Streptococcus pneumoniae is a common resident in the human nasopharynx. However, carriage can result in severe diseases due to a unique repertoire of pathogenicity factors that are rare in closely related commensal streptococci. We investigated a penicillin-resistant S. pneumoniae clone of serotype 23F isolated from a cystic fibrosis patient on multiple occasions over an unusually long period of over 3 years that was present without causing disease. Genome comparisons revealed an apparent nonfunctional pneumococcus-specific gene encoding a hyaluronidase, supporting the view that this enzyme adds to the virulence potential of the bacterium. The 23F clone harbored unique mosaic genes encoding penicillin resistance determinants, the product of horizontal gene transfer involving the commensal S. mitis as donor species. Sequences identical to one such mosaic gene were identified in an S. mitis strain from the same patient, suggesting that in this case S. pneumoniae played the role of donor.
KEYWORDS: 23F clone, Streptococcus pneumoniae, cystic fibrosis, hyaluronidase, penicillin-binding proteins, persistence
ABSTRACT
Streptococcus pneumoniae isolates of serotype 23F with intermediate penicillin resistance were recovered on seven occasions over a period of 37 months from a cystic fibrosis patient in Berlin. All isolates expressed the same multilocus sequence type (ST), ST10523. The genome sequences of the first and last isolates, D122 and D141, revealed the absence of two phage-related gene clusters compared to the genome of another ST10523 strain, D219, isolated earlier at a different place in Germany. Genomes of all three strains carried the same novel mosaic penicillin-binding protein (PBP) genes, pbp2x, pbp2b, and pbp1a; these genes were distinct from those of other penicillin-resistant S. pneumoniae strains except for pbp1a of a Romanian S. pneumoniae isolate. All PBPs contained mutations that have been associated with the penicillin resistance phenotype. Most interestingly, a mosaic block identical to an internal pbp2x sequence of ST10523 was present in pbp2x of Streptococcus mitis strain B93-4, which was isolated from the same patient. This suggests interspecies gene transfer from S. pneumoniae to S. mitis within the host. Nearly all genes expressing surface proteins, which represent major virulence factors of S. pneumoniae and are typical for this species, were present in the genome of ST10523. One exception was the hyaluronidase gene hlyA, which contained a 12-nucleotide deletion within the promoter region and an internal stop codon. The lack of a functional hyaluronidase might contribute to the ability to persist in the host for an unusually long period of time.
IMPORTANCE Streptococcus pneumoniae is a common resident in the human nasopharynx. However, carriage can result in severe diseases due to a unique repertoire of pathogenicity factors that are rare in closely related commensal streptococci. We investigated a penicillin-resistant S. pneumoniae clone of serotype 23F isolated from a cystic fibrosis patient on multiple occasions over an unusually long period of over 3 years that was present without causing disease. Genome comparisons revealed an apparent nonfunctional pneumococcus-specific gene encoding a hyaluronidase, supporting the view that this enzyme adds to the virulence potential of the bacterium. The 23F clone harbored unique mosaic genes encoding penicillin resistance determinants, the product of horizontal gene transfer involving the commensal S. mitis as donor species. Sequences identical to one such mosaic gene were identified in an S. mitis strain from the same patient, suggesting that in this case S. pneumoniae played the role of donor.
INTRODUCTION
Streptococcus pneumoniae is a common member of the commensal flora of the nasopharynx, particularly in children. Carriage rates between 5% and 20% have been observed in healthy children in Europe and the United States (1–3); however, high rates of over 80% are reported occasionally (4–6) and especially in developing countries (7, 8). Carriage may lead to a variety of diseases, such as otitis media, pneumonia, septicemia, and meningitis, especially in young children, elderly people, and immunocompromised patients (for a review, see reference 9). In fact, pneumococcal infections cause more deaths than other infectious diseases worldwide. The pathogenic potential distinguishes S. pneumoniae from other members of the group of viridans streptococci (10).
Numerous virulence factors of S. pneumoniae have been described (for a review, see references 11 and 12), but most of them are also present in the closely related commensal Streptococcus species S. mitis, S. pseudopneumoniae, and S. oralis (13–15). Typical for S. pneumoniae is the polysaccharide capsule, which is crucial for the pathogenicity of this species. Over 90 capsular types have been reported, based on biochemical and genetic analyses (16), and the potential to cause disease depends on the serotype (ST) (17). Before introduction of pneumococcal conjugate vaccines (PCVs), only a few serotypes accounted for the majority of invasive diseases, including types 4, 6B, 9V, 14, 18C, 19F, and 23F. The prevalence of serotypes changed after introduction of the first seven-valent conjugated vaccine (PCV7) in 2000, which covered serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F, followed later by PSV10, which also included serotypes 1, 5, and 7F, and by PCV13, with the additional serotypes 3, 6A, and 19A. Vaccination was accompanied by the appearance of antibiotic-resistant clones expressing nonvaccine serotypes (18). Other important virulence factors present in most S. pneumoniae strains are the pneumolysin Ply, choline-binding proteins (CBPs), which include the autolysin LytA as well as the variable CBPs PspA, PspC, and PspA, and the hyaluronidase HlyA (11, 12).
Due to its ability for genetic transformation, the genomes of S. pneumoniae isolates are highly diverse and include a large accessory genome. The increasing number of available genome sequences has provided an insight into the astounding repertoire of genes available in the pan-genome of pneumococcus (19, 20). The current standard for the definition of clones is based on comparative sequence analysis of housekeeping genes, which are part of the core genome common to all strains of the species; the methods is termed multilocus sequence typing (MLST) (21). The Streptococcus pneumoniae MLST database (https://pubmlst.org/spneumoniae) listed 13,126 STs in February 2017. Different capsular serotypes may be found within one ST due to capsule switching (22, 23). Genomes of an identical ST may vary considerably in their accessory genome content (24).
We report here on a rare clone of serotype 23F S. pneumoniae representing isolates with intermediate penicillin resistance which have been collected over a period of over 3 years from a patient in Berlin, Germany, with cystic fibrosis (CF); presence of the clone was not associated with disease. The genome sequences of three isolates of the same clone, including one isolate obtained from a different hospital in Germany, were used for comparative analysis of penicillin resistance determinants, the penicillin-binding proteins PBP2x, PBP1a, and PBP2b, and the main pneumococcal virulence factors.
RESULTS
Twenty-nine S. pneumoniae isolates were obtained between 1992 and 1995 from the Wannsee-Lungenklinik-Heckeshorn in Berlin. Seven of these isolates were recovered from one CF patient over a period of 37 months and were not associated with disease (Table 1). All seven strains expressed serotype 23F and showed identical antibiotic resistance patterns that were distinct from patterns of the other 22 strains (data not shown). MLST revealed that these seven isolates were members of the same clone of a new ST, ST10523. Screening of our strain collection detected another member of ST10523, strain D219, which was isolated in Leipzig, Germany, in 1989 (25). In order to see whether special virulence factors are associated with this clone and whether the isolates from the CF patient differed from D219, the genomes of the first (D122) and last (D141) isolate from Berlin and of D219 from Jena were sequenced.
TABLE 1 .
Bacterial strainsa
| Species and isolate no.b | Date of isolation (day/mo/yr) | Site | ST | MIC (µg/ml) |
TET/CLO/ERY susceptibility | ||
|---|---|---|---|---|---|---|---|
| PEN-G | CTX | OXA | |||||
| S. pneumoniae (ST23F) | |||||||
| D122 | 27/07/1992 | Nasopharynx | 10523 | 0.19–0.25 | 0.125–0.19 | 4–6 | S/S/S |
| D127 | 8/4/1994 | Nasopharynx | 10523 | 0.19–0.25 | 0.125–0.19 | 4–6 | S/S/S |
| D128 | 25/07/1994 | Nasopharynx | 10523 | 0.19–0.25 | 0.125–0.19 | 4–6 | S/S/S |
| D134 | 31/10/1994 | Nasopharynx | 10523 | 0.19–0.25 | 0.125–0.19 | 4–6 | S/S/S |
| D136 | 9/1/1995 | Nasopharynx | 10523 | 0.19–0.25 | 0.125–0.19 | 4–6 | S/S/S |
| D139 | 9/5/1995 | Sputum | 10523 | 0.19–0.25 | 0.125–0.19 | NDd | S/S/S |
| D141 | 1/8/1995 | Nasopharynx | 10523 | 0.19–0.25 | 0.125–0.19 | 4–6 | S/S/S |
| D219 | 1989 | Throat | 10523 | 0.19–0.25 | 0.125–0.19 | ND | S/S/S |
| S. mitis | |||||||
| B8 | 1995 | Oral cavity | 0.12–0.2 | 0.006–0.03 | 0.5 | S/S/S | |
| B93-4c | 1995 | Oral cavity | ND | ND | ND | ND | |
| B10 | 1995 | Oral cavity | 0.47 | 0.23 | 0.38 | S/S/S | |
| S. oralis | |||||||
| B11 | 1995 | Oral cavity | 8 | 2 | 96 | R/S/(R) | |
S, sensitive; R, resistant; (R) intermediate resistant. Drug abbreviations: PEN-G, benzylpenicillin; CTX, cefotaxime; OXA, oxacillin; TET, tetracycline; CLO, chloramphenicol; ERY, erythromycin.
All isolates were obtained from the Wannsee-Lungenklinik-Heckeshorn, Berlin, and from the same patient, except for D219, which was isolated in Leipzig (25).
The strain could not be recovered after DNA isolation.
ND, not determined.
Antibiotic resistance and penicillin-binding proteins.
All seven ST10523 strains had intermediate resistance to beta-lactam antibiotics but were sensitive to tetracycline, erythromycin, and chloramphenicol (Table 1). Since PBP2x, PBP2b, and PBP1a play key roles in penicillin resistance, these genes were analyzed in detail to see whether they are related to PBP alleles of other penicillin-resistant S. pneumoniae (PRSP) strains. The PBP sequences of D219, D122, and D141 were identical and contained sequence blocks that diverged from PBP genes of the penicillin-sensitive R6 laboratory strain by ~20%. They did not match any other PBP sequences in the NCBI database except for pbp1a (see below). Interestingly, the genomic regions containing pbp2x (spr302 to spr307; yllC to clpL) and pbp2b (spr1513 to spr1517; mutT to pbp2b) contained a significantly larger amount of single nucleotide polymorphisms (SNPs; >5.3%) in the ST10523 strains than in the entire genome of the R6 strain, excluding variable genes (<0.7%), indicating transfer of large sequence blocks flanking the PBP genes. Similar observations have been reported previously (26). In contrast, flanking genes of pbp1a showed no signs of recombination events.
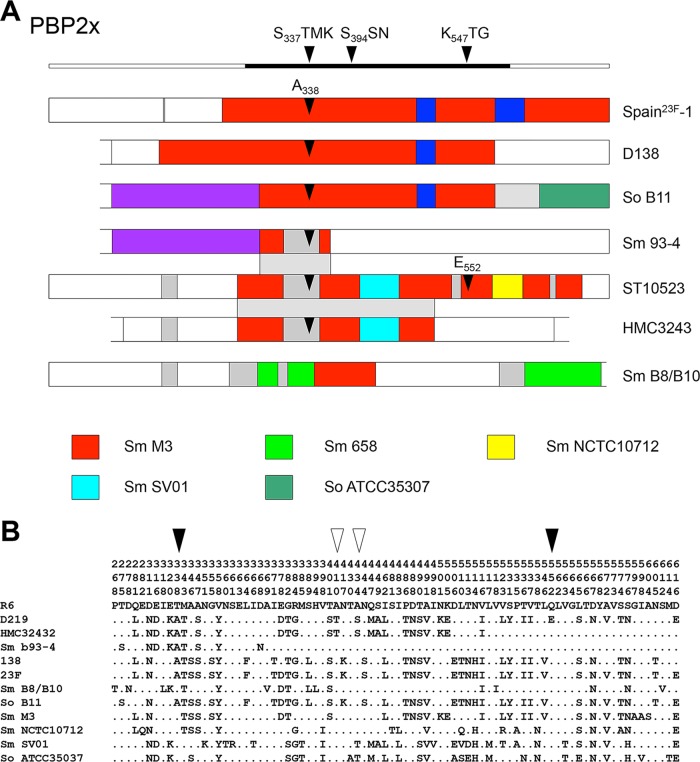
The PBP2x gene of ST10523 has a complex mosaic structure (Fig. 1). It contains sequence blocks that are highly related (<3% difference on the DNA level) to pbp2x of putative penicillin-sensitive donor S. mitis strains M3, SV01, and NCTC10712 (27), in addition to other divergent sequences of unknown origin (Fig. 1A). The deduced protein sequence included mutations A338 and E552, which are known to confer resistance to beta-lactams; both amino acid changes are frequent in clinical PRSP isolates. The only other 2 amino acids that did not match any of the sensitive reference S. mitis strains were T410, present in pbp2x of low-level-resistant viridans streptococci, and T434, which occurs in some penicillin-sensitive strains (Fig. 1B) (27).
FIG 1 .
Schematic representation of the mosaic genes for PBP2x, PBP1a, and PBP2b from S. pneumoniae and S. mitis strains. (A) Mosaic sequence blocks in PBP2x. PBP2x genes of ST10523 (S. pneumoniae strains D122, D139, D141, and D219) are identical; pbp2x of S. pneumoniae Spain23F-1 is included for comparison. The pbp2x gene of S. pneumoniae HMC3243 from Romania, which is partially identical to pbp2x of ST10523, was also included. The colors indicate closely related sequence portions with >95% identity to pbp2x reference sequences from penicillin-sensitive S. mitis (Sm.) strains M3, 658, NCTC10712, and SV01 and S. oralis (So) ATCC 35307 (76), as indicated by the color coded boxes. White indicates sequences related to the penicillin-sensitive strain S. pneumoniae R6, and gray sequences are highly divergent from R6 sequences and other pbp2x reference sequences. The domain structure of PBP2x with the three homology boxes is shown above; the central penicillin-binding domain is indicated by the black bar. Black arrows represent mutations A338 and E552 in pbp2x. The regions that are identical between pbp2x of ST10523, S. mitis B93-4, and S. pneumoniae HMC3243 are indicated by the shaded area. (B) Deduced amino acid sequences of the transpeptidase domain of PBP2x; PBP2x of S. pneumoniae R6 was used as a reference. The positions of the amino acids are indicated vertically in the top three rows. Only positions with altered residues are shown; residues identical to those in strain R6 are indicated by dots. The mutations A338 and E552 are highlighted by black arrowheads; white arrowheads indicate amino acids which are not present in PBP2x of the penicillin-sensitive reference strains (see text for details).
Furthermore, we obtained pbp2x sequences from four commensal streptococcus isolates from the same patient, isolates D122 and D141 (Table 1). The PBP2x genes of S. mitis strains B8 and B10 were identical and were distinct from pbp2x of S. oralis B11 and that of S. pneumoniae ST10523. The gene pbp2xB11 contained a central sequence block almost identical to that of Spain23F-1, and 3′-sequences were related to S. oralis ATCC 35307 (Fig. 1A). BLAST searches revealed one Romanian isolate, HMC3243, of unknown serotype (28) which was partially identical to D219 up to codon 517, whereas the C-terminal part represented R6 sequences (Fig. 1A and B). Interestingly, pbp2x of S. mitis B93-4 contained a 284-nucleotide (nt) sequence block almost identical to PBP2xD219 (codons 284 to 377), including two SNPs resulting in 1 amino acid change, D368N, suggesting that this block was acquired from S. pneumoniae ST10523 (Fig. 1A). The flanking 5′-sequence block was identical to S. oralis B11, and the 3′-end was identical to the S. pneumoniae R6 sequence, documenting multiple interspecies gene transfer events.
The internal mosaic blocks of ST10523 pbp2b and pbp1a were highly related to the respective genes in S. mitis NCTC10712 (see Fig. S1 in the supplemental material). PBP2b contained the mutation A446 close to the conserved S443SN motif, which mediates low resistance levels (29) and which is present in most penicillin-resistant isolates. Moreover, it had one more amino acid, Y430, that resulted in a deduced protein of 681 residues (Fig. S1). Regarding PBP1a of ST10523, the change of four consecutive residues, T574SQF to NTGY, has been associated with penicillin resistance, but the mutation A371 within the active site motif S470TMK, which also has been implicated in penicillin resistance, was not present (30–32). The PBP1a gene of ST10523 was identical to that of S. pneumoniae strain HMC3243 (Fig. S1) except for three silent SNPs. In contrast, the mosaic structure of pbp2b was entirely different. This clearly indicated that the three PBP genes were acquired from different sources or occasions in S. pneumoniae HMC3243 compared to ST10523. In summary, all three PBPs contained amino acid mutations that have been associated with the resistance phenotype corresponding to the intermediate penicillin resistance of ST10523.
Mosaic structure of PBP1a and PBP2b of S. pneumoniae clone ST10523 and strain HMC3243. The homology boxes of PBP1a and PBP2b are shown above the mosaic genes. The mutation A446 in PBP2b is represented by the black arrow; the white arrow shows an additional amino acid. The colors indicate closely related sequences with >95% identity to the pbp2x reference sequences from penicillin-sensitive S. mitis strains M3 and NCTC10712, as indicated by the color coded boxes. Download FIG S1, TIF file, 6.6 MB (6.7MB, tif) .
Copyright © 2017 Rieger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic comparison of S. pneumoniae genomes.
Since ST10523 is a new sequence type, the 2,050,063-nt draft genome of strain D219 was first compared to the R6 genome. Genes with no match in R6 were then used in a BLAST search of the NCBI database. Excluding transposases, the genome of D219 differed from the R6 genome by ~8%, including parts of a bacteriocin cluster (SPND219_00557 to SPND219_00567), the cps biosynthesis cluster encoding the 23F capsule (SPND219_00380 to SPND219_00398), and two clusters encoding phage-related genes (SPND219_00003 to SPND219_00023 [phage relict] and SPND219_01526 to SPND219_01585 [prophage]). This percentage corresponds to data obtained in comparative genomic hybridization on an oligonucleotide microarray representing the TIGR4 genome (33). Large parts of the phage relict were present in several genomes of S. pneumoniae. The prophage shows high similarity to S. pneumoniae phage 040922 (GenBank accession number FR671406), which is associated with a Tn916-like element in one S. pneumoniae strain, 18C/3 (34). The prophage contains two large genes (fragments SPND219_01501-3 and SPND219_01535) that encode the surface-expressed tail fiber PblB and the tape measure protein PblA. Homologues of PblA and PblB of S. mitis phage SM1 have been shown to be involved in the platelet-binding activity of S. mitis SF100 (35) and for its virulence in an animal model of infective endocarditis (36). No genes exclusively carried by ST10523 could be detected in BLAST searches.
Between the genome of D219 on one hand and D122 and D141 on the other hand, little difference regarding gene content was noted. The only exceptions were the phage relict and the prophage mentioned above, which were absent in D122 and D141. One gene cluster, SPND122_00705 to SPND122_00709 and SPND141_00707 to SPND141_00711 related to the R6 genes spr0623 to spr0627 (ABC transporter, lactate monooxygenase, 2-lysyl-tRNA synthetase) were not found in D219. However, since these genes are all located on small contigs, including repeat elements such as BOX and RUP (37, 38), which result in problems during the genome assembly process, verification of their absence in D219 will require further analyses.
The genomes of D122 and D141 differed from each other by approximately 0.01% SNPs in 49 genes, less than that found in D219 (0.02 to 0.024% affecting a total of 178 genes in D219), i.e., the isolates from the patient are more closely related to each other than to D219, in agreement with their distinct place of isolation. One clear difference was observed in the gene encoding the two-component sensor kinase HK07 (39), which is not essential in S. pneumoniae (40). It was intact in strains D122 and D219 (SPND122_00180 and SPND219_00200), whereas an ISL3 family transposase fragment was inserted into the D141 gene, resulting in three incomplete gene fragments, SPND141_00182 to SPND141_00184.
Virulence genes in ST10523.
All genes encoding the main S. pneumoniae specific virulence factors lytA, ply, Nana, and nanB and the CBPs pspC, pspA, and pcpC were present in the three ST10523 genomes. The deduced protein sequences were identical to each other and identical or highly similar to R6 proteins except for pspA, which represented a distinct genetic variant in ST10523. Differences in the repeat regions in genes encoding CBPs were not considered, since they are most likely caused by assembly problems of the choline-binding repeat regions. Other genes encoding surface proteins of important biological function, pavA and, for the IgA proteases, zmpB and zmpA were also present and identical in all three strains, except for the IgA protease, which contained gaps in the sequence of D219 and two SNPs plus a single-nucleotide deletion in D122, resulting in a premature stop codon. No pilus cluster was detectable. The capsule cluster SPND219_00380 to SPND219_00394 differed from that of S. pneumoniae strain Spain23F-1 by 20 SNPs, as expected for genes of distinct clonal origins. The last four-gene rml operon encoding enzymes involved in dTDP-rhamnose synthesis and which is common to several serotypes (41) included two short divergent regions in rmlB identical to rmlB of many serotype 19F strains.
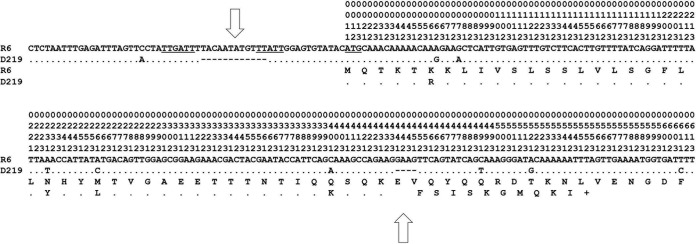
However, all three ST10523 genomes contained significant differences in hlyA, which encodes the hyaluronidase (SPND219_00348, SPND122_00329, and SPND141_00331) and which are unusual in other S. pneumoniae genomes. Within the HlyA gene, a 4-bp deletion corresponding to the position 128 nt downstream of the putative ATG start codon of R6 resulted in a premature stop codon. Moreover, a 12-nt deletion within the promoter region 10 nt upstream of the ATG start codon was present (Fig. 2). This strongly suggests that no functional hyaluronidase is expressed in ST10523 strains. Searches in the whole-genome contig NCBI database revealed another six genomes which contained this peculiarity (Table 2), but they differed by up to four SNPs from the D219 region shown in Fig. 2.
FIG 2 .
The HlyA gene of S. pneumoniae R6 and ST10523. The HlyA gene of S. pneumoniae R6, including upstream sequences (57 nt) to codon 72, is indicated. The −35 region, the −10 region, and the start codon ATG are underlined. The deduced amino acid sequence is indicated below. In the hlyA region of D219 (ST10523), only nucleotides and deduced amino acids that differ from the reference R6 sequence are shown. The 12-nt deletion upstream of the ATG start codon and the 4-nt deletion in the D219 sequence are indicated by open arrows. Vertical numbers in the first three rows refer to codons; numbers 1, 2, and 3 in the fourth row indicate the first, second, and third positions in the respective codon.
TABLE 2 .
S. pneumoniae genome sequences with an incomplete hlyA
| Accession no. | Strain | Serotype | Source | Date (day/mo/yr) | Country (region or city) |
|---|---|---|---|---|---|
| LJVO01000185 | NTPn 4 | NT | Blood | 2004 | South Africa (KwaZulu-Natal) |
| CVHP01000011 | 0338 | NT | Blood | 2001 | USA (Alaska) |
| CKDL01000005 | Type strain | NT | Nasopharynx | 14/4/2008 | Thailand (Maela) |
| CPLS01000001 | LMG205 | 6B | Not known | 2008 | Thailand |
| CRPU01000001 | SMRU824 | NT | Nasopharynx | 10/10/2008 | Thailand (Maela) |
| AGOE01000004 | GA16531 | NAa | NA | 2001 | USA (metropolitan Atlanta) |
| CFNW01000018 | 6378-99 | 19F | Not known | 1999 | USA (Tennessee) |
NA, not available.
DISCUSSION
Prolonged carriage of the same S. pneumoniae clone for a period of 37 months, as observed for serotype 23F isolates obtained from a CF patient in our study, is unusual. S. pneumoniae is not considered a persistent colonizer in CF patients, unlike Pseudomonas aeruginosa and Staphylococcus aureus (42–44). Long-term persistence has been reported for Staphylococcus aureus for up to 70 months (45). The duration of pneumococcal carriage in healthy children is a few weeks, ranging from 2 days and in rare cases up to 6 to 12 months, depending on the age of the carrier and the serotype of the isolate (2, 3, 46–48). Some serogroups, including serotype 23F, are generally carried for longer periods than other serogroups (46, 49) and have a low propensity to cause invasive disease (50). Interestingly, an inverse relationship between the attack rate of a given capsular serotype and its duration of carriage has been noted (3).
Carriage rates for S. pneumoniae isolated from CF patients are similar, with colonization rates ranging between 3 and 20% (51–56). No special serotypes appear to be associated with CF (57), but some serotypes may be more common, depending on the geographic area. Serotype 23F isolates were prevalent in a CF unit in Madrid, mainly due to the clone Spain23F-1 of a varied multiresistance phenotype (52). Serotype 3 prevailed in another study which reported no 23F serotype isolates in a CF center in Rome, probably because all patients had received vaccination (58). In both studies, S. pneumoniae was recovered more than once from some patients. However, only three strains of serotype 23F isolated over a period of 3 months showed identical SmaI restriction patterns, revealed by pulsed-field gel electrophoresis (PFGE), and the same antibiotic resistance profile, and thus most likely represented members of the same clone (52). Three patients carried S. pneumoniae with the same serotype and identical SmaI PFGE pattern for 1 to 8 months (58). In these cases, S. pneumoniae was considered a colonizer, since at the time of isolation the patients showed no evidence of pulmonary exacerbation.
Genomic comparisons showed that the two strains, D122 and D141, from the CF patient are more closely related to each other than to D219, which was isolated 3 years before from a different geographical site. They differed from D219 by the absence of two large gene clusters encoding a prophage and a phage relict, by the presence of a five-gene cluster, including an amino acid ABC transporter, and by the estimated number of SNPs. The prophage carries two genes encoding large proteins PblA and PblB. Homologues of these proteins are frequent in S. pneumoniae phages (59) and have been shown to play a role in adhesion and virulence in S. mitis (35, 36). It these proteins play a similar role in S. pneumoniae, it is conceivable that their absence in D122 and D141 supports extended carriage.
The variation between D122 and D141 concerned only 49 genes, and D141 contained an insertion of an ISL3 family transposase fragment into the gene encoding the histidine protein kinase HK07, which was absent in the D122 gene. This element was present at another three sites in the D141 genome and at one site in the D122 genome, whereas it could not be detected in D219. Other studies have supported little genomic variation during carriage of the same S. pneumoniae clone. Minimal variation was observed during carriage established experimentally with a single serotype 6B strain of ST138 (60). The maximum SNP distance between any of the 229 isolates obtained over a period of 35 days versus the reference strain was three SNPs (60). It should be noted that the genomic comparison between two isolates of strain D39, a historically important serotype 2 isolate from the early 1940s (61) and which have been cultivated separately for at least 21 years, revealed only five mutations (62). Similarly, some strains isolated during a 7-month period from a child with chronic pneumococcal infection varied by only ≤30 SNPs (63). However, those authors noted there were also multiple events of horizontal gene transfer in some strains, which most likely occurred during polyclonal infection. In contrast, we saw no evidence of gene acquisition in D141 versus D122, and the S. mitis strain B93-4 contained a pbp2x fragment identical to pbp2x of ST10523. The mosaic PBP2x and PBP2b genes of ST10523 represent new gene variants and are distinct from all others found in the NCBI database. Therefore, this finding indicates interspecies gene transfer from S. pneumoniae to S. mitis in the same host.
No genes specifically associated with ST10523 genomes were identified. This is not astounding, given the vast number of actually available genomes of S. pneumoniae. Based on a pan-genome analysis of 158 S. pneumoniae genomes, it has been predicted that only 0.3 new genes will be discovered in a new genome if a data set from 1,000 genomes is already available, and only 0.06 new genes will be discovered from 5,000 genomes (24). However, an unusual hyaluronidase gene, hlyA, was present in the ST10523 genomes which contained a stop codon (Fig. 2) distinct from that described in a serotype 3 clone, ST180, where an SNP at position 376 of the hyaluronidase coding sequence resulted in a stop codon and truncation of the protein after 125 amino acids (24). Moreover, a 12-nt deletion within the promoter region 10 nt upstream of the ATG was detected. Hyaluronidase is produced by almost all clinical isolates of pneumococci (64). It is one of the genes which have not been found in closely related viridans streptococci, except for some S. oralis isolates, but not in S. mitis strains (65), i.e., it represents a typical component of the species S. pneumoniae. The enzyme depolymerizes hyaluronic acid, which is an important component of the host connective tissue and extracellular matrix (66). No significant impact on virulence was found in a mouse intraperitoneal infection model when a single hlyA mutant was used (67); however, when a double mutant that was also deficient for the pneumolysin gene ply was tested, virulence was significantly decreased (68). Hyaluronidase significantly potentiated pneumolysin-mediated ciliary slowing and epithelial damage in an in vitro model, suggesting that its presence favors colonization and subsequently extrapulmonary dissemination of the pneumococcus (69). It is therefore tempting to assume that the lack of a functional hyaluronidase predestines ST10523 to survive within the host for an extended period without causing disease. In conclusion, the unusually long carriage rate observed for S. pneumoniae isolates D122 and D141 might not be related to mutations or genetic variants acquired during persistence in the human host, but rather to the loss of a large prophage carrying potential virulence factors and to the absence of a complete hyaluronidase gene.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. pneumoniae strains D122 to D141, S. mitis B8, B93-4, and B10, and S. oralis B11 (Table 1) were part of a strain collection obtained from the Wannsee-Lungenklinik-Heckeshorn in Berlin; strain D219, isolated in Leipzig, has been described elsewhere (25). Strains were grown at 37°C without aeration in complex C medium (70) supplemented with 0.1% yeast extract (C + Y). MICs were determined by the agar dilution method (for beta-lactams) or by using E-test strips for all other antibiotics (Oxoid GmbH, Basingstoke, United Kingdom) on d-agar plates supplemented with 3% sheep blood (71).
DNA sequencing and analysis.
Chromosomal DNA from streptococci was isolated as described previously (72). Internal sequences of the seven housekeeping genes were obtained with primers described on the Streptococcus pneumoniae MLST homepage (https://pubmlst.org/spneumoniae). PBP2x gene fragments were amplified with the primers pn2xup and pn2xdown (72), and direct sequencing of PCR products was performed with consecutive primers. PCR products were purified using a JetQuick DNA purification kit (GenoMed). PCRs were performed using either Goldstar Red Taq polymerase (Eurogentec) or DreamTaq polymerase (Fermentas), according to the manufacturer instructions. The genomes of D219, D122, and D144 were sequenced using a 454 Life Sciences FLX sequencer, and reads were assembled by the 454 Newbler Assembler version 2.6. Contigs were aligned to the S. pneumoniae R6 genome sequence (73). The rapid annotation subsystem technology (RAST) server (74) designed for annotation of bacterial and archaeal genomes was applied to obtain EMBL-formatted files containing protein, tRNA, and rRNA annotations from a large set of several output formats.
DNA analysis and bioinformatic tools.
For the analysis of SNPs in the three ST10523 genomes, only sequences that were 350 nt from contig ends were included, to avoid potential errors generated by the 454-generated sequences. Individual open reading frames were investigated manually and compared with other genome sequences by using BLAST analyses and the NCBI database (nucleotides and whole-genome contigs). As a reference for the serotype 23F capsule cluster, genes of strain ATCC (Spain23F-1) were used (75). Alignments were prepared using Clustal X2 (76). Codon sites were included manually and trimmed by using the program Clustal Formatter 3 (http://nbc11.biologie.uni-kl.de/sequence_analysis/ClustalFormatter3/documentation.html) to reveal only sites that differ from the reference sequence shown in Fig. 1B.
Accession number(s).
The following sequences have been submitted to GenBank and assigned the following accession numbers (shown in parentheses): for genomes, D219 (CP016227), D122 (CP016632), D141 (CP016633); for PBP genes of S. pneumoniae strain HMC3243, pbp2x (FJ439546), pbp1a (FJ439538), and pbp2b (FJ439554) (28); for PBP2x genes, S. mitis strain B8 (KY292528), strain SV01 (KY292540), and strain B93-4 (KY783589), and S. oralis strain B11 (KY783587).
ACKNOWLEDGMENTS
We thank Shwan Rachid for help with the MLST analysis and Jennifer Loewe for DNA sequencing of PBP genes.
This work was supported by the Bundesministerium für Bildung und Forschung (grant 0313801 1) and the Deutsche Forschungsgemeinschaft (grant Ha 1011/11-3 to R.H.).
REFERENCES
- 1.Short KR, Diavatopoulos DA. 2015. Nasopharyngeal colonization with Streptococcus pneumoniae, p 279–291. In Brown J, Hammerschmidt S, Orihuela C (ed), Streptococcus pneumoniae molecular mechanisms of host-pathogen interactions. Academic Press, London, United Kingdom. [Google Scholar]
- 2.Högberg L, Geli P, Ringberg H, Melander E, Lipsitch M, Ekdahl K. 2007. Age- and serogroup-related differences in observed durations of nasopharyngeal carriage of penicillin-resistant pneumococci. J Clin Microbiol 45:948–952. doi: 10.1128/JCM.01913-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sleeman KL, Griffiths D, Shackley F, Diggle L, Gupta S, Maiden MC, Moxon ER, Crook DW, Peto TE. 2006. Capsular serotype-specific attack rates and duration of carriage of Streptococcus pneumoniae in a population of children. J Infect Dis 194:682–688. doi: 10.1086/505710. [DOI] [PubMed] [Google Scholar]
- 4.Nunes S, Sá-Leão R, Carriço J, Alves CR, Mato R, Avô AB, Saldanha J, Almeida JS, Sanches IS, de Lencastre H. 2005. Trends in drug resistance, serotypes, and molecular types of Streptococcus pneumoniae colonizing preschool-age children attending day care centers in Lisbon, Portugal: a summary of 4 years of annual surveillance. J Clin Microbiol 43:1285–1293. doi: 10.1128/JCM.43.3.1285-1293.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ercibengoa M, Arostegi N, Marimón JM, Alonso M, Pérez-Trallero E. 2012. Dynamics of pneumococcal nasopharyngeal carriage in healthy children attending a day care center in northern Spain. Influence of detection techniques on the results. BMC Infect Dis 12:69. doi: 10.1186/1471-2334-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyllie AL, Chu ML, Schellens MH, van Engelsdorp Gastelaars J, Jansen MD, van der Ende A, Bogaert D, Sanders EA, Trzciński K. 2014. Streptococcus pneumoniae in saliva of Dutch primary school children. PLoS One 9:e102045. doi: 10.1371/journal.pone.0102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jebaraj R, Cherian T, Raghupathy P, Brahmadathan KN, Lalitha MK, Thomas K, Steinhoff MC. 1999. Nasopharyngeal colonization of infants in southern India with Streptococcus pneumoniae. Epidemiol Infect 123:383–388. doi: 10.1017/S0950268899003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill PC, Cheung YB, Akisanya A, Sankareh K, Lahai G, Greenwood BM, Adegbola RA. 2008. Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian infants: a longitudinal study. Clin Infect Dis 46:807–814. doi: 10.1086/528688. [DOI] [PubMed] [Google Scholar]
- 9.Henriques-Normark B, Tuomanen EI. 2013. The pneumococcus: epidemiology, microbiology, and pathogenesis. Cold Spring Harb Perspect Med 3:a010215. doi: 10.1101/cshperspect.a010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogaert D, de Groot R, Hermans PW. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell TJ. 2003. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat Rev Microbiol 1:219–230. doi: 10.1038/nrmicro771. [DOI] [PubMed] [Google Scholar]
- 12.Hammerschmidt S. 2007. Pneumococcal virulence factors and adhesion proteins targeting the host, p 141–203. In Hakenbeck R, Chhatwal GS (ed), Molecular biology of streptococci. Horizon Press, Wymondham, Norfolk. [Google Scholar]
- 13.Denapaite D, Brückner R, Nuhn M, Reichmann P, Henrich B, Maurer P, Schähle Y, Selbmann P, Zimmermann W, Wambutt R, Hakenbeck R. 2010. The genome of Streptococcus mitis B6: what is a commensal? PLoS One 5:e9426. doi: 10.1371/journal.pone.0009426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shahinas D, Thornton CS, Tamber GS, Arya G, Wong A, Jamieson FB, Ma JH, Alexander DC, Low DE, Pillai DR. 2013. Comparative genomic analyses of Streptococcus pseudopneumoniae provide insight into virulence and commensalism dynamics. PLoS One 8:e65670. doi: 10.1371/journal.pone.0065670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denapaite D, Rieger M, Köndgen S, Brückner R, Ochigava I, Kappeler P, Mätz-Rensing K, Leendertz F, Hakenbeck R. 2016. Highly variable Streptococcus oralis strains are common among viridans streptococci isolated from primates. mSphere 1:e00041-15. doi: 10.1128/mSphere.00041-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hausdorff WP, Feikin DR, Klugman KP. 2005. Epidemiological differences among pneumococcal serotypes. Lancet Infect Dis 5:83–93. doi: 10.1016/S1473-3099(05)01280-6. [DOI] [PubMed] [Google Scholar]
- 18.Torres A, Bonanni P, Hryniewicz W, Moutschen M, Reinert RR, Welte T. 2015. Pneumococcal vaccination: what have we learnt so far and what can we expect in the future? Eur J Clin Microbiol Infect Dis 34:19–31. doi: 10.1007/s10096-014-2208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muzzi A, Donati C. 2011. Population genetics and evolution of the pan-genome of Streptococcus pneumoniae. Int J Med Microbiol 301:619–622. doi: 10.1016/j.ijmm.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Dunning Hotopp JC, Hu FZ, Riley DR, Covacci A, Mitchell TJ, Bentley SD, Kilian M, Ehrlich GD, Rappuoli R, Moxon ER, Masignani V. 2010. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol 11:R107. doi: 10.1186/gb-2010-11-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brueggemann AB, Pai R, Crook DW, Beall B. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog 3:e168. doi: 10.1371/journal.ppat.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crisafulli G, Guidotti S, Muzzi A, Torricelli G, Moschioni M, Masignani V, Censini S, Donati C. 2013. An extended multi-locus molecular typing schema for Streptococcus pneumoniae demonstrates that a limited number of capsular switch events is responsible for serotype heterogeneity of closely related strains from different countries. Infect Genet Evol 13:151–161. doi: 10.1016/j.meegid.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Tettelin H, Chancey S, Mitchell T, Denapaite D, Schähle Y, Rieger M, Hakenbeck R. 2015. Genomics, genetic variation, and regions of differences, p 81–107. In Brown J, Hammerschmidt S, Orihuela C (ed), Streptococcus pneumoniae molecular mechanisms of host-pathogen interactions. Academic Press, London, United Kingdom. [Google Scholar]
- 25.Reichmann P, Varon E, Günther E, Reinert RR, Lüttiken R, Marton A, Geslin P, Wagner J, Hakenbeck R. 1995. Penicillin-resistant Streptococcus pneumoniae in Germany: genetic relationship to clones from other European countries. J Med Microbiol 43:377–385. doi: 10.1099/00222615-43-5-377. [DOI] [PubMed] [Google Scholar]
- 26.Chewapreecha C, Marttinen P, Croucher NJ, Salter SJ, Harris SR, Mather AE, Hanage WP, Goldblatt D, Nosten FH, Turner C, Turner P, Bentley SD, Parkhill J. 2014. Comprehensive identification of single nucleotide polymorphisms associated with beta-lactam resistance within pneumococcal mosaic genes. PLoS Genet 10:e1004547. doi: 10.1371/journal.pgen.1004547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Linden M, Otten J, Bergmann C, Latorre C, Linares J, Hakenbeck R. 2017. PBP2x in Streptococcus pseudopneumoniae: an insight into the diversity of PBP2x alleles and mutations in viridans streptococci. Antimicrob Agents Chemother 61:e02646-16. doi: 10.1128/AAC.02646-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosowska-Shick K, McGhee P, Appelbaum PC. 2009. Binding of faropenem and other beta-lactam agents to penicillin-binding proteins of pneumococci with various beta-lactam susceptibilities. Antimicrob Agents Chemother 53:2176–2180. doi: 10.1128/AAC.01566-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grebe T, Hakenbeck R. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob Agents Chemother 40:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith AM, Klugman KP. 2003. Site-specific mutagenesis analysis of PBP 1A from a penicillin-cephalosporin-resistant pneumococcal isolate. Antimicrob Agents Chemother 47:387–389. doi: 10.1128/AAC.47.1.387-389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith AM, Klugman KP. 1998. Alterations in PBP 1A essential for high-level penicillin resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 42:1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Job V, Di Guilmi AM, Martin L, Vernet T, Dideberg O, Dessen A. 2003. Structural studies of the transpeptidase domain of PBP1a from Streptococcus pneumoniae. Acta Crystallogr D Biol Crystallogr 59:1067–1069. doi: 10.1107/S0907444903006954. [DOI] [PubMed] [Google Scholar]
- 33.Hakenbeck R, Balmelle N, Weber B, Gardès C, Keck W, de Saizieu A. 2001. Mosaic genes and mosaic chromosomes: intra- and interspecies genomic variation of Streptococcus pneumoniae. Infect Immun 69:2477–2486. doi: 10.1128/IAI.69.4.2477-2486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyres KL, van Tonder A, Lambertsen LM, Hakenbeck R, Parkhill J, Bentley SD, Brueggemann AB. 2013. Evidence of antimicrobial resistance-conferring genetic elements among pneumococci isolated prior to 1974. BMC Genomics 14:500. doi: 10.1186/1471-2164-14-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bensing BA, Rubens CE, Sullam PM. 2001. Genetic loci of Streptococcus mitis that mediate binding to human platelets. Infect Immun 69:1373–1380. doi: 10.1128/IAI.69.3.1373-1380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell J, Siboo IR, Takamatsu D, Chambers HF, Sullam PM. 2007. Mechanism of cell surface expression of the Streptococcus mitis platelet binding proteins PblA and PblB. Mol Microbiol 64:844–857. doi: 10.1111/j.1365-2958.2007.05703.x. [DOI] [PubMed] [Google Scholar]
- 37.Oggioni MR, Claverys JP. 1999. Repeated extragenic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology 145:2647–2653. doi: 10.1099/00221287-145-10-2647. [DOI] [PubMed] [Google Scholar]
- 38.Martin B, Humbert O, Càmara M, Guenzi E, Walker J, Mitchell T, Andrew P, Prudhomme M, Alloing G, Hakenbeck R, Morrison DA, Boulnois GJ, Claverys J-P. 1992. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res 20:3479–3483. doi: 10.1093/nar/20.13.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lange R, Wagner C, de Saizieu A, Flint N, Molnos J, Stieger M, Caspers P, Kamber M, Keck W, Amrein KE. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223–234. doi: 10.1016/S0378-1119(99)00266-8. [DOI] [PubMed] [Google Scholar]
- 40.Throup JP, Koretke KK, Bryant AP, Ingraham KA, Chalker AF, Ge Y, Marra A, Wallis NG, Brown JR, Holmes DJ, Rosenberg M, Burnham MK. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol Microbiol 35:566–576. doi: 10.1046/j.1365-2958.2000.01725.x. [DOI] [PubMed] [Google Scholar]
- 41.Morona JK, Miller DC, Coffey TJ, Vindurampulle CJ, Spratt BG, Morona R, Paton JC. 1999. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 23F. Microbiology 145:781–789. doi: 10.1099/13500872-145-4-781. [DOI] [PubMed] [Google Scholar]
- 42.Renders N, Verbrugh H, Van Belkum A. 2001. Dynamics of bacterial colonisation in the respiratory tract of patients with cystic fibrosis. Infect Genet Evol 1:29–39. doi: 10.1016/S1567-1348(01)00004-1. [DOI] [PubMed] [Google Scholar]
- 43.Iacocca VF, Sibinga M, Barbero GJ. 1963. Respiratory tract bacteriology in cystic fibrosis. Am J Dis Child 106:315–324. doi: 10.1001/archpedi.1963.02080050317012. [DOI] [PubMed] [Google Scholar]
- 44.Høiby N. 1982. Microbiology of lung infections in cystic fibrosis patients. Acta Paediatr 71:33–54. doi: 10.1111/j.1651-2227.1982.tb09640.x. [DOI] [Google Scholar]
- 45.Kahl BC, Duebbers A, Lubritz G, Haeberle J, Koch HG, Ritzerfeld B, Reilly M, Harms E, Proctor RA, Herrmann M, Peters G. 2003. Population dynamics of persistent Staphylococcus aureus isolated from the airways of cystic fibrosis patients during a 6-year prospective study. J Clin Microbiol 41:4424–4427. doi: 10.1128/JCM.41.9.4424-4427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gray BM, Converse GM III, Dillon HC Jr. 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis 142:923–933. doi: 10.1093/infdis/142.6.923. [DOI] [PubMed] [Google Scholar]
- 47.Ekdahl K, Ahlinder I, Hansson HB, Melander E, Mölstad S, Söderström M, Persson K. 1997. Duration of nasopharyngeal carriage of penicillin-resistant Streptococcus pneumoniae: experiences from the South Swedish Pneumococcal Intervention Project. Clin Infect Dis 25:1113–1117. doi: 10.1086/516103. [DOI] [PubMed] [Google Scholar]
- 48.Sá-Leão R, Nunes S, Brito-Avô A, Alves CR, Carriço JA, Saldanha J, Almeida JS, Santos-Sanches I, de Lencastre H. 2008. High rates of transmission of and colonization by Streptococcus pneumoniae and Haemophilus influenzae within a day care center revealed in a longitudinal study. J Clin Microbiol 46:225–234. doi: 10.1128/JCM.01551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith T, Lehmann D, Montgomery J, Gratten M, Riley ID, Alpers MP. 1993. Acquisition and invasiveness of different serotypes of Streptococcus pneumoniae in young children. Epidemiol Infect 111:27–39. doi: 10.1017/S0950268800056648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sá-Leão R, Pinto F, Aguiar S, Nunes S, Carriço JA, Frazão N, Gonçalves-Sousa N, Melo-Cristino J, de Lencastre H, Ramirez M. 2011. Analysis of invasiveness of pneumococcal serotypes and clones circulating in Portugal before widespread use of conjugate vaccines reveals heterogeneous behavior of clones expressing the same serotype. J Clin Microbiol 49:1369–1375. doi: 10.1128/JCM.01763-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foweraker J. 2009. Recent advances in the microbiology of respiratory tract infection in cystic fibrosis. Br Med Bull 89:93–110. doi: 10.1093/bmb/ldn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.del Campo R, Morosini MI, de la Pedrosa EG, Fenoll A, Muñoz-Almagro C, Máiz L, Baquero F, Cantón R, Spanish Pneumococcal Infection Study Network . 2005. Population structure, antimicrobial resistance, and mutation frequencies of Streptococcus pneumoniae isolates from cystic fibrosis patients. J Clin Microbiol 43:2207–2214. doi: 10.1128/JCM.43.5.2207-2214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bauernfeind A, Bertele RM, Harms K, Hörl G, Jungwirth R, Petermüller C, Przyklenk B, Weisslein-Pfister C. 1987. Qualitative and quantitative microbiological analysis of sputa of 102 patients with cystic fibrosis. Infection 15:270–277. doi: 10.1007/BF01644137. [DOI] [PubMed] [Google Scholar]
- 54.May JR, Herrick NC, Thompson D. 1972. Bacterial infection in cystic fibrosis. Arch Dis Child 47:908–913. doi: 10.1136/adc.47.256.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esposito S, Colombo C, Tosco A, Montemitro E, Volpi S, Ruggiero L, Lelii M, Bisogno A, Pelucchi C, Principi N, Italian Pneumococcal Study Group on Cystic Fibrosis . 2016. Streptococcus pneumoniae oropharyngeal colonization in children and adolescents with cystic fibrosis. J Cyst Fibros 15:366–371. doi: 10.1016/j.jcf.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 56.Thornton CS, Brown EL, Alcantara J, Rabin HR, Parkins MD. 2015. Prevalence and impact of Streptococcus pneumoniae in adult cystic fibrosis patients: a retrospective chart review and capsular serotyping study. BMC Pulm Med 15:49. doi: 10.1186/s12890-015-0041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoiby N, Hoff GE, Jensen K, Lund E. 1976. Serological types of Diplococcus pneumoniae isolated from the respiratory tract of children with cystic fibrosis and children with other diseases. Scand J Respir Dis 57:37–40. [PubMed] [Google Scholar]
- 58.Pimentel de Araujo F, D’Ambrosio F, Camilli R, Fiscarelli E, Di Bonaventura G, Baldassarri L, Visca P, Pantosti A, Gherardi G. 2014. Characterization of Streptococcus pneumoniae clones from paediatric patients with cystic fibrosis. J Med Microbiol 63:1704–1715. doi: 10.1099/jmm.0.072199-0. [DOI] [PubMed] [Google Scholar]
- 59.Romero P, Croucher NJ, Hiller NL, Hu FZ, Ehrlich GD, Bentley SD, García E, Mitchell TJ. 2009. Comparative genomic analysis of ten Streptococcus pneumoniae temperate bacteriophages. J Bacteriol 191:4854–4862. doi: 10.1128/JB.01272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gladstone RA, Gritzfeld JF, Coupland P, Gordon SB, Bentley SD. 2015. Genetic stability of pneumococcal isolates during 35 days of human experimental carriage. Vaccine 33:3342–3345. doi: 10.1016/j.vaccine.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Avery OT, MacLeod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79:137–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:83–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hiller NL, Ahmed A, Powell E, Martin DP, Eutsey R, Earl J, Janto B, Boissy RJ, Hogg J, Barbadora K, Sampath R, Lonergan S, Post JC, Hu FZ, Ehrlich GD. 2010. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog 6:e1001108. doi: 10.1371/journal.ppat.1001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyer K, Chaffee E, Hobby GL, Dawson MH. 1941. Hyaluronidases of bacterial and animal origin. J Exp Med 73:309–326. doi: 10.1084/jem.73.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kilian M, Poulsen K, Blomqvist T, Håvarstein LS, Bek-Thomsen M, Tettelin H, Sørensen UBS. 2008. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One 3:e2683. doi: 10.1371/journal.pone.0002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li S, Kelly SJ, Lamani E, Ferraroni M, Jedrzejas MJ. 2000. Structural basis of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. EMBO J 19:1228–1240. doi: 10.1093/emboj/19.6.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paton JC, Berry AM, Lock RA. 1997. Molecular analysis of putative pneumococcal virulence proteins. Microb Drug Resist 3:1–10. doi: 10.1089/mdr.1997.3.1. [DOI] [PubMed] [Google Scholar]
- 68.Berry AM, Paton JC. 2000. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect Immun 68:133–140. doi: 10.1128/IAI.68.1.133-140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feldman C, Cockeran R, Jedrzejas MJ, Mitchell TJ, Anderson R. 2007. Hyaluronidase augments pneumolysin-mediated injury to human ciliated epithelium. Int J Infect Dis 11:11–15. doi: 10.1016/j.ijid.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 70.Lacks S, Hotchkiss RD. 1960. A study of the genetic material determining an enzyme activity in Pneumococcus. Biochim Biophys Acta 39:508–518. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 71.Alloing G, Granadel C, Morrison DA, Claverys JP. 1996. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol Microbiol 21:471–478. doi: 10.1111/j.1365-2958.1996.tb02556.x. [DOI] [PubMed] [Google Scholar]
- 72.Sibold C, Henrichsen J, König A, Martin C, Chalkley L, Hakenbeck R. 1994. Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol Microbiol 12:1013–1023. doi: 10.1111/j.1365-2958.1994.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 73.Hoskins J, Alborn WE, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, Estrem ST, Fritz L, Fu DJ, Fuller W, Geringer C, Gilmour R, Glass JS, Khoja H, Kraft AR, Lagace RE, LeBlanc DJ, Lee LN, Lefkowitz EJ, Lu J, Matsushima P, McAhren SM, McHenney M, McLeaster K, Mundy CW, Nicas TI, Norris FH, O’Gara M, Peery RB, Robertson GT, Rockey P, Sun PM, Winkler ME, Yang Y, Young-Bellido M, Zhao G, Zook CA, Baltz RH, Jaskunas SR, Rosteck PR, Skatrud PL, Glass JI. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J Bacteriol 183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mosaic structure of PBP1a and PBP2b of S. pneumoniae clone ST10523 and strain HMC3243. The homology boxes of PBP1a and PBP2b are shown above the mosaic genes. The mutation A446 in PBP2b is represented by the black arrow; the white arrow shows an additional amino acid. The colors indicate closely related sequences with >95% identity to the pbp2x reference sequences from penicillin-sensitive S. mitis strains M3 and NCTC10712, as indicated by the color coded boxes. Download FIG S1, TIF file, 6.6 MB (6.7MB, tif) .
Copyright © 2017 Rieger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.