. 10.1038/ismej.2010.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denner E. B., Smith G. W., Busse H. J., Schumann P., Narzt T., Polson S. W., et al. (2003). Aurantimonas coralicida gen. nov., sp. nov., the causative agent of white plague type II on Caribbean scleractinian corals. Int. J. Syst. Evol. Microbiol. 53 1115–1122. 10.1099/ijs.0.02359-0 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edwards A. J., Clark S., Zahir H., Rajasuriya A., Naseer A. (2001). Coral bleaching and mortality on artificial and natural reefs in Maldives in 1998, sea surface temperature anomalies and initial recovery. Mar. Pollut. Bull. 42 7–15. 10.1016/S0025-326X(00)00200-9 [DOI] [PubMed] [Google Scholar]
- Fiore C. L., Jarett J. K., Olson N. D., Lesser M. P. (2010). Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 18 455–463. 10.1016/j.tim.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Geiser D. M., Taylor J. W., RitchieandAmp K. B., Smith G. W. (1998). Cause of sea fan death in the West Indies. Nature 394 137–138. 10.1038/280799671296 [DOI] [Google Scholar]
- Glasl B., Herndl G. J., Frade P. R. (2016). The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 10 2280–2292. 10.1038/ismej.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn P. W. (1993). Coral reef bleaching: ecological perspectives. Coral Reefs 12 1–17. 10.1007/BF00303779 [DOI] [Google Scholar]
- Hoegh-Guldberg O., Fine M. (2004). Low temperatures cause coral bleaching. Coral Reefs 23:444 10.1007/s00338-004-0401-2 [DOI] [Google Scholar]
- Hong M.-J., Yu Y.-T., Chen C. A., Chiang P.-W., Tang S.-L. (2009). Influence of species specificity and other factors on bacteria associated with the coral Stylophora pistillata in Taiwan. Appl. Environ. Microbiol. 75 7797–7806. 10.1128/AEM.01418-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushmaro A., Loya Y., Fine M., Rosenberg E. (1996). Bacterial infection and coral bleaching. Nature 380:396 10.1038/380396a0 [DOI] [Google Scholar]
- Lee O. O., Yang J., Bougouffa S., Wang Y., Batang Z., Tian R., et al. (2012). Spatial and species variations in bacterial communities associated with corals from the Red Sea as revealed by pyrosequencing. Appl. Environ. Microbiol. 78 7173–7184. 10.1128/AEM.01111-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser M. P., Mazel C. H., Gorbunov M. Y., Falkowski P. G. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305 997–1000. 10.1126/science.1099128 [DOI] [PubMed] [Google Scholar]
- Li J., Chen Q., Long L.-J., Dong J.-D., Yang J., Zhang S. (2014). Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci. Rep. 4:7320 10.1038/srep07320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen Q., Zhang S., Huang H., Yang J., Tian X.-P., et al. (2013). Highly heterogeneous bacterial communities associated with the South China Sea reef corals Porites lutea, Galaxea fascicularis and Acropora millepora. PLoS ONE 8:e71301 10.1371/journal.pone.0071301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Yu K., Shi Q., Chen T., Zhao M. (2009). Low water temperature tolerance and responding mode of scleractinian corals in Sanya Bay. Chin. J. Appl. Ecol. 20 2289–2295. [PubMed] [Google Scholar]
- Li S., Yu K., Shi Q., Chen T., Zhao M., Yan H. (2008). Experimental study of stony coral response to the high temperature in Luhuitou of Hainan Island. Trop. Geogr. 28 534–539. [Google Scholar]
- Lin S., Cheng S., Song B., Zhong X., Lin X., Li W., et al. (2015). The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350 691–694. 10.1126/science.aad0408 [DOI] [PubMed] [Google Scholar]
- Mahmoud H. M., Kalendar A. A. (2016). Coral-associated actinobacteria: diversity, abundance, and biotechnological potentials. Front. Microbiol. 7:204 10.3389/fmicb.2016.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao-Jones J., Ritchie K. B., Jones L. E., Ellner S. P. (2010). How microbial community composition regulates coral disease development. PLoS Biol. 8:e1000345 10.1371/journal.pbio.1000345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall P. A., Baird A. H. (2000). Bleaching of corals on the Great Barrier Reef: differential susceptibilities among taxa. Coral Reefs 19 155–163. 10.1007/s003380000086 [DOI] [Google Scholar]
- McDermott A. (2016). Coral bleaching event is longest on record. Science News, June 22, 2016. [Google Scholar]
- McKew B. A., Dumbrell A. J., Daud S. D., Hepburn L., Thorpe E., Mogensen L., et al. (2012). Characterization of geographically distinct bacterial communities associated with coral mucus produced by Acropora spp. and Porites spp. Appl. Environ. Microbiol. 78 5229–5237. 10.1128/AEM.07764-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meron D., Atias E., Kruh L. I., Elifantz H., Minz D., Fine M., et al. (2011). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J. 5 51–60. 10.1038/ismej.2010.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H., Maruyama F., Kato H., Toyoda A., Dozono A., Ohtsubo Y., et al. (2014). Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 21 217–227. 10.1093/dnares/dst052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow K. M., Moss A. G., Chadwick N. E., Liles M. R. (2012). Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl. Environ. Microbiol. 78 6438–6449. 10.1128/AEM.01162-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K. L., Porter J. W., Ritchie K. B., Polson S. W., Mueller E., Peters E. C., et al. (2002). The etiology of white pox, a lethal disease of the Caribbean elkhorn coral, Acropora palmata. Proc. Natl. Acad. Sci. U.S.A. 99 8725–8730. 10.1073/pnas.092260099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41 D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef L., Koren O., Loya Y., Zilber-Rosenberg I., Rosenberg E. (2006). The coral probiotic hypothesis. Environ. Microbiol. 8 2068–2073. 10.1111/j.1462-2920.2006.01148.x [DOI] [PubMed] [Google Scholar]
- Ritchie K. B., Smith G. W. (1995). Preferential carbon utilization by surface bacterial communities from water mass, normal, and white-band diseased Acropora cervicornis. Mol. Mar. Biol. Biotechnol. 4 345–352. [Google Scholar]
- Rosenberg E., Ben-Haim Y. (2002). Microbial diseases of corals and global warming. Environ. Microbiol. 4 318–326. 10.1046/j.1462-2920.2002.00302.x [DOI] [PubMed] [Google Scholar]
- Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5 355–362. 10.1038/nrmicro1635 [DOI] [PubMed] [Google Scholar]
- Saxby T. A., Dennison W. C., Hoegh-Guldberg O. (2003). Photosynthetic responses of the coral Montipora digitata to cold temperature stress. Mar. Ecol. Prog. Ser. 248 85–97. 10.3354/meps248085 [DOI] [Google Scholar]
- Schloss P. D., Gevers D., Westcott S. L. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6:e27310 10.1371/journal.pone.0027310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G., Rosenberg E. (2010). Healthy corals maintain Vibrio in the VBNC state. Environ. Microbiol. Rep. 2 116–119. 10.1111/j.1758-2229.2009.00113.x [DOI] [PubMed] [Google Scholar]
- Shinzato C., Shoguchi E., Kawashima T., Hamada M., Hisata K., Tanaka M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476 320–323. 10.1038/nature10249 [DOI] [PubMed] [Google Scholar]
- Sun Z., Li G., Wang C., Jing Y., Zhu Y., Zhang S., et al. (2014). Community dynamics of prokaryotic and eukaryotic microbes in an estuary reservoir. Sci. Rep. 4:6966 10.1038/srep06966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S., Woodley C. M., Medina M. (2010). Threatened corals provide underexplored microbial habitats. PLoS ONE 5:e9554 10.1371/journal.pone.0009554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet M. J., Croquer A., Bythell J. C. (2011). Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30 39–52. 10.1007/s00338-010-0695-1 [DOI] [Google Scholar]
- Tout J., Siboni N., Messer L. F., Garren M., Stocker R., Webster N. S., et al. (2015). Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front. Microbiol. 6:432 10.3389/fmicb.2015.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegley L., Edwards R., Rodriguez-Brito B., Liu H., Rohwer F. (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9 2707–2719. 10.1111/j.1462-2920.2007.01383.x [DOI] [PubMed] [Google Scholar]
- Xu N., Tan G., Wang H., Gai X. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74 1–8. 10.1016/j.ejsobi.2016.02.004 [DOI] [Google Scholar]
- Yu K. (2012). Coral reefs in the South China Sea: their response to and records on past environmental changes. Sci. Chin. Ear. Sci. 55 1217–1229. 10.1007/s11430-012-4449-5 [DOI] [Google Scholar]
- Yu K.-F., Zhao J.-X., Liu T.-S., Wei G.-J., Wang P.-X., Collerson K. D. (2004). High-frequency winter cooling and reef coral mortality during the Holocene climatic optimum. Earth Planet. Sci. Lett. 224 143–155. 10.1016/j.epsl.2004.04.036 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.