Abstract
The inositol polyphosphates are a group of multifunctional signaling metabolites whose synthesis is catalyzed by a family of inositol kinases that are evolutionarily conserved from yeast to humans. Inositol polyphosphate multikinase (IPMK) was first identified as a subunit of the arginine-responsive transcription complex in budding yeast. In addition to its role in the production of inositol tetrakis- and pentakisphosphates (IP4 and IP5), IPMK also exhibits phosphatidylinositol 3-kinase (PI3-kinase) activity. Through its PI3-kinase activity, IPMK activates Akt/PKB and its downstream signaling pathways. IPMK also regulates several protein targets non-catalytically via protein-protein interactions. These non-catalytic targets include cytosolic signaling factors and transcription factors in the nucleus. In this review, we highlight the many known functions of mammalian IPMK in controlling cellular signaling networks and discuss future challenges related to clarifying the unknown roles IPMK plays in physiology and disease.
Keywords: inositol, inositol phosphates, inositol polyphosphate multikinase, IPMK, PI3-kinase
INTRODUCTION
Myo-inositol, a glucose isomer with one axial and five equatorial hydroxyl groups, is a key nutrient in the human diet (Holub, 1986). Fatty acyl modifications to cellular inositol partition it into two biochemically distinct pools: membrane lipid phosphatidylinositol (PI) and water-soluble inositol phosphate (IP). Inositol 1,4,5-trisphosphate (IP3), one of the most well-known inositol-derived metabolites, is a second messenger that mediates cytosolic calcium release from the endoplasmic reticulum (Berridge et al., 2000; Streb et al., 1983). IP3 is synthesized by the hydrolysis of phosphatidylinositol 4,5-bisphosphates (PIP2) upon growth factor stimulation and by the activation of phospholipases. Further investigation of the metabolic fate of IP3 revealed the family of enzymes known as the inositol phosphate kinases. Their phosphorylation of the inositol ring at its different hydroxyl groups generates higher inositol polyphosphates, which include the inositol pyrophosphates IP7 and IP8. These harbor diphosphate groups containing one or more high energy pyrophosphate bonds. These inositol poly- and pyrophosphates have recently drawn significant attention for their regulatory roles in major cellular signaling events like growth and apoptosis (Chakraborty et al., 2011). In this review, we will focus particularly on recent breakthroughs in inositol polyphosphate multikinase (IPMK)-mediated signaling.
A ROLE OF IPMK IN INOSITOL PHOSPHATE METABOLISM
IPMK was initially cloned as Arg82 from yeast as a regulator of arginine-responsive transcription, but we now refer to it as Ipk2 or yeast IPMK (Bechet et al., 1970; Hatch et al., 2017). In 1999, Snyder and his colleagues discovered that IPMK is essential for the synthesis of IP4 (both Ins(1,3,4,5)P4 and Ins(1,4,5,6)P4) and IP5 (Ins(1,3,4,5,6)P5) (Odom et al., 2000; Saiardi et al., 1999) (Fig. 1). IPMK is highly conserved and is the only enzyme capable of generating IP5 from IP4. Its inositol 6-kinase activity also makes IPMK particularly unique and indispensable in inositol polyphosphate biosynthesis. In yeast, the kinase activity of IPMK also regulates the activities of the chromatin remodeling SWI/SNF and Ino80 complexes in response to phosphate availability (Shen et al., 2003; Steger et al., 2003).
Fig. 1.
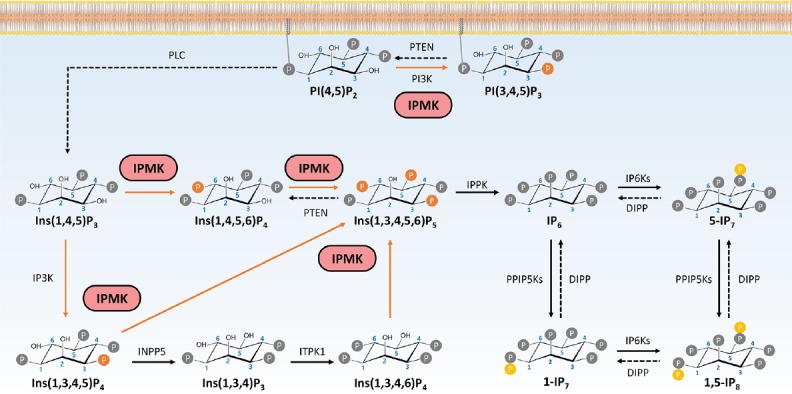
The functions of IPMK in inositol biosynthesis pathway.
IPMK possesses the activity of phosphorylating IP3 to IP4 as well as IP4 to IP5. IPMK also acts as a phosphoinositide kinase, which produces PIP3 from PIP2.
Genetic deletion of IPMK in mouse embryonic fibroblasts (MEFs) eliminates IP5 and the higher phosphorylated inositol phosphates IP6 and IP7 (Frederick et al., 2005). In addition to its inositol phosphate kinase activity, IPMK acts as a phosphoinositide kinase; it can produce phosphatidylinositol 3,4,5-trisphosphate (PIP3) by phosphorylating PIP2 at the 3 position (Resnick et al., 2005). In response to growth stimuli such as serum or growth factors, IPMK-deficient MEFs show a 50% reduction in PIP3 as well as significantly reduced PIP3-dependent Akt phosphorylation. These suggest IPMK acts as a physiological PI3-kinase (Maag et al., 2011).
IPMK-DEPENDENT INOSITOL POLYPHOSPHATES AS CELL SIGNALS
Studies focused on the actions of IP3 in controlling cytosolic calcium levels have established the different water-soluble IPs as signaling messengers that mediate diverse cellular processes such as growth (Chakraborty et al., 2011). Of those IPs, Ins(1,4,5,6)P4 and Ins(1,3,4,5,6)P5 are produced only via the inositol 6-kinase activity of IPMK (Lee et al., 2012). IPs are structurally similar to the head groups of phosphoinositides and are capable of binding specifically to the PH (pleckstrin homology) domains of various signaling proteins such as Akt/PKB (Razzini et al., 2000). The exogenous supply of IPMK products like Ins(1,4,5,6)P4 can suppress human cancer cell growth by inhibiting activation of Akt/PKB (Jackson et al., 2011; Piccolo et al., 2004; Razzini et al., 2000). Ins(1,4,5,6)P4 also plays an unexpected role in the regulation of histone deacetylase 3 (HDAC3). Ins(1,4,5,6)P4 was found to bind tightly to the highly basic interface between HDAC3 and the SMRT-DAD domain when they form a complex (Watson et al., 2012). Ins(1,4,5,6)P4 is actually essential for HDAC3 deacetylase activity—HDAC3 loses its activity in the absence of Ins(1,4,5,6)P4, and its activity is restored upon addition of exogenous Ins(1,4,5,6)P4 (Millard et al., 2013; Watson et al., 2016). In addition, the incubation of Ins(1,4,5,6)P4 with other HDAC isoforms such as the HDAC1-MTA1 complex significantly stimulates their histone acetylase activity. Since IPMK is the only enzyme capable of synthesizing Ins(1,4,5,6)P4 from Ins(1,4,5)P3, it is reasonable to speculate that IPMK acts as the primary point of control over class I HDACs in mammalian cells. Ins(1,3,4,5,6)P5, which is also produced by IPMK, is important in Wnt/β-catenin signaling for triggering the translocation of β-catenin to the nucleus (Gao and Wang, 2007; Wang and Wang, 2012). In fact, full activation of the Wnt pathway seems to require Ins(1,3,4,5,6)P5, because upon Wnt stimulation, IPMK is recruited to the plasma membrane via its interaction with Dishevelled-3 (Wang and Wang, 2012). Another recent report defining the role IPMK plays in promoting phospholipase Cβ1-dependent myogenic differentiation in C2C12 cells further corroborates the physiological significance of IPMK’s catalytic activity in controlling β-catenin activation (Ramazzotti et al., 2016; 2017).
IPMK ACTS AS A PI3-KINASE
IPMK is a promiscuous kinase, being unusually flexible in its specificity for inositol substrates. IPMK can function as a nuclear PI3-kinase, specifically generating PIP3 from PIP2 (Maag et al., 2011; Resnick et al., 2005). The importance of IPMK’s PI3-kinase function was recently highlighted in a study of Huntington’s disease (HD), a neurodegenerative disorder characterized by motor abnormalities and caused by an expansion of glutamine repeats in mutant huntingtin (mHtt). Although Akt deficits have been well-documented in the striatum and lymphoblasts of HD patients (Colin et al., 2005), in 2015, Ahmed et al. found severely reduced levels of IPMK in the striatum of HD patients and HD animal models that was linked to subsequent reductions in Akt phosphorylation (Ahmed et al., 2015). In normal striatal cells, the transcription factor COUP-TF-interacting protein 2 (Ctip2), enhances IPMK expression. In HD striatal cells, mHtt inhibits the transcriptional activity of Ctip2, thereby, reducing IPMK expression. Moreover, IPMK’s interaction with mHtt disrupts its stability. Overall, the reduced levels of IPMK protein in striatal neurons overexpressing mHtt lead to hypoactivation of Akt. This subsequently contributes to reduced activity in key neuroprotective signaling pathways.
In addition to IPMK’s role in coordinating with the p85/p110 PI3-kinase for full activation of Akt, IPMK-dependent synthesis of PIP3 has been observed inside the nucleus. Steroidogenic factor 1 (SF-1; also known as NR5A1) is a nuclear receptor that interacts with PIP2 to control the expression levels of steroidogenic enzymes and peptide hormones (Parker and Schimmer, 1997; De Santa Barbara et al., 1998). IPMK interacts directly with the SF-1–PIP2 complex and phosphorylates PIP2, converting it to PIP3. This is why IPMK depletion reduces SF-1 target gene induction (Blind, 2013; Blind et al., 2012; Malabanan and Blind, 2016). Because SF-1–PIP2 is not phosphorylated by the classical PI3-kinase (e.g., p110 PI3-kinase), they cannot compensate for the loss of IPMK. In other words, SF-1–dependent transcriptional activation requires IPMK-dependent PIP3 production.
In addition to its function in SF-1–dependent transcriptional control, IPMK also functions in the export of mRNA from the nucleus to the cytoplasm (Carmody and Wente, 2009; Kim et al., 2016). An interaction between the mRNA export factor ALY and PIP3 has long been thought to regulate the subcellular localization of ALY. This, in turn, mediates the release of nuclear mRNAs from the nuclear speckle domain into the cytosol. In the absence of IPMK, ALY no longer binds the RAD51 3′UTR—an event that is critical for the export of target mRNAs into the cytosol. Importantly, exogenous PIP3 rescues the interaction between ALY and the RAD51 3′UTR in IPMK-depleted cell extracts. Functionally speaking, IPMK deficiency reduces DNA damage responses because these responses require the nuclear export of mRNAs encoding proteins like RAD51 and BRCA1 (Wickramasinghe et al., 2013). Thus, IPMK, in its role as a PI3-kinase, regulates transcript-selective mRNA export by controlling the levels of nuclear PIP3.
NON-CATALYTIC FUNCTIONS OF IPMK
Through its physical interactions with various target proteins, IPMK can also regulate cellular signaling in non-catalytic ways. IPMK’s maintenance of the integrity of mTOR complex 1 (mTORC1) was the first of these non-catalytic functions to be identified (Kim et al., 2011). mTOR is a protein kinase that interacts with raptor to form the mTORC1 complex, which itself is activated by amino acids (Sancak et al., 2010; Zoncu et al., 2011). Genetic depletion of IPMK in MEFs reduces amino acid-stimulated mTOR activation and disrupts the stability of the mTOR-raptor complex. Because catalytically-inactive IPMK activates mTORC1 just as efficiently as wild-type IPMK (Kim and Snyder, 2011; Kim et al., 2011), IPMK likely modulates amino acid-dependent mTORC1 signaling by acting as an mTORC1 co-factor rather than as a kinase.
IPMK also binds the energy-sensing protein kinase AMPK in the cytoplasm. AMPK is a heterotrimeric kinase comprising a catalytic subunit (α) and two regulatory (β, γ) subunits (Hawley et al., 1996; Scott et al., 2004; Xiao et al., 2011). The AMP and ADP-bound forms of the AMPK γ subunit promote phosphorylation, while its ATP-bound form inhibits phosphorylation. AMPK activity is also regulated by the upstream kinase liver kinase B1 (LKB1) (Alessi et al., 2006). In the presence of AMP (i.e., in the low energy state), LKB1 constitutively phosphorylates AMPK. In 2012, Bang et al. reported the dynamic regulation of IPMK mRNA and protein levels in the mouse hypothalamus according to the feeding status of the animal (Bang et al., 2012). IPMK was discovered to dynamically regulate AMPK in response to glucose levels. IPMK-deficient MEFs show increased AMPK phosphorylation in low glucose conditions. During glucose depravation, IPMK dissociates from AMPK, allowing upstream kinase LKB1 enough access to phosphorylate AMPK. Interestingly, IPMK also directly interacts with LKB1 to mediate the metformin-induced activation of AMPK. Disruption of the LKB1-IPMK interaction via a dominant-negative peptide was discovered to attenuate metformin-induced AMPK activation (Bang et al., 2014).
Recently, Kim et al. (2017) revealed IPMK as an important regulator of Toll-like receptor (TLR) signaling in myeloid cells like macrophages. As microbial sensors, the TLRs are responsible for activating host defense systems in response to invading pathogens (Akira et al., 2001; Lee and Kim, 2007). TLR activation in immune cells triggers the production of pro-inflammatory cytokines that help protect the host from pathogens (Kondo et al., 2012; Liew et al., 2005). The uncontrolled production of cytokines, however, can lead the host’s immune system to attack itself. TRAF6 is a E3 ubiquitin ligase that activates TLR downstream effectors such as IκB kinase, mitogen-activated protein kinase, and NFκB (Deng et al., 2000; Lamothe et al., 2007). Independent of its catalytic activity, IPMK regulates TLR-induced inflammatory cytokine production by controlling TRAF6 protein stability. It does so by interacting directly with TRAF6 and preventing its K48-linked ubiquitination. In the absence of this interaction, ubiquitinated TRAF6 is degraded by the proteasome (Hjerpe and Rodriguez, 2008; Pickart, 1997). Thus, myeloid cell-specific IPMK deletion in mice reduces excessive inflammation thereby protecting mice against polymicrobial sepsis and endotoxemic shock. Together, these findings suggest IPMK is a cytoplasmic signaling hub that coordinates cellular growth, energy metabolism, and inflammatory signaling networks.
Since IPMK was discovered as an essential regulator of arginine-responsive transcription in yeast, many studies have focused on identifying nuclear functions for mammalian IPMK (Dubois et al., 1987; Odom et al., 2000). Just as Ipk2 binds and stabilizes the transcription factor MCM1 in budding yeast (Bercy et al., 1987; Christ and Tye, 1991; Messenguy and Dubois, 1993), IPMK binds the mammalian ortholog of MCM1, serum response factor (SRF) (Kim et al., 2013, 2016). SRF is a transcription factor in mammals that is essential for the induction of a wide range of immediate early genes (IEGs) like c-jun and c-fos, which are rapidly induced in response to various stimuli like serum (Hill and Treisman, 1995). IPMK-deficient MEFs show reduced SRF target gene expression and protein levels in response to serum stimulation (Cole et al., 1995; Curran and Morgan, 1985; Hope et al., 1992; Hunt et al., 1987). The fact that overexpression of either wild-type or catalytically inactive IPMK in IPMK-deficient MEFs restores SRF target gene expression suggests IPMK regulates SRF and its transcriptional activity in a non-catalytic manner. SRF shows reduced binding to the promoter serum response element (SRE) in the absence of IPMK, suggesting IPMK helps maintain stable interactions between SRF and its target promoters. These findings extend beyond cultured cells, as mice whose excitatory neurons lack IPMK also show reduced SRF-SRE binding and fail to induce IEG expression. IPMK’s role as a key transcriptional co-activator of neural IEG expression was further expanded when IPMK was identified as a CREB-binding protein (CBP) (Xu et al., 2013a). Histone acetyltransferase CBP is an epigenetic regulator essential for the induction of IEGs in synaptic plasticity and SRF’s regulation of cognition (Bartsch et al., 1998; Bourtchuladze et al., 1994; Kandel, 2001; Matynia et al., 2002; Silva et al., 1998; Yin et al., 1994). The interaction between IPMK and CBP seems to be dynamically induced by neuronal stimulation. Mechanistically, IPMK is a key factor in recruiting CBP to the promoters of IEGs. As with IPMK-SRF regulation, the expression of a catalytically inactive IPMK mutant can rescue the phenotypes associated with CBP dysfunction in IPMK-null neurons, confirming that this function for nuclear IPMK is a non-catalytic one.
IPMK was also identified as a potential regulator of p53 in a loss-of-function screen designed to identify genes required for p53-dependent oncogene-induced senescence (Drost et al., 2010). p53 is a well-known tumor suppressor that senses diverse stress signals and regulates gene expression to cause cell cycle arrest, cellular senescence, and apoptosis (Brady and Attardi, 2010; Chen et al., 2005; Collado et al., 2005; Vousden and Prives, 2009). IPMK regulates p53-dependent gene expression (e.g., PUMA, Bax, and p21) via direct protein-protein interactions (Xu and Snyder, 2013; Xu et al., 2013b). IPMK non-catalytically stimulates p53’s transcriptional activity by recruiting the histone acetyl transferase p300 to acetylate both the p53 promoter and its associated histones.
CONCLUSIONS
A growing body of compelling evidence implicates IPMK as a multifunctional regulator of cellular signaling networks in mammals. The development of a variety of tissue-specific IPMK knockout mouse models will undoubtedly uncover new dimensions to the IPMK-dependent control of cellular signaling, but even the known functions of IPMK are still not fully understood in mechanistic detail. For example, what is the molecular switch that shifts IPMK’s function among its many catalytic specificities? In other words, what molecular changes determine when IPMK acts as a PI3-kinase, an inositol 3-kinase, or an inositol 6-kinase? As Akt and synaptotagmin are known to bind IP7 with high-affinity, it is likely that IP5 and the other IPMK-dependent products also have cognate binding partners (Lee et al., 2016). What are these binding partners? Moreover, the biochemical relationship between the PI3-kinase activities of IPMK and the more classical p85/p110-PI3K also awaits further investigation.
Recently, human IPMK has been associated with specific pathophysiological phenomena. A germ line mutation in IPMK was discovered among familial small intestinal carcinoid patients (Sei et al., 2015). In these patients, the inheritance of a truncated IPMK allele is associated with reduced p53 signaling. This suggests a role for IPMK in suppressing intestinal epithelial tumorigenesis. In addition, the tumor suppressor miR-18a was recently found to downregulate IPMK mRNA levels, thereby contributing to the inhibition of ovarian tumor growth (Liu et al., 2017). Thus, a further dissection of the roles IPMK plays in regulating the different types of cancer will be both interesting and important. The IPMK gene has also been identified in genome-wide association studies of Alzheimer disease and immune-mediated diseases, suggesting a potential role for IPMK in the pathogenesis of inflammation-associated neurodegeneration (Yokoyama et al., 2016). As further efforts are made to detect novel mutations and single nucleotide polymorphisms in the human IPMK gene and as more comprehensive analyses are made of conditional knockout mouse models, all of the many in vivo functions of IPMK will become more clear. We fully expect the resulting understanding of IPMK’s multifunctional regulation of cellular signaling will direct the development of future therapeutics aimed at alleviating a broad range of ailments.
Fig. 2.
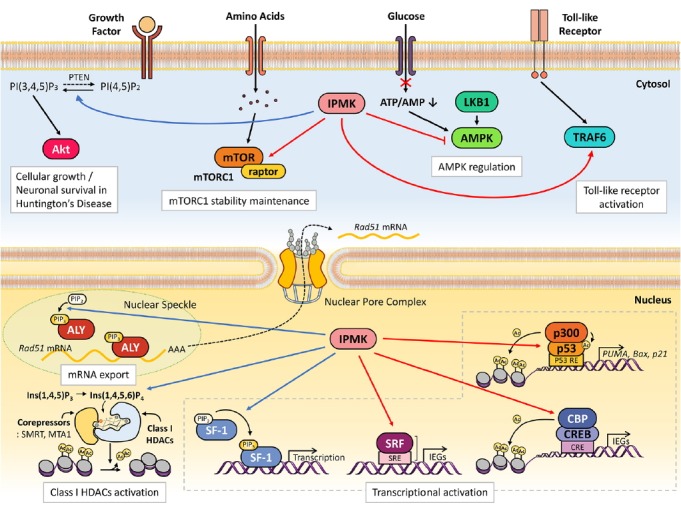
Versatile signaling actions of mammalian IPMK.
As a PI3-kinase, IPMK activates Akt signaling. Water-soluble, inositol phosphates produced from IPMK such as Ins(1,4,5,6)P4 appears essential for class I HDAC function. Independent of its catalytic activity, IPMK directly binds to various cytosolic signaling factors, thereby regulating multiple signaling networks. Inside the nucleus, IPMK is a key node of molecular events (e.g., selective mRNA export, transcription). It acts as a transcription coactivator via forming complexes with transcription factors as well as epigenetic regulators. See main text for further details. Blue lines indicate catalytic activity-dependent IPMK actions. Red lines designate IPMK-binding partners which are non-catalytically regulated by IPMK.
ACKNOWLEDGMENTS
We thank all the members of the Kim lab for discussion and helpful comments. This work was supported by National Research Foundation of Korea (NRF-2013M3C7A1056102 to S.K.).
REFERENCES
- Ahmed I., Sbodio J.I., Harraz M.M., Tyagi R., Grima J.C., Albacarys L.K., Hubbi M.E., Xu R., Kim S., Paul B.D., et al. Huntington’s disease: Neural dysfunction linked to inositol polyphosphate multikinase. Proc Natl Acad Sci USA. 2015;112:9751–9756. doi: 10.1073/pnas.1511810112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Takeda K., Kaisho T. Toll-like receptors : critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–80. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., Sakamoto K., Bayascas J.R. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- Bang S., Kim S., Dailey M.J., Chen Y., Moran T.H., Snyder S.H., Kim S.F. AMP-activated protein kinase is physiologically regulated by inositol polyphosphate multikinase. Proc Natl Acad Sci USA. 2012;109:616–620. doi: 10.1073/pnas.1119751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang S., Chen Y., Ahima R.S., Kim S.F. Convergence of IPMK and LKB1-AMPK signaling pathways on metformin action. Mol Endocrinol. 2014;28:1186–1193. doi: 10.1210/me.2014-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch D., Casadio A., Karl K.A., Serodio P., Kandel E.R. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95:211–223. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- Bechet J., Greenson M., Wiame J.M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Bercy J., Dubois E., Messenguy F. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene. 1987;55:277285. doi: 10.1016/0378-1119(87)90287-3. [DOI] [PubMed] [Google Scholar]
- Berridge M.J., Lipp P., Bootman M.D. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:1121. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Blind R.D. Disentangling biological signaling networks by dynamic coupling of signaling lipids to modifying enzymes. Adv Biol Regul. 2013;54:114. doi: 10.1016/j.jbior.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blind R.D., Suzawa M., Ingraham H.A. Direct modification and regulation of a nuclear receptor-PIP2 complex by the nuclear inositol-lipid kinase IPMK. Sci Signal. 2012;5:110. doi: 10.1126/scisignal.2003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchuladze R., Frenguelli B., Blendy J., Cioffi D., Schutz G., Silva A.J. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:5968. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- Brady C.A., Attardi L.D. p53 at a glance. J Cell Sci. 2010;123:25272532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody S.R., Wente S.R. mRNA nuclear export at a glance. J Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A., Kim S., Snyder S.H. Inositol pyrophosphates as mammalian cell signals. Sci Signal. 2011;4:1–11. doi: 10.1126/scisignal.2001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Trotman L.C., Shaffer D., Lin H.-K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ C., Tye B.K. Functional domains of the yeast transcription/replication factor MCM1. Genes Dev. 1991;5:751–763. doi: 10.1101/gad.5.5.751. [DOI] [PubMed] [Google Scholar]
- Cole R.L., Konradi C., Douglass J., Hyman S.E. Neuronal adaptation to amphetamine and dopamine: molecular mechanisms of prodynorphin gene regulation in rat striatum. Neuron. 1995;14:813–823. doi: 10.1016/0896-6273(95)90225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin E., Regulier E., Perrin V., Durr A., Brice A., Aebischer P., Deglon N., Humbert S., Saudou F. Akt is altered in an animal model of Huntington’s disease and in patients. Eur J Neurosci. 2005;21:1478–1488. doi: 10.1111/j.1460-9568.2005.03985.x. [DOI] [PubMed] [Google Scholar]
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Curran T., Morgan J.I. Superinduction of c-fos by nerve growth factor in the presence of peripherally active benzodiazepines. Science. 1985;229:1265–1268. doi: 10.1126/science.4035354. [DOI] [PubMed] [Google Scholar]
- Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z.J. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- Drost J., Mantovani F., Tocco F., Elkon R., Comel A., Holstege H., Kerkhoven R., Jonkers J., Voorhoeve P.M., Agami R., et al. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol. 2010;12:380–389. doi: 10.1038/ncb2038. [DOI] [PubMed] [Google Scholar]
- Dubois E., Bercy J., Messenguy F. Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast. Mol Gen Genet. 1987;207:142–148. doi: 10.1007/BF00331501. [DOI] [PubMed] [Google Scholar]
- Frederick J.P., Mattiske D., Wofford J.A., Megosh L.C., Drake L.Y., Chiou S.-T., Hogan B.L.M., York J.D. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc Natl Acad Sci USA. 2005;102:8454–8459. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Wang H. Inositol pentakisphosphate mediates Wnt/beta-catenin signaling. J Biol Chem. 2007;282:26490–26502. doi: 10.1074/jbc.M702106200. [DOI] [PubMed] [Google Scholar]
- Hatch A.J., Odom A.R., York J.D. Inositol phosphate multikinase dependent transcriptional control. Adv Biol Regul. 2017;64:9–19. doi: 10.1016/j.jbior.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley S.A., Davison M., Woods A., Davies S.P., Beri R.K., Carling D., Hardie D.G. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem. 1996;271:27879–27887. doi: 10.1074/jbc.271.44.27879. [DOI] [PubMed] [Google Scholar]
- Hill C.S., Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- Hjerpe R., Rodriguez M. Alternative UPS drug targets upstream the 26S proteasome. Int J Biochem Cell Biol. 2008;40:1126–1140. doi: 10.1016/j.biocel.2007.11.021. [DOI] [PubMed] [Google Scholar]
- Holub B.J. Metabolism and function of myo-inositol and inositol phospholipids. Annu Rev Nutr. 1986;6:563–597. doi: 10.1146/annurev.nu.06.070186.003023. [DOI] [PubMed] [Google Scholar]
- Hope B., Kosofsky B., Hyman S.E., Nestler E.J. Regulation of immediate early gene expression and AP-1 binding in the rat nucleus accumbens by chronic cocaine. Proc Natl Acad Sci USA. 1992;89:5764–5768. doi: 10.1073/pnas.89.13.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S.P., Pini A., Evan G. Induction of c-fos-like protein in spinal cord neurons following sensory stimulation. Nature. 1987;328:632–634. doi: 10.1038/328632a0. [DOI] [PubMed] [Google Scholar]
- Jackson S.G., Al-Saigh S., Schultz C., Junop M.S. Inositol pentakisphosphate isomers bind PH domains with varying specificity and inhibit phosphoinositide interactions. BMC Struct Biol. 2011;11:11. doi: 10.1186/1472-6807-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel E.R. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kim S., Snyder S.H. Nutrient amino acids signal to mTOR via inositol polyphosphate multikinase. Cell Cycle. 2011;10:1708–1710. doi: 10.4161/cc.10.11.15559. [DOI] [PubMed] [Google Scholar]
- Kim S., Kim S.F., Maag D., Maxwell M.J., Resnick A.C., Juluri K.R., Chakraborty A., Koldobskiy M.A., Cha S.H., Barrow R., et al. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell Metab. 2011;13:215–221. doi: 10.1016/j.cmet.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Tyagi R., Lee J.-Y., Park J., Kim Y.-R., Beon J., Chen P.Y., Cha J.Y., Snyder S.H., Kim S. Inositol polyphosphate multikinase is a coactivator for serum response factor-dependent induction of immediate early genes. Proc Natl Acad Sci USA. 2013;110:19938–19943. doi: 10.1073/pnas.1320171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Beon J., Lee S., Park J., Kim S. IPMK: A versatile regulator of nuclear signaling events. Adv Biol Regul. 2016;61:25–32. doi: 10.1016/j.jbior.2015.11.005. [DOI] [PubMed] [Google Scholar]
- Kim E., Beon J., Lee S., Park S.J., Ahn H., Kim M.G., Park J.E., Kim W., Yuk J.-M., Kang S.-J., et al. Inositol polyphosphate multikinase promotes Toll-like receptor–induced inflammation by stabilizing TRAF6. Sci Adv. 2017;3:e1602296. doi: 10.1126/sciadv.1602296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Kawai T., Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol. 2012;33:449–458. doi: 10.1016/j.it.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Lamothe B., Besse A., Campos A.D., Webster W.K., Wu H., Darnay B.G. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of IκB kinase activation. J Biol Chem. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.S., Kim Y.-J. Pattern-recognition receptor signaling initiated from extracellular, membrane, and cytoplasmic space. Mol Cells. 2007;23:1–10. [PubMed] [Google Scholar]
- Lee J.-Y., Kim Y., Park J., Kim S. Inositol polyphosphate multikinase signaling in the regulation of metabolism. Ann N Y Acad Sci. 2012;1271:68–74. doi: 10.1111/j.1749-6632.2012.06725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.-S., Lee J.-Y., Kyung J.W., Yang Y., Park S.J., Lee S., Pavlovic I., Kong B., Jho Y.S., Jessen H.J., et al. Inositol pyrophosphates inhibit synaptotagmin-dependent exocytosis. Proc Natl Acad Sci USA. 2016;113:8314–8319. doi: 10.1073/pnas.1521600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew F.Y., Xu D., Brint E.K., O’Neill L.A.J. Negative regulation of Toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- Liu P., Qi X., Bian C.E., Yang F.A.N., Lin X. MicroRNA-18a inhibits ovarian cancer growth via directly targeting TRIAP1 and IPMK. Oncol Lett. 2017:1–8. doi: 10.3892/ol.2017.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D., Maxwell M.J., Hardesty D.A., Boucher K.L., Choudhari N., Hanno A.G., Ma J.F., Snowman A.S., Pietropaoli J.W., Xu R., et al. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc Natl Acad Sci USA. 2011;108:1391–1396. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabanan M.M., Blind R.D. Inositol polyphosphate multikinase (IPMK) in transcriptional regulation and nuclear inositide metabolism. Biochem Soc Trans. 2016;44:279–285. doi: 10.1042/BST20150225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matynia A., Kushner S.A., Silva A.J. Genetic approaches to molecular and cellular cognition: a focus on LTP and learning and memory. Annu Rev Genet. 2002;36:687–720. doi: 10.1146/annurev.genet.36.062802.091007. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2586–2592. doi: 10.1128/mcb.13.4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard C.J., Watson P.J., Celardo I., Gordiyenko Y., Cowley S.M., Robinson C.V., Fairall L., Schwabe J.W.R. Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol Cell. 2013;51:57–67. doi: 10.1016/j.molcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom A.R., Stahlberg A., Wente S.R., York J.D. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Parker K.L., Schimmer B.P. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev. 1997;18:361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- Piccolo E., Vignati S., Maffucci T., Innominato P.F., Riley A.M., Potter B.V.L., Pandolfi P.P., Broggini M., Iacobelli S., Innocenti P., et al. Inositol pentakisphosphate promotes apoptosis through the PI 3-K/Akt pathway. Oncogene. 2004;23:1754–1765. doi: 10.1038/sj.onc.1207296. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- Ramazzotti G., Maria Billi A., Manzoli L., Mazzetti C., Ruggeri A., Erneux C., Kim S., Suh P.-G., Cocco L., Faenza I. IPMK and β-catenin mediate PLC-β1-dependent signaling in myogenic differentiation. Oncotarget. 2016;7:84118–84127. doi: 10.18632/oncotarget.11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazzotti G., Faenza I., Fiume R., Billi A.M., Manzoli L., Mongiorgi S., Ratti S., McCubrey J.A., Suh P.-G., Cocco L., et al. PLC-beta1 and cell differentiation: An insight into myogenesis and osteogenesis. Adv Biol Regul. 2017;63:1–5. doi: 10.1016/j.jbior.2016.10.005. [DOI] [PubMed] [Google Scholar]
- Razzini G., Berrie C.P., Vignati S., Broggini M., Mascetta G., Brancaccio A., Falasca M. Novel functional PI 3-kinase antagonists inhibit cell growth and tumorigenicity in human cancer cell lines. FASEB J. 2000;14:1179–1187. doi: 10.1096/fasebj.14.9.1179. [DOI] [PubMed] [Google Scholar]
- Resnick A.C., Snowman A.M., Kang B.N., Hurt K.J., Snyder S.H., Saiardi A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc Natl Acad Sci USA. 2005;102:12783–12788. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A., Erdjument-Bromage H., Snowman A.M., Tempst P., Snyder S.H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa Barbara P., Bonneaud N., Boizet B., Desclozeaux M., Moniot B., Sudbeck P., Scherer G., Poulat F., Berta P. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene. Mol Cell Biol. 1998;18:6653–6665. doi: 10.1128/mcb.18.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.W., Hawley S.A., Green K.A., Anis M., Stewart G., Scullion G.A., Norman D.G., Hardie D.G. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sei Y., Zhao X., Forbes J., Szymczak S., Li Q., Trivedi A., Voellinger M., Joy G., Feng J., Whatley M., et al. A hereditary form of small intestinal carcinoid associated with a germline mutation in inositol polyphosphate multikinase. Gastroenterology. 2015;149:67–78. doi: 10.1053/j.gastro.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Xiao H., Ranallo R., Wu W.-H., Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Silva A.J., Kogan J.H., Frankland P.W., Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- Steger D.J., Haswell E.S., Miller A.L., Wente S.R., O’Shea E.K. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb H., Irvine R.F., Berridge M.J., Schulz I. Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature. 1983;306:67–69. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- Vousden K.H., Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang H.Y. Dvl3 translocates IPMK to the cell membrane in response to Wnt. Cell Signal. 2012;24:2389–2395. doi: 10.1016/j.cellsig.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P., Fairall L., Santos G., Schwabe J. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P.J., Millard C.J., Riley A.M., Robertson N.S., Wright L.C., Godage H.Y., Cowley S.M., Jamieson A.G., Potter B.V.L., Schwabe J.W.R. Insights into the activation mechanism of class I HDAC complexes by inositol phosphates. Nat Commun. 2016;7:11262. doi: 10.1038/ncomms11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe V., Savill J., Chavali S., Jonsdottir A., Rajendra E., Grüner T., Laskey R., Babu M.M., Venkitaraman A. Human inositol polyphosphate multikinase regulates transcriptselective nuclear mRNA export to preserve genome integrity. Mol Cell. 2013;51:737–750. doi: 10.1016/j.molcel.2013.08.031. [DOI] [PubMed] [Google Scholar]
- Xiao B., Sanders M.J., Underwood E., Heath R., Mayer F.V., Carmena D., Jing C., Walker P.A., Eccleston J.F., Haire L.F., et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472:230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Snyder S.H. Gene transcription by p53 requires inositol polyphosphate multikinase as a co-activator. Cell Cycle. 2013;12:1819–1820. doi: 10.4161/cc.25119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Paul B.D., Smith D.R., Tyagi R., Rao F., Khan A.B., Blech D.J., Vandiver M.S., Harraz M.M., Guha P., et al. Inositol polyphosphate multikinase is a transcriptional coactivator required for immediate early gene induction. Proc Natl Acad Sci USA. 2013a;110:16181–16186. doi: 10.1073/pnas.1315551110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Sen N., Paul B.D., Snowman A.M., Rao F., Vandiver M.S., Xu J., Snyder S.H. Inositol polyphosphate multikinase is a coactivator of p53-mediated transcription and cell death. Sci Signal. 2013b;6:1–17. doi: 10.1126/scisignal.2003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J.C., Wallach J.S., Del Vecchio M., Wilder E.L., Zhou H., Quinn W.G., Tully T. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- Yokoyama J.S., Wang Y., Schork A.J., Thompson W.K., Karch C.M., Cruchaga C., McEvoy L.K., Witoelar A., Chen C.-H., Holland D., et al. Association between genetic traits for immunemediated diseases and alzheimer disease. JAMA Neurol. 2016;73:691–697. doi: 10.1001/jamaneurol.2016.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A., Sabatini D.M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]


