Abstract
Anthocyanins are responsible for the different colors of ornamental plants. Grape hyacinth (Muscari armeniacum), a monocot plant with bulbous flowers, is popular for its fascinating blue color. In the present study, we functionally characterized an R2R3-MYB transcription factor gene MaAN2 from M. armeniacum. Our results indicated that MaAN2 participates in controlling anthocyanin biosynthesis. Sequence alignment and phylogenetic analysis suggested that MaAN2 belonged to the R2R3-MYB family AN2 subgroup. The anthocyanin accumulation of grape hyacinth flowers was positively correlated with the expression of MaAN2. And the transcriptional expression of MaAN2 was also consistent with that of M. armeniacum dihydroflavonol 4-reductase (MaDFR) and M. armeniacum anthocyanidin synthase (MaANS) in flowers. A dual luciferase transient expression assay indicated that when MaAN2 was co-inflitrated with Arabidopsis thaliana TRANSPARENT TESTA8 (AtTT8), it strongly activated the promoters of MaDFR and MaANS, but not the promoters of M. armeniacum chalcone synthase (MaCHS), M. armeniacum chalcone isomerase (MaCHI), and M. armeniacum flavanone 3-hydroxylase (MaF3H). Bimolecular fluorescence complementation assay confirmed that MaAN2 interacted with AtTT8 in vivo. The ectopic expression of MaAN2 in Nicotiana tabacum resulted in obvious red coloration of the leaves and much redder flowers. Almost all anthocyanin biosynthetic genes were remarkably upregulated in the leaves and flowers of the transgenic tobacco, and NtAn1a and NtAn1b (two basic helix–loop–helix anthocyanin regulatory genes) were highly expressed in the transformed leaves, compared to the empty vector transformants. Collectively, our results suggest that MaAN2 plays a role in anthocyanin biosynthesis.
Keywords: flower color, monocots, grape hyacinth, R2R3-MYB transcription factor, anthocyanin biosynthesis
Introduction
Flowers, because of their varieties of colors, have great ornamental and economic value. In ornamental plant breeding, many measures are taken to breed different cultivars with various colors, hues, and patterns (Yamagishi et al., 2010). Pigmentation in flowers is mainly attributed to anthocyanins, a type of flavonoid; the major pigments imparting a great variety of colors from light yellow, red, dark red, magenta, purple, to blue in higher plants have been investigated extensively (Holton and Cornish, 1995; Winkel-Shirley, 2001; Hichri et al., 2011). Anthocyanin biosynthetic pathway is regulated by R2R3-MYB TFs and their interacting partners bHLH TFs and WD repeat or WD40 proteins (Zhang et al., 2014). Particularly, in the three different types of proteins, the R2R3-MYB TFs play crucial roles in anthocyanin biosynthesis and are usually considered more specific, because their expression influences the spatial and temporal distribution of anthocyanins (Holton and Cornish, 1995; Schwinn et al., 2006; Allan et al., 2008; Feller et al., 2011; Davies et al., 2012; Zhang et al., 2014).
The first R2R3-MYB C1 was discovered in the monocot Zea mays; it was involved in kernel pigmentation (Paz-Ares et al., 1987). Subsequently, in the dicot Arabidopsis thaliana, PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1) and PAP2 were identified (Kranz et al., 1998); these proteins induced anthocyanin accumulation in Arabidopsis and Nicotiana tabacum (Borevitz et al., 2000). In dicot flowers, the anthocyanin-related R2R3-MYBs have been studied extensively and deeply in Petunia hybrida (Quattrocchio et al., 1999), Antirrhinum majus (Schwinn et al., 2006), Ipomoea nil (Morita et al., 2006), Gerbera hybrida (Elomaa et al., 2003; Laitinen et al., 2008), Gentiana tricolor (Nakatsuka et al., 2008a), and Malus crabapple (Jiang et al., 2014). Moreover, some studies have reported in monocot plants, such as Elaeis guineensis (Singh et al., 2014), Anthurium andraeanum (Li et al., 2016), Lilium (Yamagishi et al., 2010, 2011, 2014a,b; Yamagishi, 2011, 2015; Lai et al., 2012), Allium cepa (Schwinn et al., 2016), and Orchidaceae (Chiou and Yeh, 2008; Ma et al., 2009; Hsu et al., 2015; Li et al., 2017). In Araceae, Anthurium andraeanum AaMYB2 showed a positive correlation with pigmentation in anthurium spathes and leaves and induced anthocyanin accumulation in tobacco (Li et al., 2016). In Liliaceae, Lilium MYB6 regulated the development of anthocyanin spots in the tepals as well as the pigmentation in the leaves of the Asiatic hybrid lily ‘Montreux’ (Yamagishi et al., 2010), whereas MYB12 controlled the level of the coloration and pigmentation in the tepals of the Oriental and Asiatic hybrid lilies (Yamagishi et al., 2010, 2011, 2014a; Yamagishi, 2011, 2015; Lai et al., 2012). In Orchidaceae flowers, Oncidium ‘Gower Ramsey’ OgMYB1, produced red pigment when it was transiently overexpressed in the yellow lips (Chiou and Yeh, 2008). Purple Phalaenopsis schilleriana PsUMYB6 induced the production of anthocyanin when it was transiently co-expressed with Lc (a Z. mays bHLH TF determining anthocyanin accumulation) in the white P. amabilis petal tissue (Ma et al., 2009). PeMYB2, PeMYB11, and PeMYB12 regulated pigmentation in the petals and sepals, spots, and venation of red P. equestris, respectively (Hsu et al., 2015). Besides, Dendrobium hybrids DhMYB2 interacted with DhbHLH1 positively regulated pigmentation in the petals (Li et al., 2017).
Most R2R3-MYB TFs, as anthocyanin regulators in dicot flowers and a few monocots (oil palm, onion, anthurium, and lily), are included in the AN2 subgroup (corresponding to PhAN2 and AtPAP1 in dicots) (Yamagishi et al., 2010; Lai et al., 2012; Singh et al., 2014; Li et al., 2016; Schwinn et al., 2016). However, the anthocyanin-related MYBs in Orchidaceae are categorized into the C1 subgroup in Poaceae that includes Z. mays C1 and Oryza sativa MYB (CAA75509) (Chiou and Yeh, 2008; Ma et al., 2009; Hsu et al., 2015; Li et al., 2017). Schwinn et al. (2016) speculated that the distinct clades of AN2 and C1 subgroup in the R2R3-MYB family might occur within the monocot Asparagales in the evolution process of Allioideae and Orchidaceae. However, to date, only MYBs from Allioideae and Orchidaceae have been identified in the Asparagales (Chiou and Yeh, 2008; Ma et al., 2009; Hsu et al., 2015; Li et al., 2017). Therefore, more MYBs that control pigment synthesis should be studied in the monocots, especially in the Asparagales to address this issue.
Grape hyacinth (Muscari armeniacum), belonging to Scilloideae (Asparagales), is a monocot species used as cut flowers and garden ornamentals, and is famous for its fascinating blue color. To date, anthocyanin regulators in M. armeniacum have seldom been reported. In this study, we functionally identified an R2R3-MYB transcription factor MaAN2 from M. armeniacum, which plays a role in anthocyanin biosynthesis. And our findings might provide some information for the evolutionary divergence of R2R3-MYBs in monocots.
Results
Isolation of MaAN2 from Grape Hyacinth Petals
An anthocyanin-related R2R3-MYB unigene was screened from the transcriptome of M. armeniacum flowers (Lou et al., 2014). The 723-bp full-length MaAN2 cDNA encoding 240 amino acids was isolated from the flowers of M. armeniacum by using the RACE method. An amino acid alignment of R2R3-MYBs in grape hyacinth and other species showed that the R2 and R3 repeats were highly conserved in the N-terminal of MaAN2 (Figure 1A). However, when considering the entire protein sequence, MaAN2 showed only 48.12% identity with EgVIR (Elaeis guineensis anthocyanin-related R2R3-MYB), 39.92% identity with PhAN2, 39.12% identity with LhMYB6, and 33.55% identity with AaMYB2. Besides, in the R3 repeat, a conserved motif [D/E]LX2[R/K]X3LX6LX3R (as marked by black triangles in Figure 1A) is necessary for its interaction with the R-like bHLH protein (Zimmermann et al., 2004; Takos et al., 2006). Like LhMYB6 and LhMYB12 in Lilium, MaAN2 protein also had a conserved motif [K/R]P[Q/R]P[Q/R] in the C-terminal variable region (as marked by red box in Figure 1A), which was responsible for anthocyanin biosynthesis in eudicots (Yamagishi et al., 2010).
FIGURE 1.
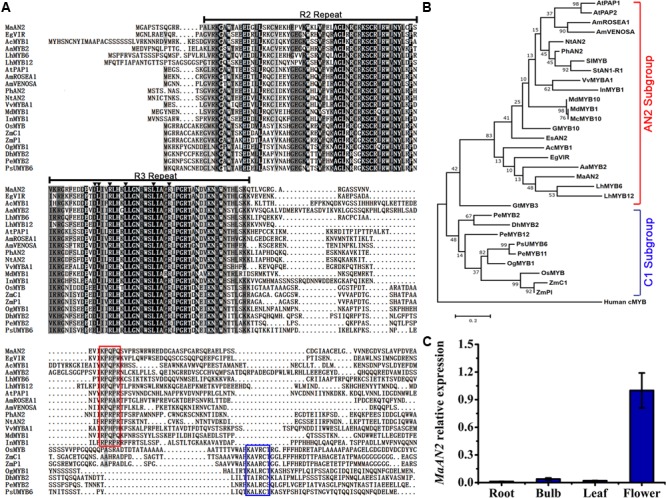
Protein sequence alignment and phylogenetic tree of MaAN2 transcription factor and its putative orthologs, and tissue-specific analysis of MaAN2. (A) Alignment of the deduced amino acid sequences of MaAN2 and other R2R3-MYBs associated with flavonoid biosynthesis from other plant species. If the identities of amino acids in each column are 100, 75, and 50%, these amino acids are indicated on a black, dark gray, and light gray background, respectively. The R2 and R3 repeats are indicated above the alignment. The motif [D/E]LX2[K/R]X3LX6LX3R in the R3 repeat necessary for interaction with a bHLH protein in the R3 repeat is indicated by dark triangles. The small motif [K/R]P[Q/R]P[Q/R] in the AN2 subgroup is shown by a red box. The short conserved sequences KAX[K/R]C[S/T], recognized in the C1 subgroup, are shown by blue box. (B) The maximum-likelihood phylogenetic tree was generated using MEGA 6.0 software. Numbers next to the nodes indicate the bootstrap values from 1000 replications. The bar indicates a genetic distance of 0.1. The gene names from various plant species and NCBI GenBank accession numbers for the sequences are as follows: Arabidopsis thaliana AtPAP1 (NP_176057), AtPAP2 (NP_176813); Petunia hybrida PhAN2 (AAF66727); Nicotiana tabacum NtAN2 (ACO52472); Solanum lycopersicum SlMYB (AAQ55181); Antirrhinum majus AmROSEA1 (ABB83826), AmVENOSA (ABB83828)); Ipomoea nil InMYB1 (BAE94391.1); Gerbera hybrida GMYB10 (CAD87010); Vitis vinifera VvMYBA1 (BAD18977); Malus domestica MdMYB1 (ABK58138) and MdMYB10 (ABB84753); Malus crabapple McMYB10 (AFP89357); Epimedium sagittatum EsAN2 (ALO24362); Solanum tuberosum StAN1-R1 (AKA95392); Elaeis guineensis EgVIR (KJ789862); Allium cepa AcMYB1 (KX785130); Anthurium andraeanum AaMYB2 (KU726561); Lilium hybrid division I LhMYB6 (BAJ05399) and LhMYB12 (BAJ05398); Gentiana triflora GtMYB3 (BAF96933); Oncidium Gower Ramsey OgMYB1 (ABS58501); Zea mays ZmPl (NP_001105885) and ZmC1 (NP_001106010); Oryza sativa OsMYB (CAA75509); Phalaenopsis schilleriana PsUMYB6 (ACH95792); Phalaenopsis equestris PeMYB2 (AIS35919), PeMYB11 (AIS35928), and PeMYB12 (AIS35929); Dendrobium hybrid DhMYB2 (AQS79852); Human cMYB (M15024). (C) The expression profile of MaAN2 in the roots, bulbs, leaves, and flowers of Muscari armeniacum.
The phylogenetic analysis of MaAN2 and other R2R3-MYBs from different species showed that MaAN2 was clustered closely with the sequences of monocots AaMYB2 and LhMYB6 in the AN2 subgroup, while the MYBs from monocot Poaceae and Orchidaceae were in C1 subgroup (Figure 1B). In addition, the transcription expression of MaAN2 in roots, bulbs, leaves, and flowers were analyzed by Quantitative Real-Time PCR (qRT-PCR) assay. The result showed that the expression of MaAN2 was very low in roots, bulbs, and leaves, but very high in flowers, which indicated that MaAN2 was predominantly expressed in flowers (Figure 1C).
MaAN2 Is a Transcription Factor
In order to detect whether MaAN2 is located in cell nucleus, the plasmid pCambia1304-MaAN2 (MaAN2-GFP) was transformed into Arabidopsis mesophyll protoplasts, and the nuclear localization of MaAN2 in Arabidopsis protoplasts was thus confirmed. In the positive control 35S:GFP (pCambia1304), the expression of GFP was found to be distributed throughout the entire cell (Figure 2A).
FIGURE 2.
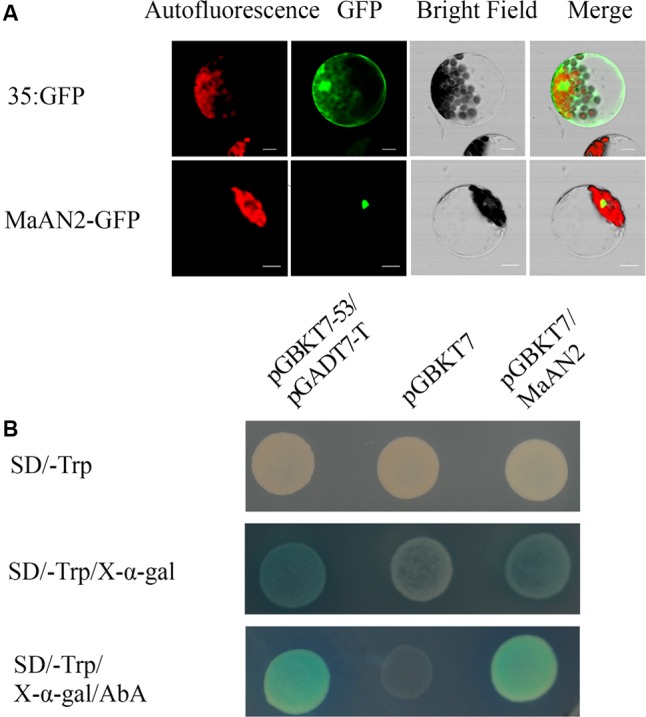
Subcellular localization and transcription activation ability of MaAN2. (A) Transient expression of the MaAN2-GFP fusion protein in A. thaliana mesophyll protoplasts showed MaAN2 located in the nucleus. Autofluorescence: chloroplast autofluorescence; GFP: GFP fluorescence; Merge is merged with chloroplast autofluorescence, GFP fluorescence, and bright field images. Bars, 10 μm. (B) Transcription activation ability of MaAN2 in Y2Hgold yeast. The positive control was pGBKT7-53 plus pGADT7-T and the negative control was pGBKT7. Yeasts transformed with the positive and negative controls, and pGBKT7-MaAN2 vectors were cultivated in SD/-Trp medium, SD/-Trp medium with 40 μg/ml X-α-Gal, and SD/-Trp medium plus 40 μg/ml X-α-Gal and 200 ng/ml AbA, respectively. The positive control and pGBKT7-MaAN2 exhibited blue yeast plaques, while the negative control did not grow in SD/-Trp medium plus 40 μg/ml X-α-Gal and 200 ng/ml AbA.
To determine whether MaAN2 has the transcriptional activity, we conducted a transactivation assay in yeast. Yeasts transformed with pGBKT7-MaAN2 vector and the positive control pGBKT7-53 plus pGADT7-T exhibited blue yeast plaques, while the negative control pGBKT7 did not grow in SD/-Trp medium plus 40 μg/ml X-α-Gal and 200 ng/ml AbA (Figure 2B). These results showed that MaAN2 possessed transcriptional activity. Therefore, MaAN2 was thought to be a transcription factor.
MaAN2 Expression Correlates with Anthocyanin Biosynthetic Gene Expression and Anthocyanin Accumulation during Grape Hyacinth Flowering
The inflorescence and the petals of different flower developmental stages (S1–S5) of M. armeniacum are shown in Figure 3A. The total anthocyanin content of M. armeniacum at the five flowering stages was significantly different. The anthocyanin content first increased, and then decreased, during the flower development period, peaking at stage S4 (Figure 3B). Specifically, the anthocyanin was seldom detected at stage S1, and accumulated at stage S2. The anthocyanin content showed a rapid increase from stage S2 to S4, but decreased from stage S4 to S5.
FIGURE 3.
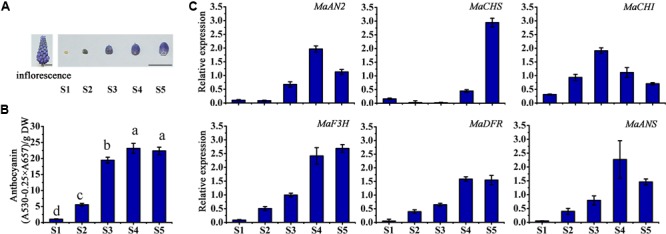
The anthocyanin content and expression profiles of anthocyanin regulatory and structural genes at different flower developmental stages in M. armeniacum. (A) The inflorescence and the petals of five flower developmental stages of M. armeniacum. Bars, 1 cm. (B) The total anthocyanin content of petals at the five flower developmental stages. DW: dry weight. Different lower case letters represent significant difference calculated using Least–Significant Difference (LSD) analysis at the level of P < 0.05. (C) The expression profiles of MaAN2, MaCHS, MaCHI, MaF3H, MaDFR, and MaANS in flowers during the five developmental stages (S1–S5) of M. armeniacum. MaActin was the reference gene to normalize the expression of these genes. Each column represents means ± SD from three independent experiments.
In order to detect the expression levels of anthocyanin biosynthetic genes during flower development, the anthocyanin biosynthetic genes: chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), and anthocyanidin synthase (ANS) (as markedby red color in Supplementary Figures S1–S4), were screened from the M. armeniacum flower transcriptome (Lou et al., 2014), as well as M. armeniacum dihydroflavonol 4-reductase (MaDFR; Jiao et al., 2014) were used in this study. And then, the transcription levels of MaAN2 and pigment biosynthetic genes were analyzed using the qRT-PCR for stage S1–S5 (Figure 3C). The transcriptional expression of MaAN2 was correlated with that of MaDFR and MaANS (Figure 3C). The expression levels of MaDFR, MaANS, and MaAN2 first increased and peaked at stage S4, and then decreased with flower development; this expression pattern was similar to the anthocyanin content changes during progression from stage S1 to S5 (Figures 3B,C). Furthermore, the mRNA levels of MaCHS and MaCHI were not correlated with anthocyanin biosynthesis during flower blossoming. MaF3H expression increased from stages S1 to S4, coordinating with that of MaAN2, but the expression of MaF3H continued to increase until stage S5.
MaAN2 Interacts with a bHLH Protein to Activate the Promoters of Anthocyanin Structural Genes
To investigate whether MaAN2 could interact with the bHLH TF to regulate the expression of anthocyanin pathway genes, the different TFs and promoters were co-infiltrated in N. benthamiana leaves, and subsequently, a dual-luciferase assay was conducted (Figure 4). Firstly, we cloned 871, 1113, 692, and 1044 bp of promoters of anthocyanin biosynthetic genes (MaCHS, MaCHI, MaF3H, and MaANS; as marked by red color in Supplementary Figures S1–S4), respectively; these genes were screened from the M. armeniacum flower transcriptome (Lou et al., 2014). Moreover, we also cloned 1523 bp promoter of MaDFR (Jiao et al., 2014). Next, we analyzed the cis-elements of these gene promoters (Figure 4A) using the online website PlantCARE. All promoters contained the MYB-binding sites: MYB core element (5′-CNGTTR-3′) or the AC-rich element ([A/C]CC[A/T]A[A/C]) (Xu et al., 2015) and the bHLH-binding site: G-box (5′-CACGTG-3′) (Hichri et al., 2011) (Figure 4A). Thus, these genes might be regulated by MYB and bHLH TFs.
FIGURE 4.
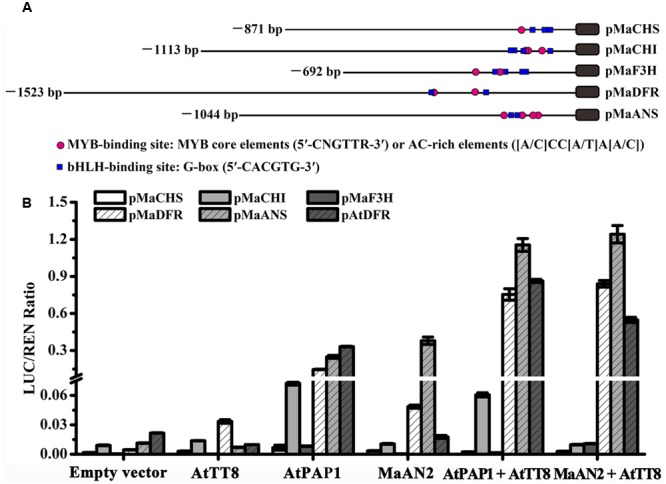
Schematics of M. armeniacum anthocyanin biosynthetic gene promoters and dual luciferase transient expression assay. (A) The lengths, MYB binding sites, and bHLH binding sites of MaCHS, MaCHI, MaF3H, MaDFR, and MaANS promoters are marked by black solid lines, red circles, and blue rectangles, respectively. (B) Dual luciferase transient expression assay showed the effects of MaAN2, AtPAP1, and AtTT8 TFs on MaCHS, MaCHI, MaF3H, MaDFR, MaANS, and A. thaliana AtDFR promoters. All TFs were co-infiltrated with promoters in N. benthamiana leaves for transient transformation assays. The results show the promoter activity expressed as the ratio of the activities of these promoters: luciferase (LUC) to 35S: Renilla (REN). Each column represents means ± SD from four biological replicates.
Further, the R2R3-MYB protein PAP1 and bHLH protein TT8 involved in the regulation of flavonoid synthesis in Arabidopsis (Nesi et al., 2000) along with the AtDFR promoter were cloned and used as positive control. The result showed that the promoter activities of MaDFR, MaANS, and AtDFR indicated a considerable increase when MaAN2 or AtPAP1 was co-infiltrated with AtTT8, compared with the treatment with MYBs alone (Figure 4B). The promoter activities of MaCHS, MaCHI, and MaF3H were very low when MaAN2 or AtPAP1 co-infiltrated with or without AtTT8 (Figure 4B). MaAN2 could not promote the expression of MaCHS, MaCHI, MaF3H, MaDFR, and AtDFR, but could slightly activate the MaANS promoter without the co-expression of AtTT8. In brief, MaAN2 depended on the presence of bHLH protein AtTT8 to activate the expression of MaDFR, MaANS, and AtDFR.
In order to confirm the interaction between MaAN2 and AtTT8 in vivo, a BiFC assay was conducted in A. thaliana mesophyll protoplasts (Figure 5). The YFP expression was detected only when pSPYNE/MaAN2 was co-expressed with pSPYCE/AtTT8, but not when pSPYNE/MaAN2 was co-expressed with pSPYCE and pSPYCE/AtTT8 was co-expressed with pSPYNE. Thus, MaAN2 interacted with bHLH TF AtTT8 in vivo.
FIGURE 5.
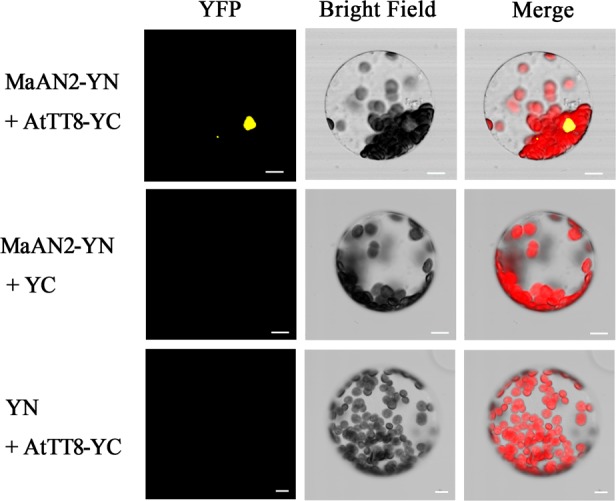
Bimolecular fluorescence complementation of MaAN2 and AtTT8 interaction in A. thaliana mesophyll protoplasts. YFP: fluorescence of YFP; Merge is digital image merged with bright field and fluorescent images. Bars, 10 μm.
Overexpression of MaAN2 Promotes Anthocyanin Accumulation in Tobacco
To characterize the function of MaAN2, we obtained the overexpressing MaAN2 transgenic tobaccos (OE-MaAN2) by the method of tobacco leaf disks. The result showed that MaAN2 promoted pigmentation in the corolla of flowers (Figures 6A,B). Besides, the pistil, anther, sepal, ovary, and seeds showed the strong accumulation of anthocyanins (Figures 6C–E); the whole tobacco leaves of OE-MaAN2 turned red (Figures 6F–H).
FIGURE 6.
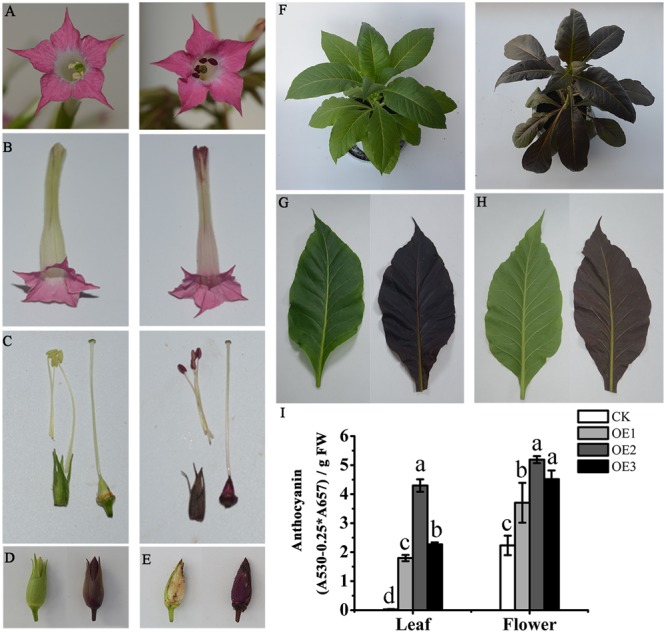
Phenotypic observation and anthocyanin content in empty vector (CK) and OE-MaAN2 tobacco transplants. The front (A) and lateral (B) parts of flower; anthers, stigma, and ovary (C); the sepal (C,D); and seeds (E), total plant (F), the adaxial side (G) and abaxial side (H) of a leaf. The left sides in each block show the transgenic tobacco of CK, whereas the right sides in each block show the transgenic tobacco of OE-MaAN2 (i.e., OE2). (I) Total anthocyanin content of the leaf and flower of CK and three independent OE-MaAN2 transgenic lines (OE1, OE2, and OE3). Each column represents means ± SD from three independent experiments. Different lower case letters represent significant difference which is calculated using LSD analysis at the level of P < 0.05.
Additionally, in the leaves and flowers of three OE-MaAN2 lines (OE1, OE2, and OE3), the total anthocyanin contents were significantly increased compared to those in the empty vector transgenic tobaccos (CK) (Figure 6I). Next, we used HPLC to analyze the anthocyanin composition in the transgenic tobacco leaves and corollas. Previous studies reported that cyanidin-3-rutinoside mainly existed in the petal of wild-type tobacco (Deluc et al., 2006; Nakatsuka et al., 2008b). Even in our study, cyanidin-3-rutinoside was mainly detected in the leaves and flowers of OE-MaAN2 (Supplementary Figure S5).
Overexpression of MaAN2 Promotes the Expression of Anthocyanin Pathway Genes in Tobacco
By qRT-PCR assay, the presence of MaAN2 was confirmed in transgenic tobacco (Figures 7B,D). We found that MaAN2 positively regulated the transcriptional expression of anthocyanin biosynthetic genes in tobacco leaves and flowers. The anthocyanin biosynthetic genes, NtCHS, NtCHI, NtF3H, NtF3′H, NtDFR, NtANS, and NtUFGT, were upregulated in the leaves of OE1, OE2, and OE3 (Figure 7A). Specifically, the transcripts of NtCHS, NtCHI, and NtANS were remarkably increased in the three transgenic lines, especially in OE2 and OE3. In the flowers of transgenic tobacco, the expressions of NtCHS, NtCHI, NtF3H, NtF3’H, NtDFR, NtANS, and NtUFGT, were increased in OE2 (Figure 7C). However, in the flowers of OE1, the expressions of NtCHS, NtCHI, NtF3’H, and NtANS were slightly decreased, and the expressions of the other pigment genes maintained or slightly increased compared to that of CK (Figure 7C), which might be because the expression of MaAN2 in OE1 was much weaker than that in OE2 and OE3 (Figure 7D). The endogenous bHLH TF genes, NtAn1a and NtAn1b, homologous to AtTT8, which regulated anthocyanin biosynthesis in tobacco (Bai et al., 2011), were upregulated in the leaves of OE1, OE2, and OE3 (Figure 7B). Further, the transcription level of NtAn1a in the transplant flowers was considerably higher in OE2 than that in CK (Figure 7D). However, the transcription level of NtAn1b in transgenic flowers was similar to that in CK (Figure 7D).
FIGURE 7.
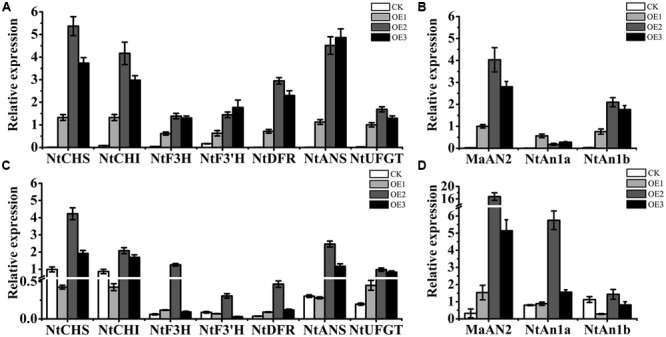
Expression profiles of anthocyanin biosynthetic genes and endogenous transcription factors in the leaves and flowers of the empty vector transgenic tobaccos (CK) and three independent OE-MaAN2 transplants (OE1, OE2, and OE3). Expression patterns of NtCHS, NtCHI, NtF3H, NtF3′H, NtDFR, NtANS, and NtUFGT genes in the leaves (A) and flowers (C) of CK and OE1, OE2, and OE3. Expression patterns of MaAN2, NtAn1a, and NtAn1b in the leaves (B) and flowers (D) of CK and OE1, OE2, and OE3. NtTubA1 was the reference gene to normalize the expression of these genes. Each column represents means ± SD from three independent experiments.
Discussion
MaAN2 Belongs to the AN2 Subgroup
R2R3-MYBs play vital roles in plant anthocyanin biosynthesis; and they usually have a conserved DNA-binding domain (R2 and R3 repeats) in the N-terminal and a variable C-terminal region (Dubos et al., 2010). In dicots, the [K/R]P[Q/R]P[Q/R] motif in the C-terminal is the typically characteristic of the AN2 subgroup in R2R3-MYB TFs family (Yamagishi et al., 2010). MaAN2, like other monocot MYBs, such as LhMYBs, EgVIR, AaMYB2, and AcMYB1 (Allium cepa anthocyanin activator), contained the motif [K/R]P[Q/R]P[Q/R] (Figure 1A), not the conserved motif KAx[K/R]C[S/T] (as marked by blue box in Figure 1A), which is the characteristic of the C1 subgroup and usually appears in monocot maize (Cone et al., 1993).
Muscari armeniacum belongs to the Scilloideae subfamily, which is included in the family Asparagaceae of the order Asparagales (Angiosperm Phylogeny Group III, 2009). The anthocyanin-related MYBs have also been studied in the family Orchidaceae (Oncidium, Phalaenopsis, and Dendrobium) (Chiou and Yeh, 2008; Ma et al., 2009; Hsu et al., 2015; Li et al., 2017) and Allioideae (Allium cepa) (Schwinn et al., 2016), which also belong to the order Asparagales (Angiosperm Phylogeny Group III, 2009). Besides, some other anthocyanin MYB regulators have been reported in the monocots, including Arecaceae (E. guineensis; Singh et al., 2014), Araceae (Anthurium andraeanum; Li et al., 2016), and Liliaceae (Lilium; Yamagishi et al., 2010), as well as Poaceae (Z. mays; Paz-Ares et al., 1987). In these reported MYBs in monocots, the MYBs from Orchidaceae and Poaceae were clustered in the C1 subgroup, and the others were included in the AN2 subgroup. Hence, the anthocyanin-related MYBs showed divergence among the monocot species. However, they are conserved in the reported dicot species (Davies et al., 2012).
MaAN2 Interacts with a bHLH Protein AtTT8 and Targets MaDFR and MaANS to Control Anthocyanin Pigmentation
In the flower development of grape hyacinth, the expression profile of MaAN2, as well as those of MaDFR and MaANS, was positively concomitant with the flower pigmentation in M. armeniacum (Figure 3). Furthermore, via the dual luciferase transient expression assay, we found that MaAN2 was highly dependent on AtTT8 as a partner to activate the promoters of MaDFR and MaANS (Figure 4B), and the BiFC assay also confirmed that MaAN2 interacted with AtTT8 in vivo (Figure 5). Additionally, the expression profile of MaF3H was similar to those of MaDFR, MaANS, and MaAN2 in flowers at stages S1–S4 (Figure 3C), but it was not activated by MaAN2 (Figure 4B). In plant, anthocyanin biosynthesis is mainly catalyzed by a set of enzymes encoded by anthocyanin structural genes (Dooner et al., 1991; Grotewold, 2006; Katsumoto et al., 2007; Tanaka et al., 2008); these structural genes are usually classified into early and late biosynthetic genes (EBGs and LBGs) depending on different species (Martin et al., 1991; Quattrocchio et al., 1993; Boss et al., 1996; Pelletier et al., 1997; Gonzalez et al., 2008). In Z. mays, the EBGs and LBGs are usually regulated by the MYB-bHLH (i.e., ZmC1-ZmLc) complex (Dooner et al., 1991). However, in dicot Arabidopsis, these genes are controlled separately by different TFs (Dubos et al., 2010); LBGs are regulated by the complex of AtPAP1-AtTT8 (Xu et al., 2014). PhAN2 interaction with the bHLH PhAN1 is able to activate the PhDFR promoter (Spelt et al., 2000). GtMYB3 interaction with GtbHLH1 could activate the expressions of LBGs: flavonoid 3′, 5′-hydroxylase (F3′5′H) and hydroxycinnamoyl-CoA: anthocyanin 5-O-acyltransferase (5AT) (Nakatsuka et al., 2008a). In M. armeniacum, MaF3H might be an EBG; its expression was not activated by MaAN2 but possibly by some other TF(s). In Arabidopsis, F3H, as the EBG, is regulated by subgroup 7 R2R3-MYBs (Dubos et al., 2010). Moreover, in Antirrhinum majus, Lilium spp., and Phalaenopsis spp., there are two or three MYBs responsible for their flower pigmentation (Schwinn et al., 2006; Yamagishi et al., 2010; Hsu et al., 2015). In the present study, MaAN2 expression at stage S2 was similar to that at stage S1, but the expression levels of MaDFR and MaANS were higher at stage S2 than at stage S1 (Figure 3C), which indicated that the likely involvement of other TFs in flower pigmentation. In brief, our study suggested that MaAN2 interacted with AtTT8 to control LBGs (MaDFR and MaANS) expression. However, in Lilium spp., LhMYB12 activated LhCHSa (EBG) and LhDFR (LBG) expression in the presence of LhbHLH2 in Asiatic hybrid lily tepals (Lai et al., 2012). In Phalaenopsis spp., PeMYBs could activate the expression of F3H5 (EBG), DFR1 and ANS3 (LBGs) in flowers to influence the patterns and quantity of color. When Oncidium ‘Gower Ramsey’ OgMYB1 was overexpressed in its yellow lip, the transcripts of OgCHI and OgDFR were increased (Chiou and Yeh, 2008).
Overall, the regulation pattern of anthocyanin-related MYBs is conserved in dicots (i.e., the interaction of MYB with bHLH-activating LBGs) (Dubos et al., 2010; Xu et al., 2014, 2015; Li et al., 2017). Moreover, we proposed that the anthocyanin-related MYBs originating from the monocots except Poaceae and Orchidaceae were included in the AN2 subgroup and depended on the bHLH TFs for inducing anthocyanin biosynthesis. However, R2R3-MYBs from Gramineae and Orchidaceae belong to the C1 subgroup (Hsu et al., 2015; Li et al., 2017). Whether these MYBs regulated EBGs and LBGs or only LBGs varies with different species in monocots (Chiou and Yeh, 2008; Lai et al., 2012; Hsu et al., 2015). Further, whether the distinction in activating EBGs or LBGs is caused by the MYB subgroups or by the divergence between monocots and dicots is not yet known (Lai et al., 2012), thus more evidence is needed to be provided for finding the regularity.
MaAN2 Promotes Anthocyanin Accumulation by Upregulating the Expression of Anthocyanin Biosynthetic and bHLH TF Genes
Many studies showed that ectopically expressing R2R3-MYB genes of the AN2 subgroup, such as AtPAP1 and AtPAP2 (Borevitz et al., 2000; Xie et al., 2006), Epimedium sagittatum EsAN2 (Huang et al., 2016b), Solanum tuberosum StAN1-R1 (Liu et al., 2016), AaMYB2 (Li et al., 2016), and LhMYB12-Lat (Yamagishi et al., 2014b) could regulate anthocyanin biosynthesis to various extent in tobacco. In this study, the OE-MaAN2 accumulated anthocyanin in the flowers and leaves and showed distinct phenotypes compared with CK (Figures 6A–H). The anthocyanin contents were significantly higher in the pigmented leaves and flowers of OE-MaAN2 than in those of CK (Figure 6I).
Zhang et al. (2014) considered that both MYB and bHLH partners can likely induce the entire biosynthetic pathway for anthocyanin accumulation. Our study showed that the overexpression of MaAN2 in tobacco could differentially activate the expression of anthocyanin biosynthetic genes. The EBGs (NtCHS, NtCHI, and NtF3H) as well as the LBGs (NtDFR, NtANS, and NtUFGT) were activated in leaves of three transgenic lines and flowers of OE2 and OE3 (Figures 7A,C). This result is consistent with that of overexpression of StAN1-R1 (a potato R2R3-MYB anthocyanin regulator) in tobacco (Liu et al., 2016); the result was also observed in the leaves of N. tabacum ectopically expressing EsAN2 or AaMYB2 (Huang et al., 2016b; Li et al., 2016). Two bHLH TF genes, NtAn1a and NtAn1b, controlled anthocyanin biosynthesis in the corollas of tobacco plants (Bai et al., 2011); they were also activated in the transgenic leaves when ectopically expressing EsAN2, StAN1-R1, or AaMYB2 in tobacco (Huang et al., 2016b; Li et al., 2016; Liu et al., 2016). The similar result was also found in the leaves of OE-MaAN2 tobacco (Figure 7B). However, in flowers, the transcripts of NtAn1a and NtAn1b were not influenced by EsAN2 (Huang et al., 2016b), but strongly upregulated by StAN1-R1 (Liu et al., 2016). In this study, MaAN2 activated the expression of NtAn1a, not NtAn1b in transplant flowers (Figure 7D). Therefore, the bHLH gene expression activated by MYBs were different between leaves and flowers in tobacco. Overall, the heterogeneous R2R3-MYBs could activate the expression of endogenous bHLH genes in N. tabacum leaves. Then these AN2 subgroup MYBs might interact with the endogenous bHLH TF to enhance anthocyanin contents. However, in the tobacco leaves of overexpressing LhMYB12-Lat (an anthocyanin regulatory R2R3-MYB gene), it is showed higher levels of the transcripts of NtCHS, NtCHI, NtF3H, NtDFR, and NtANS. Whereas, LhMYB12-Lat could not activate the transcription of bHLH in a heterologous system (Yamagishi et al., 2014b). Therefore, MYBs from different species might exert diverse effects.
Conclusion
The newly identified R2R3-MYB MaAN2 was possibly an anthocyanin activator involved in grape hyacinth flower coloration. It belongs to the AN2 subgroup of R2R3-MYB family, and interacts with a bHLH to regulate the expression of LBGs. The regulatory pattern of anthocyanin biosynthesis is similar to that in dicot. The finding might provide a sight into elucidating of the anthocyanin biosynthesis in grape hyacinth and suggest the evidence for the evolutionary divergence of R2R3-MYB TFs in monocot lineages.
Materials and Methods
Plants Materials
Plants of grape hyacinth M. armeniacum were cultivated in an experimental field of the Northwest A&F University in the Yangling District of the Shaanxi Province in China. The flower development is divided into five stages mainly based on petal pigmentation: S1, no pigmentation; S2, pigmentation visible on the basal part; S3, pigmentation beginning to turn blue; S4, flowers were completely blue, but had not opened; and S5, flowers completely opened. Besides, the roots, bulbs, and leaves of M. armeniacum were reserved. N. tabacum ‘NC89’ was cultured in a light incubator with a 16/8 h day/night and used for genetic transformation at the four-leaf stage; the transplant tobaccos were transferred from aseptic culture room to a greenhouse in natural light and artificial light extension to 16 h. The fully expanded tobacco leaves were collected from the mature transplants, and the flower limbs were picked when the corollas opened at an angle of 90°. All samples collected from grape hyacinth and tobacco were frozen in liquid nitrogen and stored at -80°C.
qRT-PCR Assay
Total RNA was isolated from M. armeniacum flowers, roots, bulbs, and leaves as well as from the leaves and flowers of tobacco using the Omega Total RNA Kit (Omega, United States). For this, 1 μg RNA aliquots were treated with the PrimeScript RT reagent Kit with gDNA Eraser (Takara, Japan) to remove the residual genomic DNA and then synthesize cDNA. The cDNA was diluted five times and used as the template, and the BioEasy Master Mix Plus (SYBR Green) (Bioer, China) was used as the fluorochrome for qPCR assay. Specifically, the cDNA templates from five flower stages were mixed equally and used for qRT-PCR analysis to detect the tissue-specific expression of MaAN2. The assay was conducted by using the iQ5 RT-PCR detection system (Bio-Rad, United States). The qRT-PCR primers of grape hyacinth and tobacco are listed in Supplementary Table S1. MaActin and NtTubA1 were used as the internal control genes in each grape hyacinth and tobacco sample, respectively. All analyses were conducted with three independent experiments.
Isolation of Full-Length Coding Sequence of MaAN2
We obtained an anthocyanin-related R2R3-MYB unigene from the transcriptome of M. armeniacum flowers (Lou et al., 2014). This DNA fragment lacked the 5′- and 3′-untranslated regions; thus, we isolated full-length cDNA of MaAN2 from the flowers in M. armeniacum followed the method described by Huang et al. (2016b). The primers used for the 5′- and 3′-RACE PCR, as well as full-length gene-specific primers are listed in Supplementary Table S2. Finally, this cDNA sequence was submitted to the NCBI GenBank database with an accession number KY781168. Besides, the R2R3-MYB was clustered with the AN2 subgroup; therefore, it was considered as MaAN2.
Multiple alignments were analyzed using DNAMAN 8.0. The R2 and R3 repeats as well as the two conserved motifs were highlighted with differently colored lines and boxes. The listed R2R3-MYBs were downloaded from the GenBank database (accession numbers listed in Figure 1). The program MEGA 6.0 was used for constructing a phylogenetic tree by maximum likelihood method.
Subcellular Localization of MaAN2
For subcellular localization analysis, the ORF of MaAN2 was introduced into the pCambia1304 vector using the Seamless Cloning and Assembly Kit (Novoprotein, Shanghai; primers are listed in Supplementary Table S3). The C-terminal of MaAN2 protein was fused to GFP, which allowed the transient expression of these proteins in planta. The polyethylene glycol-mediated transfection of A. thaliana mesophyll protoplasts was conducted according to previously described protocols (Yoo et al., 2007). 35S:GFP (pCambia1304) alone (positive control), chlorophyll fluorescence and MaAN2-GFP (pCambia1304-MaAN2) were observed 16 h after transformation using a FV1000 confocal microscope (Olympus, Japan). Images were analyzed using the FV1000 Viewer software.
Transcription Activation Ability of MaAN2
To determine the transcription activation ability of MaAN2, a yeast expression vector was constructed by fusing the ORF PCR product into a pGBKT7 vector using the Seamless Cloning and Assembly Kit (Novoprotein, Shanghai; primers are listed in Supplementary Table S3). The vectors pGBKT7-53 plus pGADT7-T (positive control), pGBKT7 (negative control), and pGBKT7-MaAN2 were introduced into the yeast strain Y2Hgold according to the YeastmakerTM Yeast Transformation System 2 User Manual (Clontech, Japan). These transformants were cultivated at 30°C for about 3 d on SD/-Trp medium, SD/-Trp medium with 40 μg/ml X-α-Gal, and SD/-Trp medium plus 40 μg/ml X-α-Gal and 200 ng/ml AbA.
Isolation of MaCHS, MaCHI, MaF3H, MaDFR, and MaANS Promoters by Genome Walking
In order to explore whether MaAN2 regulates the expression of anthocyanin biosynthetic genes, the promoters of anthocyanin biosynthetic genes were needed to clone. We screened the unigenes of each anthocyanin biosynthetic gene from the M. armeniacum flower transcriptome (Lou et al., 2014), including MaCHS, MaCHI, MaF3H, and MaANS. The unigene of each anthocyanin biosynthetic gene which showed higher expression among its respective candidate genes and meanwhile could be clustered with its respective orthologs was used in this study (as marked by red color in Supplementary Figures S1–S4). Then these anthocyanin biosynthetic genes used in this study were submitted to NCBI GenBank database. The accession numbers of MaCHS, MaCHI, MaF3H, and MaANS were MF041820, MF041821, MF041822, and MF041819, respectively. Notably, the cDNA of M. armeniacum DFR (KJ619963) reported by Jiao et al. (2014) also used in this study. The promoter regions of these genes were cloned from M. armeniacum genomic DNA by conducting a nested PCR with the genome walker kit (Clontech, United States). The nested PCR assays were performed using the degenerate primer from the kit and three gene-specific primers, as described in Supplementary Table S4. The obtained sequences were analyzed using DNAMAN 8.0. Finally, we isolated 871, 1113, 692, 1523, and 1044 bp promoter sequences of MaCHS, MaCHI, MaF3H, MaDFR, and MaANS, with accession numbers KY781171, KY781172, KY781173, KY781169, KY781170 in NCBI GenBank database, respectively. In addition, the AtDFR (AT5G42800) promoter sequence, which was used as the positive control, was downloaded from TAIR1 and obtained from the Arabidopsis genomic DNA. The cis-elements of these promoters were speculated using the online website PlantCARE2.
Dual Luciferase Assay
For dual luciferase assay, the full-length sequences of TF genes: MaAN2, AtPAP1 (AT1G56650), AtTT8 (AT4G09820), and the promoters: MaCHS, MaCHI, MaF3H, MaDFR, MaANS, and AtDFR, were amplified using primers listed in Supplementary Table S5, and jointed with the pGreenII 62-SK and pGreenII 0800-LUC vector (Hellens et al., 2005) using Seamless Cloning and Assembly Kit (Novoprotein, China), respectively. N. benthamiana were grown in a light incubator until four to six leaf stage when infiltration with Agrobacterium tumefaciens GV3101 can be performed (Yin et al., 2010). The enzyme activities of LUC and REN were determined using a previously reported protocol (Palapol et al., 2009). The measurements were performed using a luminometer Tecan Infinite M200 (Männedorf, Switzerland) from at least four biological replicates for each assay. The promoter activities were expressed as the ratios of the activities of these promoters: luciferase (LUC) to 35S: Renilla (REN).
BiFC Assay
To verify the interaction between MaAN2 and AtTT8 in vivo, a BiFC assay was conducted in A. thaliana mesophyll protoplasts by the polyethylene glycol-mediated method. The ORF of MaAN2 was inserted into the pSPYNE (R) 173 vector, and the coding sequence of AtTT8 without the stop codon was inserted into pSPYCE (M) (Waadt et al., 2008). The primers are listed in Supplementary Table S6. In brief, MaAN2 and AtTT8 were located adjacent to the N-terminal and C-terminal fragments of YFP, respectively. The YFP fluorescence was visualized 16 h after transformation using a confocal microscope (Olympus, Japan). The pSPYNE/MaAN2 plus pSPYCE and pSPYNE plus pSPYCE/AtTT8 were used as negative controls. Images were analyzed using the FV1000 Viewer software.
Tobacco Stable Transformation
The function of MaAN2 was determined by constructing a pCambia1304 vector containing the ORF of MaAN2. Tobacco leaf disk transformation was conducted using the previously described protocol (Horsch et al., 1985). The OE-MaAN2 transgenic lines showing obvious color changes in leaves and flowers were used for further analysis.
Anthocyanin Content Measurement in Grape Hyacinth Petals and Transgenic Tobacco Leaves and Flowers
The total anthocyanin content in grape hyacinth petals and transgenic tobacco leaves and flowers was determined as previously described (Mancinelli et al., 1991; Huang et al., 2016a). The petals of grape hyacinth were freeze-dried for 72 h, ground to powder, and then extracted with acidic methanolic solution (methanol, H2O, formic acid, and trifluoroacetic acid, 70:27:2:1, v/v) (Gao and Mazza, 1995; Hashimoto et al., 2000). The fresh leaves and petals of tobacco were ground in liquid nitrogen and then extracted. The extraction and measurement of all samples were followed by the protocol described (Huang et al., 2016a) using a UV-Visible Spectrophotometer (UV2600, Shimadzu). The subtracted absorbance was calculated as A530 (peak absorption of anthocyanins) – 0.25 × A657 (maximum absorption of chlorophyll-degradation products). The total anthocyanin content of grape hyacinth flowers and the tobacco leaves and flowers was computed as the ratio of the subtracted absorbance to dry weight and fresh weight, respectively.
In order to analyze the anthocyanin composition of tobacco leaves and flowers, we used the above-mentioned solutions for performing the reverse HPLC method. The chromatographic analysis was conducted using an Agilent 1100 series HPLC system (TC-C18 column, 5 μm, 4.6 mm × 250 mm) with the detection wavelength at 530 nm and the column was maintained at 30°C. The eluent was aqueous solution A (0.1% formic acid in water) and organic solvent B (acetonitrile). The gradient elution program was modified as described previously (Xie et al., 2006): 0 min, 10% B; 15 min, 17% B; 20 min, 23% B; 25 min, 23% B; and 30 min, 10% B; the eluent flow rate was 1.0 ml/min with 10 μl injection volume.
Author Contributions
YL conceived and designed the research. KC and HL conducted experiments. KC drafted the manuscript. QL critically revised the manuscript. All authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 305021405). We thank Wenkong Yao for revised the manuscript and Weizhong An, Peiwen Liang, and Shimeng Cao for care for plant materials.
Abbreviations
- AbA
aureobasidin A
- BIFC
bimolecular fluorescence complementation
- bHLH
basic helix–loop–helix
- CK
the empty vector transgenic tobaccos
- EBGs and LBGs
early and late biosynthetic genes
- GFP
green fluorescent protein
- HPLC
high-performance liquid chromatography
- OE-MaAN2,
overexpressing MaAN2 transgenic tobaccos
- ORF
open reading frame
- TFs
transcription factors
- YFP
yellow fluorescent protein.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00965/full#supplementary-material
References
- Allan A. C., Hellens R. P., Laing W. A. (2008). MYB transcription factors that colour our fruit. Trends Plant Sci. 13 99–102. 10.1016/j.tplants.2007.11.012 [DOI] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group III (2009). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 161 105–121. 10.1111/j.1095-8339.2009.00996.x [DOI] [Google Scholar]
- Bai Y., Pattanaik S., Patra B., Werkman J. R., Xie C. H., Yuan L. (2011). Flavonoid-related basic helix-loop-helix regulators, NtAn1a and NtAn1b, of tobacco have originated from two ancestors and are functionally active. Planta 234 363–375. 10.1007/s00425-011-1407-y [DOI] [PubMed] [Google Scholar]
- Borevitz J. O., Xia Y., Blount J., Dixon R. A., Lamb C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2393. 10.1105/tpc.12.12.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss P. K., Davies C., Robinson S. P. (1996). Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv. Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 111 1059–1066. 10.1104/pp.111.4.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou C.-Y., Yeh K.-W. (2008). Differential expression of MYB gene (OgMYB1) determines color patterning in floral tissue of Oncidium gower ramsey. Plant Mol. Biol. 66 379–388. 10.1007/s11103-007-9275-3 [DOI] [PubMed] [Google Scholar]
- Cone K. C., Cocciolone S. M., Burr F. A., Burr B. (1993). Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5 1795–1805. 10.1105/tpc.5.12.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K. M., Albert N. W., Schwinn K. E. (2012). From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 39 619–638. 10.1071/FP12195 [DOI] [PubMed] [Google Scholar]
- Deluc L., Barrieu F., Marchive C., Lauvergeat V., Decendit A., Richard T., et al. (2006). Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140 499–511. 10.1104/pp.105.067231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H. K., Robbins T. P., Jorgensen R. A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25 173–199. 10.1146/annurev.ge.25.120191.001133 [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15 573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Elomaa P., Uimari A., Mehto M., Albert V. A., Laitinen R. A., Teeri T. H. (2003). Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiol. 133 1831–1842. 10.1104/pp.103.026039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E. L., Grotewold E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66 94–116. 10.1111/j.1365-313X.2010.04459.x [DOI] [PubMed] [Google Scholar]
- Gao L., Mazza G. (1995). Characterization, quantitation, and distribution of anthocyanins and colorless phenolics in sweet cherries. J. Agric. Food Chem. 43 343–346. 10.1021/jf00050a015 [DOI] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J. M., Lloyd A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53 814–827. 10.1111/j.1365-313X.2007.03373.x [DOI] [PubMed] [Google Scholar]
- Grotewold E. (2006). The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 57 761–780. 10.1146/annurev.arplant.57.032905.105248 [DOI] [PubMed] [Google Scholar]
- Hashimoto F., Tanaka M., Maeda H., Shimizu K., Sakata Y. (2000). Characterization of cyanic flower color of Delphinium cultivars. J. Jpn. Soc. Hort. Sci. 69 428–434. 10.2503/jjshs.69.428 [DOI] [Google Scholar]
- Hellens R. P., Allan A. C., Friel E. N., Bolitho K., Grafton K., Templeton M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1 13–26. 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri I., Barrieu F., Bogs J., Kappel C., Delrot S., Lauvergeat V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62 2465–2483. 10.1093/jxb/erq442 [DOI] [PubMed] [Google Scholar]
- Holton T. A., Cornish E. C. (1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7 1071–1083. 10.1105/tpc.7.7.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R., Fry J. E., Hoffmann N. L., Eichholtz D. Z., Rogers S. G., Fraley R. T. (1985). A simple and general method for transferring genes into plants. Science 227 1229–1232. 10.1126/science.227.4691.1229 [DOI] [PubMed] [Google Scholar]
- Hsu C.-C., Chen Y.-Y., Tsai W.-C., Chen W.-H., Chen H.-H. (2015). Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 168 175–191. 10.1104/pp.114.254599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Khaldun A., Chen J., Zhang C., Lv H., Yuan L., et al. (2016a). A R2R3-MYB transcription factor regulates the flavonol biosynthetic pathway in a traditional Chinese medicinal plant. Epimedium sagittatum. Front. Plant Sci. 7:1089 10.3389/fpls.2016.01089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Khaldun A. B., Lv H., Du L., Zhang C., Wang Y. (2016b). Isolation and functional characterization of a R2R3-MYB regulator of the anthocyanin biosynthetic pathway from Epimedium sagittatum. Plant Cell Rep. 35 883–894. 10.1007/s00299-015-1929-z [DOI] [PubMed] [Google Scholar]
- Jiang R., Tian J., Song T., Zhang J., Yao Y. (2014). The Malus crabapple transcription factor McMYB10 regulates anthocyanin biosynthesis during petal coloration. Sci. Hortic. 166 42–49. 10.1016/j.scienta.2013.12.002 [DOI] [Google Scholar]
- Jiao S. Z., Liu Y. L., Lou Q., Jiang L. (2014). Cloning and expression analysis of dihydroflavonol 4-reductase gene (DFR) from grape hyacinth (Muscari armeniacum). J. Agric. Biotechnol. 22 529–540 (in Chinese). [Google Scholar]
- Katsumoto Y., Fukuchi-Mizutani M., Fukui Y., Brugliera F., Holton T. A., Karan M., et al. (2007). Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 48 1589–1600. 10.1093/pcp/pcm131 [DOI] [PubMed] [Google Scholar]
- Kranz H. D., Denekamp M., Greco R., Jin H., Leyva A., Meissner R. C., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16 263–276. 10.1046/j.1365-313x.1998.00278.x [DOI] [PubMed] [Google Scholar]
- Lai Y.-S., Shimoyamada Y., Nakayama M., Yamagishi M. (2012). Pigment accumulation and transcription of LhMYB12 and anthocyanin biosynthesis genes during flower development in the Asiatic hybrid lily (Lilium spp.). Plant Sci. 193 136–147. 10.1016/j.plantsci.2012.05.013 [DOI] [PubMed] [Google Scholar]
- Laitinen R. A., Ainasoja M., Broholm S. K., Teeri T. H., Elomaa P. (2008). Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida. J. Exp. Bot. 59 3691–3703. 10.1093/jxb/ern216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Qiu J., Ding L., Huang M., Huang S., Yang G., et al. (2017). Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals. Plant Physiol. Biochem. 112 335–345. 10.1016/j.plaphy.2017.01.019 [DOI] [PubMed] [Google Scholar]
- Li C., Qiu J., Yang G., Huang S., Yin J. (2016). Isolation and characterization of a R2R3-MYB transcription factor gene related to anthocyanin biosynthesis in the spathes of Anthurium andraeanum (Hort.). Plant Cell Rep. 35 2151–2165. 10.1007/s00299-016-2025-8 [DOI] [PubMed] [Google Scholar]
- Liu Y., Lin-Wang K., Espley R. V., Wang L., Yang H., Yu B., et al. (2016). Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 67 2159–2176. 10.1093/jxb/erw014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Q., Liu Y., Qi Y., Jiao S., Tian F., Jiang L., et al. (2014). Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J. Exp. Bot. 65 3157–3164. 10.1093/jxb/eru168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Pooler M., Griesbach R. (2009). Anthocyanin regulatory/structural gene expression in Phalaenopsis. J. Am. Soc. Hortic. Sci. 134 88–96. [Google Scholar]
- Mancinelli A. L., Rossi F., Moroni A. (1991). Cryptochrome, phytochrome, and anthocyanin production. Plant Physiol. 96 1079–1085. 10.1104/pp.96.4.1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Prescott A., Mackay S., Bartlett J., Vrijlandt E. (1991). Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1 37–49. 10.1111/j.1365-313X.1991.00037.x [DOI] [PubMed] [Google Scholar]
- Morita Y., Saitoh M., Hoshino A., Nitasaka E., Iida S. (2006). Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol. 47 457–470. 10.1093/pcp/pcj012 [DOI] [PubMed] [Google Scholar]
- Nakatsuka T., Haruta K. S., Pitaksutheepong C., Abe Y., Kakizaki Y., Yamamoto K., et al. (2008a). Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol. 49 1818–1829. 10.1093/pcp/pcn163 [DOI] [PubMed] [Google Scholar]
- Nakatsuka T., Sato K., Takahashi H., Yamamura S., Nishihara M. (2008b). Cloning and characterization of the UDP-glucose: anthocyanin 5-O-glucosyltransferase gene from blue-flowered gentian. J. Exp. Bot. 59 1241–1252. 10.1093/jxb/ern031 [DOI] [PubMed] [Google Scholar]
- Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12 1863–1878. 10.1105/tpc.12.10.1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palapol Y., Ketsa S., Lin-Wang K., Ferguson I. B., Allan A. C. (2009). A MYB transcription factor regulates anthocyanin biosynthesis in mangosteen (Garcinia mangostana L.) fruit during ripening. Planta 229 1323–1334. 10.1007/s00425-009-0917-3 [DOI] [PubMed] [Google Scholar]
- Paz-Ares J., Ghosal D., Wienand U., Peterson P., Saedler H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M. K., Murrell J. R., Shirley B. W. (1997). Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis (Further evidence for differential regulation of “early” and “late” genes). Plant Physiol. 113 1437–1445. 10.1104/pp.113.4.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J., Van Der Woude K., Souer E., De Vetten N., Mol J., et al. (1999). Molecular analysis of the anthocyanin 2 gene of petunia and its role in the evolution of flower color. Plant Cell 11 1433–1444. 10.1105/tpc.11.8.1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J. F., Leppen H. T., Mol J. N., Koes R. E. (1993). Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5 1497–1512. 10.1105/tpc.5.11.1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn K., Venail J., Shang Y., Mackay S., Alm V., Butelli E., et al. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18 831–851. 10.1105/tpc.105.039255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn K. E., Ngo H., Kenel F., Brummell D. A., Albert N. W., Mccallum J. A., et al. (2016). The onion (Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis. Front. Plant Sci. 7:1865 10.3389/fpls.2016.01865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Low E.-T. L., Ooi L. C.-L., Ong-Abdullah M., Nookiah R., Ting N.-C., et al. (2014). The oil palm VIRESCENS gene controls fruit colour and encodes a R2R3-MYB. Nat. Commun. 5:4106 10.1038/ncomms5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C., Quattrocchio F., Mol J. N., Koes R. (2000). anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12 1619–1631. 10.1105/tpc.12.9.1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos A. M., Jaffé F. W., Jacob S. R., Bogs J., Robinson S. P., Walker A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142 1216–1232. 10.1104/pp.106.088104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Sasaki N., Ohmiya A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54 733–749. 10.1111/j.1365-313X.2008.03447.x [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L. K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56 505–516. 10.1111/j.1365-313X.2008.03612.x [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126 485–493. 10.1104/pp.126.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D. Y., Sharma S. B., Wright E., Wang Z. Y., Dixon R. A. (2006). Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J. 45 895–907. 10.1111/j.1365-313X.2006.02655.x [DOI] [PubMed] [Google Scholar]
- Xu W., Dubos C., Lepiniec L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20 176–185. 10.1016/j.tplants.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Xu W., Grain D., Bobet S., Le Gourrierec J., Thevenin J., Kelemen Z., et al. (2014). Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 202 132–144. 10.1111/nph.12620 [DOI] [PubMed] [Google Scholar]
- Yamagishi M. (2011). Oriental hybrid lily Sorbonne homologue of LhMYB12 regulates anthocyanin biosyntheses in flower tepals and tepal spots. Mol. Breed. 28 381–389. 10.1007/s11032-010-9490-5 [DOI] [Google Scholar]
- Yamagishi M. (2015). A novel R2R3-MYB transcription factor regulates light-mediated floral and vegetative anthocyanin pigmentation patterns in Lilium regale. Mol. Breed. 36:3 10.1007/s11032-015-0426-y [DOI] [Google Scholar]
- Yamagishi M., Ihara H., Arakawa K., Toda S., Suzuki K. (2014a). The origin of the LhMYB12 gene, which regulates anthocyanin pigmentation of tepals, in Oriental and Asiatic hybrid lilies (Lilium spp.). Sci. Hortic. 174 119–125. 10.1016/j.scienta.2014.05.017 [DOI] [Google Scholar]
- Yamagishi M., Toda S., Tasaki K. (2014b). The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.). New Phytol. 201 1009–1020. 10.1111/nph.12572 [DOI] [PubMed] [Google Scholar]
- Yamagishi M., Shimoyamada Y., Nakatsuka T., Masuda K. (2010). Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol. 51 463–474. 10.1093/pcp/pcq011 [DOI] [PubMed] [Google Scholar]
- Yamagishi M., Yoshida Y., Nakayama M. (2011). The transcription factor LhMYB12 determines anthocyanin pigmentation in the tepals of Asiatic hybrid lilies (Lilium spp.) and regulates pigment quantity. Mol. Breed. 30 913–925. 10.1007/s11032-011-9675-6 [DOI] [Google Scholar]
- Yin X. R., Allan A. C., Chen K. S., Ferguson I. B. (2010). Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol. 153 1280–1292. 10.1104/pp.110.157081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Butelli E., Martin C. (2014). Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 19 81–90. 10.1016/j.pbi.2014.05.011 [DOI] [PubMed] [Google Scholar]
- Zimmermann I. M., Heim M. A., Weisshaar B., Uhrig J. F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40 22–34. 10.1111/j.1365-313X.2004.02183.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


