Abstract
Regulation of feeding is essential for animal survival. The pharyngeal sense organs can act as a second checkpoint of food quality, due to their position between external taste organs such as the labellum which initially assess food quality, and the digestive tract. Growing evidence provides support that the pharyngeal sensory neurons regulate feeding, but much is still unknown. We found that a pair of gustatory receptor neurons in the LSO, a Drosophila adult pharyngeal organ which expresses four gustatory receptors, is involved in feeding inhibition in response to high concentrations of sodium ions. RNAi experiments and mutant analysis showed that the gustatory receptor Gr2a is necessary for this process. This feeding preference determined by whether a food source is perceived as appetizing or not is influenced by nutritional conditions, such that when the animal is hungry, the need for energy dominates over how appealing the food source is. Our results provide experimental evidence that factors involved in feeding function in a context-dependent manner.
Keywords: Drosophila melanogaster, feeding, gustatory receptor, pharyngeal sense organ
INTRODUCTION
The search for food and feeding is essential for the survival of all animals. Feeding can be regulated by various conditions, and food palatability is the main factor that determines feeding initiation.
Drosophila melanogaster is a genetic model system that has been useful for the study of the molecular and cellular mechanisms of taste (Freeman and Dahanukar, 2015). In particular, following the identification of the gustatory receptor (Gr) gene family, multiple studies have addressed the functions of the sugar and bitter receptors, mainly through Gr gene mutant analyses (Dahanukar et al., 2007; Fujii et al., 2015; Jiao et al., 2007; 2008; Lee et al., 2009; 2015; Miyamoto et al., 2012; Moon et al., 2009; Shim et al., 2015). 60 genes in the gustatory receptor gene family encode 68 receptor proteins (Clyne et al., 2000; Robertson et al., 2003; Scott et al., 2001). The Grs are expressed in various organs such as the labellum, legs, anterior margin of wings, pharynx, and internal organs such as the intestine in adult Drosophila (Park and Kwon, 2011; Stocker, 1994; Vosshall and Stocker, 2007). Grs are also expressed at the larval stage in Drosophila, in organs such as the terminal organ and pharyngeal sense organs (Kwon et al., 2011), and are involved in sugar and bitter sensing (Choi et al., 2016; Kim et al., 2016; Mishra et al., 2013). During metamorphosis, almost the entire larval peripheral nervous system disappears to be rebuilt into the adult peripheral nervous system (Tissot and Stocker, 2000). As an example, 21 pairs of odorant receptor neurons are responsible for olfaction at the larval stage, while more than 1,300 pairs of odorant receptor neurons are newly generated in the antenna at the adult stage (Vosshall and Stocker, 2007). The pharyngeal sensory neurons are an exception to this rebuilding during metamorphosis (Gendre et al., 2004), and it is unclear what this means for the development and physiology of the organism. Analysis of the functions of the pharyngeal sensory neurons and what chemicals they detect may yield insight into the reason why these neurons are maintained throughout development.
Three sensory organs exist in the adult Drosophila pharynx: the labral sense organ (LSO), ventral cibarial sensory organ (VCSO), and dorsal cibarial sensory organ (DCSO) (Stocker, 1994; 2004). The DCSO and VCSO occupy the dorsal and ventral parts of the cibarium, and the LSO is located near the labellum. The pharyngeal sense organs are located between the external sensory system which senses the quality of food, and the post-ingestive internal nutrient sensing system which determines whether to continue or stop feeding, and are thus in an anatomically ideal position to act as additional regulators of feeding. Recent studies have shown that the pharyngeal sense organs indeed regulate feeding. Adult gustatory receptor neurons (GRNs) that express sugar receptors promote and maintain feeding (LeDue et al., 2015), while bitter sensing by larval pharyngeal GRNs was found to inhibit ingestion (Choi et al., 2016).
In this study, we identified an adult pharyngeal GRN that expresses a subset of four Grs of previously unknown function. Using mutant analysis and molecular genetic tools, we found that this pharyngeal neuron and the Gr2a gustatory receptor expressed in this neuron are involved in feeding inhibition. Gr2a appears to be involved in the inhibition of feeding on food with a high concentration of Na+. This feeding suppression, which reflects that flies find excessively salty food unappetizing, does not manifest when flies are starved and hungry.
MATERIALS AND METHODS
Drosophila stocks, transgenic flies, and generation of the Gr2a mutant
Flies were grown on standard cornmeal agar medium. Fly culture and behavior experiments were carried out at room temperature (23°C ± 2°C). White-CantonS (wCS) flies were used as controls. The Gr2a-, Gr23a-, Gr57a-, and Gr93d-GAL4 lines used in this study were previously described (Kwon et al., 2011). RNAi lines for the Gr genes were purchased from the Vienna Drosophila Resource Center (VDRC), and the VDRC IDs are as follows: 39570 (Gr2a), 40853 (Gr23a), 45879 (Gr57a), and 6813 (Gr93d). The Gr2a mutant (Gr2aGAL4) was generated by homologous recombination. The 5′ and 3′ homology arms for the Gr2a coding region were amplified by genomic DNA PCR using specific primer pairs (GGATCCCAGAACGAAGATCGGAGACGGATTCACTA and GGTACCTCATCCACTATCTGCCGCGCG for the 5′ homology arm, CGGGCCGCCTCTCCATGTTGATGCAGGT and CCGCGGCGTTTCCGAACAGCTGTTGC for the 3′ homology arm), and cloned into the mutant construction vector pw35GAL4. After obtaining transformants carrying the targeting vector, the transgene was mobilized and offspring were screened for targeted insertions as previously described (Gong and Golic, 2003). Gr2a cDNA was amplified from cDNA (RT06756) and inserted into pUAST attB vectors via conventional molecular cloning methods. Transgenic flies were generated using PhiC31 integrase-mediated transgenesis on the second chromosome (attP40) (BestGene Inc., USA).
Chemicals
Chemicals of the highest purity commercially available were purchased for use in the CAFE assay. Calcium chloride (C0504), potassium chloride (P0515), and sucrose (S0809) were purchased from Duchefa Biochemie, and caffeine (27600), methyl cellulose (M7140), sodium chloride (71376), and sodium gluconate (S2054) were purchased from Sigma-Aldrich.
Gr expression mapping and immunohistochemistry
Mapping of Gr-GAL4 driver expression in the labral sense organ and brain immunostaining were performed on transgenic flies containing both a Gr-GAL4 transgene and UAS-mCD8-GFP as a GFP reporter. Gr-GAL4 drivers that show expression in the DP3 neuron of the larval dorsal pharyngeal sensilla (Choi et al., 2016) were crossed into UAS-mCD8-GFP flies, and expression in the labral sense organ of resulting adult progeny were observed. Adult heads were dissected and incubated in mounting solution (70% glycerol in 1X PBS-T) for 20–30 min before direct observation of fluorescence. To determine whether two Gr-GAL4 drivers are expressed in the same cells, progeny from crosses between two Gr-GAL4;UAS-mCD8-GFP strains were examined to count whether the number of GFP-expressing cells was unchanged or increased compared to the parent strains. For immunostaining of the adult brain, adult brains were dissected and immunostained as previously described (Choi et al., 2016). Anti-GFP antibody (rabbit polyclonal; Invitrogen; 1:1,000) was used to amplify the GFP signal of UAS-mCD8-GFP in GAL4-expressing cells. The nc82 antibody (mouse monoclonal; 1:50) was used to visualize a presynaptic active zone protein that marks brain morphology. The secondary antibodies used were Alexa 488-conjugated goat anti-rabbit IgG (Invitrogen;1:200) and Alexa 568-conjugated goat anti-mouse antibody (Invitrogen; 1:500). All images were taken by a confocal microscope (Zeiss LSM 700).
Capillary feeder (CAFE) assay
CAFE assays were conducted as described (Du et al., 2015) with modifications. The 2–5 day-old female flies were used for the CAFE assay. The day before the experiment, flies were either transferred to a fresh vial containing standard food and left overnight (well-fed conditions), or subjected to various periods of starvation on 0.6% agarose (starvation conditions). Two glass capillaries (Marienfeld, No. 2920109) were inserted between the vial (AS-507, Fisher Scientific) wall and plug (AS-273, Fisher Scientific) for the assays. Flies were allowed to feed for 4 h during the CAFE assay. For well-fed conditions, 24 or 32 flies were typically used for the assay, but in some cases where the flies ate so little that it was difficult to measure consumption (for example, Gr57a>RNAi), up to 48 flies were used. For starvation conditions, the number of flies used in the assay was adjusted to 4–16 flies, since the flies would eat so much during 4 h that they would completely empty the capillaries of solution. Both capillaries were filled with 100 mM sucrose solution, with the test chemical added to one capillary, and differences in consumption were quantified. The 0.125 mg/ml brilliant blue FCF was added to the solution to aid measurement. Experiments were carried out in a humid chamber (35 W × 55 D × 20 H cm) containing about 2 L of water to prevent evaporation of solution from the capillaries while flies fed on the test solution. The amount of evaporation of solution was measured in a vial without flies. The amount of solution consumed by flies was quantified by marking the start and end points of solution contained in the capillaries with a sharp marker, measuring between the start and end points using a digital caliper (resolution: 0.01mm), and subtracting the amount of evaporation without flies. After quantifying the length of solution in both capillaries in this way, a volumetric preference index (ranging between 1 and −1) was calculated using the following equation:
Tracking cumulative preferences in CAFE assays
To measure cumulative preference to the test solution in the CAFE assays, we used TrackMate, a plug-in of ImageJ. TrackMate recognizes white objects on a black background and records changes in coordinates. Therefore, in contrast to the basic CAFE assays that used blue dye in the test solution, mineral oil mixed with the fat-soluble Oil Red O dye was added on top of the test solution to use as an indicator. Images recorded for 4 h were first changed to grayscale to aid in analysis, which resulted in the color of the indicator changing to black. The images were cropped so only the test vial and empty vial are visible. The cropped images were inverted, so the indicator was visible as a white image, and the contrast of the images was adjusted to remove background white noise. This rendered the images suitable for TrackMate analysis, with a white indicator and black background. Using the coordinates of the indicator measured by TrackMate, we were able to measure the relative changes in length per minute and calculate the cumulative preference index by cumulatively adding the preference indices for each minute.
Statistics
GraphPad Prism5 was used for statistical analysis. In figures 2 and 3, the behavior data is presented as a box plot, with the middle line representing the median, the ‘+’ the mean, and the box boundaries and whiskers representing 25%/75% and 10%/90%, respectively. Asterisks shown in figures signify statistical significance (*p < 0.05).
Fig. 2.
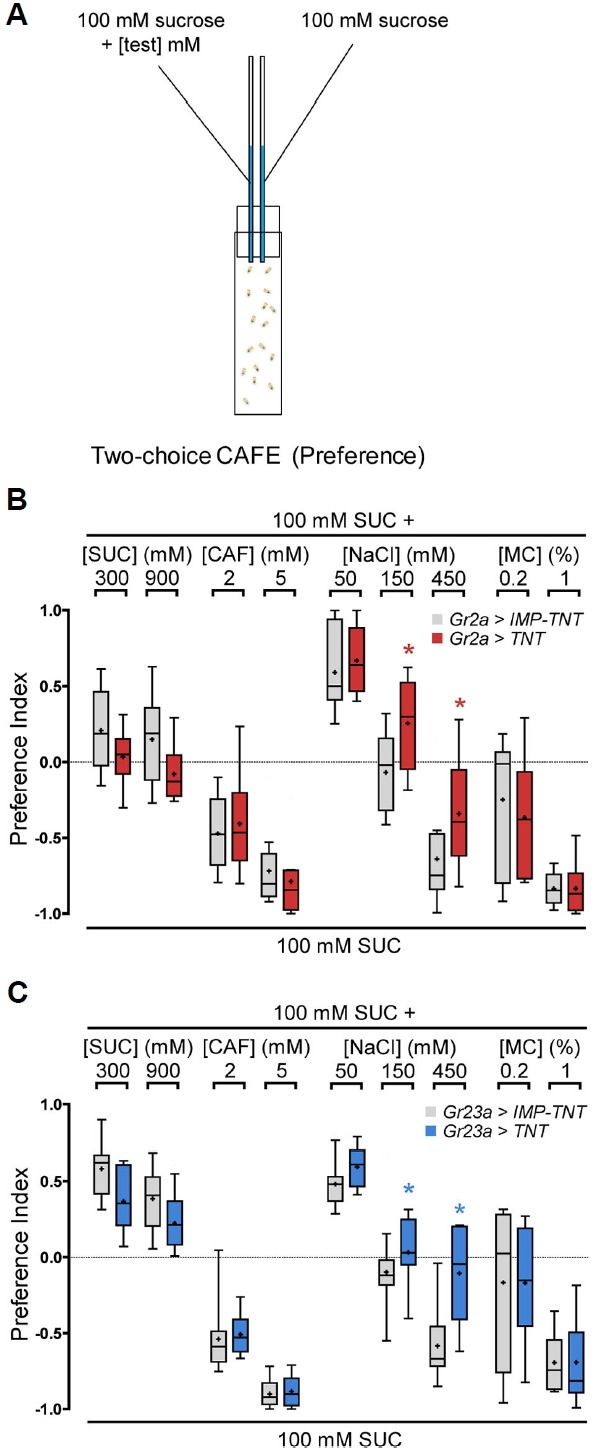
Inactivation of the Gr2a neuron impairs the feeding response to high concentrations of NaCl.
(A) Schematic of the CAFE (Capillary feeder) assay. (B, C) Feeding preference in response to the indicated tastants at various concentrations upon inhibition of the activity of Gr2a- (B) or Gr23a-GAL4-expressing GRNs (C). The Mann-Whitney U-test was used for pair-wise comparison. Asterisks signify statistical significance (*p < 0.05). For each data point, 6 < n < 18.
Fig. 3.
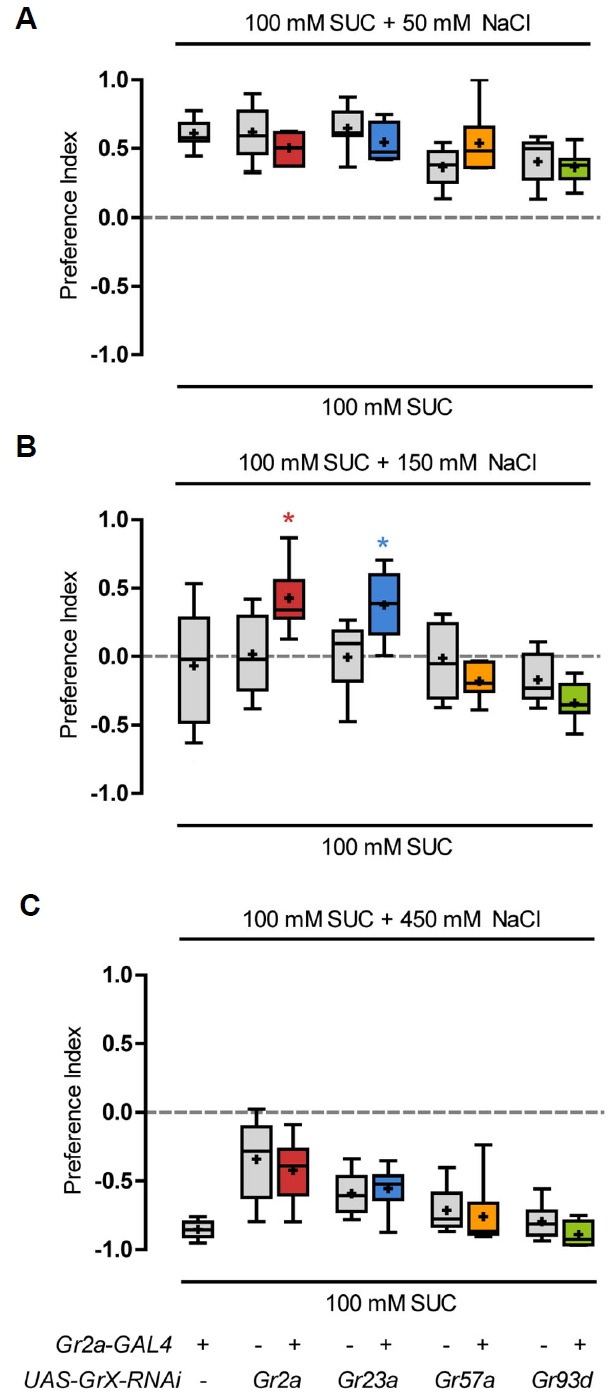
Inhibition of Gr2a and Gr23a function causes a change in feeding preference to NaCl.
Gr2a-GAL4 and the indicated Gr gene-RNAi transgenic lines were crossed, and the resulting progeny were exposed to 100 mM sucrose and 100 mM sucrose with 50 mM NaCl (A), 150 mM NaCl (B), or 450 mM NaCl (C), and feeding preferences were measured. The Kruskal-Wallis test was used for multiple comparisons of various genotypes. Dunn’s multiple comparison test was used for post hoc analysis. Asterisks signify statistical significance (*p < 0.05). For each data point, 6 < n < 14.
RESULTS AND DISCUSSION
Gr-GAL4 expressing neurons in the LSO
To examine the functions of adult fly pharyngeal GRNs and the Grs expressed in these GRNs, we consulted a previously established larval pharyngeal Gr-GRN map (Choi et al., 2016), since the larval pharyngeal sensory neurons are maintained during the transition to adulthood (Gendre et al., 2004). The 21 Gr-GAL4 drivers are expressed in five pairs of GRNs in the larval dorsal pharyngeal organ (DPS), which is one of the three larval pharyngeal organs (Choi et al., 2016). Three of these GRNs, DP1, DP2, and DP5, express Grs expected to act in bitter responses, DP4 expresses Gr43a which is known as a fructose receptor, and DP3 expresses four Grs of as yet unknown functions: Gr2a, Gr23a, Gr57a, and Gr93d (Choi et al., 2016). We decided to focus on the Grs expressed in DP3, since relatively little was known about the function of this neuron and Grs.
The first step we took was to verify that these Grs are indeed co-expressed in the same adult pharyngeal neuron that would have been maintained during the transition from larva to adulthood. All four Gr-GAL4 drivers, Gr2a-, Gr23-, Gr57a-, and Gr93d-GAL4, were expressed in single pairs of neurons in the LSO (labral sense organ) of adult flies (Fig. 1A). Since Gr43a-GAL4 is also known to express in the LSO (Fig. 1A) (LeDue et al., 2015) as well as the larval DP4 GRN (Choi et al., 2016), we examined whether the Gr2a-, Gr23-, Gr57a-, and Gr93d-GAL4 drivers were expressed in neurons independent from Gr43a-GAL4, as is the case in larva. By examining the expression of each Gr-GAL4 driver in pairwise combinations, we found that the Gr2a-, Gr23a-, Gr57a-, and Gr93d-GAL4 drivers are co-expressed in a single pair of neurons, and this neuron is independent from Gr43a-GAL4-expressing neurons (Fig. 1B). Axonal projection patterns of Gr2a-, Gr23a-, Gr57a-, and Gr93d-GAL4-expressing neurons to the subesophageal zone (SEZ), the primary gustatory center of the brain, were also very similar, with an umbrella-like projection pattern at both right and left dorsal regions of the SEZ (Fig. 1C). For Gr57a- and Gr93d-GAL4, the additional projections to the central region of the SEZ (Fig. 1C) are likely due to the additional expression of these drivers in bitter-sensitive neurons in the labellar sensilla (Weiss et al., 2011), compared to the specific expression of Gr2a- and Gr23a-GAL4 in only one pair of neurons in the peripheral nervous system. Thus, we found that at least three pairs of Gr-GAL4 expressing neurons exist in the LSO, with two pairs of GRNs acting in sugar sensing (LeDue et al., 2015) and one pair of neurons expressing four Grs of unknown function (Fig. 1D). For convenience, we will refer to this GRN of unknown function as the Gr2a neuron in the remainder of this study.
Fig. 1.
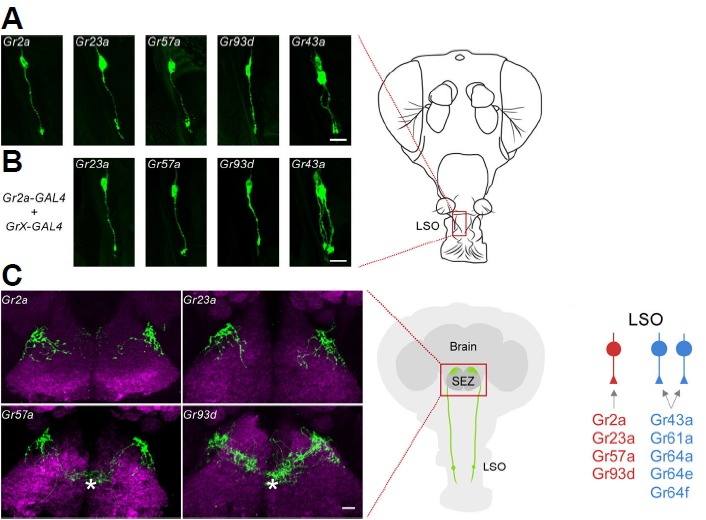
GRNs (Gustatory receptor neurons) in the labral sense organ (LSO) and Gr-GAL4 drivers expressed.
(A) Gr-GAL4 driven GFP reporter expression in the LSO. (B) Results of combinations of the Gr2a-GAL4 driver and the indicated Gr-GAL4 drivers. The position of the LSO is marked as a box on the schematic of a fly head on the right. All Gr-GAL4 expressing neurons exist as a pair in the LSO, and only one neuron of each pair is shown here. (C) Axonal projections of GRNs in the LSO to the subesophageal zone (SEZ). The brain neuropil is counterstained with the monoclonal antibody nc82 (magenta). The asterisks indicate projections due to labellar gustatory receptor neurons. On the right is a schematic of a fly head and GRNs in the LSO. (D) The three GRNs that exist in the LSO and the Gr-GAL4 drivers expressed. Scale bars, 10 μm.
The Gr2a neuron is involved in feeding suppression caused by a high concentration of salt
Since the Gr43a-expressing GRNs in the LSO were found to be important for sugar consumption (LeDue et al., 2015), it appeared highly likely that the Gr2a neuron would also regulate some aspect of feeding. To examine this hypothesis, activity of the Gr2a neuron was inhibited, and changes in feeding under various food conditions was observed. The Gr2a- and Gr23a-GAL4 drivers, which specifically express in only the Gr2a neuron, and UAS-tetanus toxin (TNT ) (Sweeney et al., 1995) were used for this purpose. A UAS construct expressing an inactive form of tetanus toxin (UAS-IMP-TNT) was used as a control. The Gr57a- and Gr93d-GAL4 drivers were not used because they express in other GRNs in addition to the Gr2a neuron. Two capillary tubes containing 100 mM sucrose solution were placed in a vial, various chemicals including sucrose (sweet) or caffeine (bitter) were added to one tube, and the amounts consumed from the test and control capillaries were compared (Fig. 2A). Both the control flies and flies with Gr2a neuron function inhibited consumed larger amounts of high concentration sucrose solution, and showed dose-dependent aversion to food containing caffeine (Figs. 2B and 2C). No difference in consumption was observed when flies were given a choice of low concentration salt, but differences were observed in the degree of aversion to food containing high salt (Figs. 2B and 2C). Addition of methyl cellulose increases viscosity and inhibits feeding, and inhibition of Gr2a neuron function did not affect this reduction in feeding (Figs. 2B and 2C). Thus, fruit flies find a low concentration of salt (50 mM) more attractive than food with no salt, and show an aversive reaction to a high concentration of salt (450 mM) (Figs. 2B and 2C) (Zhang et al., 2013). Among the conditions we tested, inhibition of Gr2a neuron activity only affected feeding upon high NaCl conditions.
Gr2a is necessary for the reduction of feeding in response to high concentration Na+
To test whether the four Grs expressed in the Gr2a neuron are involved in the reduction of feeding in response to high concentration NaCl, we used RNAi lines for each Gr gene. We examined the amounts consumed by flies with a specific Gr gene functionally inhibited in the Gr2a neuron, by crossing Gr2a-GAL4 and each individual Gr RNAi line. All control lines, including Gr2a-GAL4 only and each UAS-Gr-RNAi line, showed a feeding preference for the solution containing low concentration NaCl (100 mM sucrose + 50 mM NaCl) compared to 100 mM sucrose only solution (Fig. 3A). The control lines also showed an aversive response to food with an extremely high concentration of NaCl (450 mM), and avoided feeding (Fig. 3C). When exposed to a moderately high concentration of NaCl (150 mM), the control lines showed a neutral or slightly aversive response (Fig. 3B). RNAi knockdown of the Gr2a or Gr23a gene suppressed this reduction in feeding preference, while knockdown of Gr57a or Gr93d had no affect (Fig. 3B).
To verify the involvement of the Gr2a gene in the response to high NaCl, we generated a Gr2a mutant (Gr2aGAL4). We constructed a knock-in mutant with part of the first exon of Gr2a substituted with the GAL4 coding region (Fig. 4A). When expression of the GAL4 gene inserted into the Gr2a locus was examined by a GFP reporter, expression was observed in one pair of GRNs in the LSO, and the shape of the projections to the brain were identical to the projections observed with the Gr2a-GAL4 driver (Fig. 4B). The Gr2a mutant showed a change in response to food with a moderately high concentration of NaCl similar to flies with impaired Gr2a neuron function or flies subjected to Gr2a RNAi (Fig. 4C). Introduction of Gr2a cDNA into the Gr2a mutant fully rescued the mutant phenotype to be indistinguishable from the control (Fig. 4C). This response of the Gr2a mutant could be recapitulated in response to sodium gluconate, but not to other salts such as KCl and CaCl2 or caffeine (Fig. 4C).
Fig. 4.
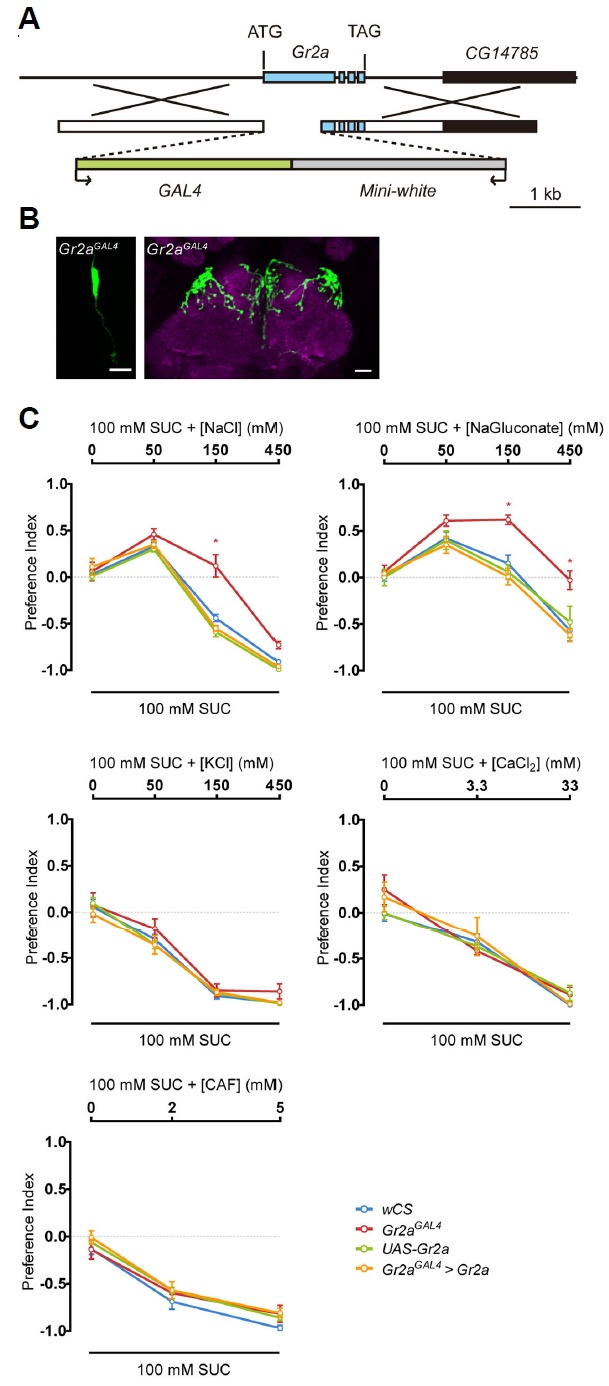
The Gr2a mutant (Gr2aGAL4) shows repression of feeding suppression specific to high concentration Na+.
(A) Schematics showing the mutant generation strategy for Gr2aGAL4. (B) Gr2aGAL4-driven GFP reporter expression. The LSO and axonal projections to the SEZ are the same regions marked in Fig. 1A and 1C, respectively. (C) Feeding preference in response to the indicated salts and caffeine (CAF) at various concentrations. Two-way ANOVA and the Bonferroni post hoc test were used for analysis of significance. Asterisks signify statistical significance (*p < 0.05). For each data point, 6 < n < 10.
In conclusion, the Gr2a neuron is involved in behavior that avoids the consumption of food with a high concentration of salt. Among the Grs that express in the Gr2a neuron, Gr2a is necessary for feeding suppression in response to a moderately high concentration of Na+, based on RNAi and mutant analyses. Changes in the response to 450 mM NaCl were observed when Gr2a neuron function was inhibited, but not in the RNAi experiments for the individual Grs or the Gr2a mutant, suggesting the existence of other unknown functionally redundant factors in the Gr2a neuron.
The hunger signal overcomes aversive behavior to unappetizing foods
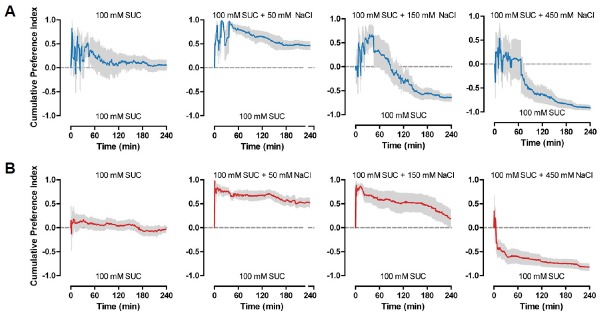
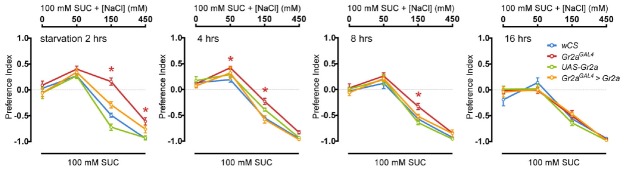
In the CAFE assay that we use as a behavioral paradigm to measure feeding, flies are allowed to feed for 4 h before measuring the amounts that were consumed. We used 4 h to minimize the effects of internal nutrient sensing on feeding. This is because, after 4 h of feeding, calories of the food, as well as taste, were shown to act as an important factor in determining feeding, with preference to non-sweet caloric sugars increasing after 4 h (Stafford et al., 2012). To analyze in detail fly feeding behavior to various salt concentrations, we used video tracking to measure cumulative feeding preference in 1 min intervals (Fig. 5). For the wCS flies that were used as controls, flies showed fluctuations in feeding during the first hour, perhaps reflecting the need for a certain amount of time to adjust. Regardless of this fluctuation during the first hour of feeding, wCS flies continuously preferred food with 50 mM NaCl added to 100 mM sucrose, showed an initial preference to 150 mM NaCl that changed to aversion, and showed aversion to 450 mM NaCl from the onset of feeding (Fig. 5A). In contrast, the change from preference to aversion to 150 mM NaCl appeared to take place at a slower rate in the Gr2a mutant flies (Fig. 5B). This subtlety in mutant phenotype suggested that experiments under different feeding conditions from the conditions that we used might result in somewhat different results. Thus, to further examine the functions of the Gr2a neuron and Gr2a, we observed feeding under various starvation conditions. A couple of results stood out from experiments subjecting Gr2a mutant flies to starvation conditions ranging from 2 h to 16 h, and exposing them to food containing various amounts of salt. First, for all genotypes tested, the enhanced feeding preference to 50 mM NaCl disappeared as starvation time increased (Fig. 6). Second, the difference in feeding preference to 150 mM NaCl observed in the Gr2a mutant also gradually disappeared as starvation time increased (Fig. 6). These results suggest that the hungrier the flies are, the less it matters whether the food is slightly more appetizing or not due to the addition of a small amount of salt, since the acquisition of an energy source would take precedence over taste.
Fig. 5.
The Gr2a mutant shows a gradual change in feeding preference to 150 mM NaCl.
Cumulative feeding preference curves for control wCS (A) and Gr2aGAL4 (B) flies during the 4 hrs of CAFE assays to various concentrations of NaCl. For each data point, 8 < n < 10.
Fig. 6.
Starvation influences feeding preference. Feeding preference to various concentrations of NaCl was measured in flies subjected to various starvation conditions prior to the CAFE assay.
Two-way ANOVA and the Bonferroni post hoc test were used for analysis of significance. Asterisks signify statistical significance (*p < 0.05). For each data point, 6 < n < 10.
In this study, we identified a pair of fly pharyngeal GRNs and the Grs that are expressed therein, and their functions as modulators of feeding. Gr2a and the Gr2a-GAL4 expressing pharyngeal GRN are involved in feeding regulation specifically in response to high concentration of Na+. IR76b, a member of the ionotropic glutamate receptor (IR) family, was previously shown to function in the detection of low salt, and to be involved in the attraction to low salt (Zhang et al., 2013). Our results show that Gr2a is involved in the aversion to high salt, but it is unclear whether Gr2a is a salt receptor per se or a modulator of the response. The subtlety of the phenotypes we observed for Gr2a illustrate that the results of behavior experiments can vary depending on experimental conditions, and also suggest that phenotypes observed for genes involved in feeding may vary depending on context, since feeding is influenced by many factors including nutritional status.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (NRF-2016R1D1A1B03932743 (JYK) and NRF-2016 R1A5A2008630 (SJM)).
REFERENCES
- Choi J., van Giesen L., Choi M.S., Kang K., Sprecher S.G., Kwon J.Y. A pair of pharyngeal gustatory receptor neurons regulates caffeine-dependent ingestion in Drosophila Larvae. Front Cell Neurosci. 2016;10:181. doi: 10.3389/fncel.2016.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne P.J., Warr C.G., Carlson J.R. Candidate taste receptors in Drosophila. Science. 2000;287:1830–1834. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- Dahanukar A., Lei Y.T., Kwon J.Y., Carlson J.R. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du E.J., Ahn T.J., Choi M.S., Kwon I., Kim H.W., Kwon J.Y., Kang K. The mosquito repellent citronellal directly potentiates drosophila TRPA1, facilitating feeding suppression. Mol Cells. 2015;38:911–917. doi: 10.14348/molcells.2015.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman E.G., Dahanukar A. Molecular neurobiology of Drosophila taste. Curr Opin Neurobiol. 2015;34:140–148. doi: 10.1016/j.conb.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Yavuz A., Slone J., Jagge C., Song X., Amrein H. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr Biol. 2015;25:621–627. doi: 10.1016/j.cub.2014.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre N., Luer K., Friche S., Grillenzoni N., Ramaekers A., Technau G.M., Stocker R.F. Integration of complex larval chemosensory organs into the adult nervous system of Drosophila. Development. 2004;131:83–92. doi: 10.1242/dev.00879. [DOI] [PubMed] [Google Scholar]
- Gong W.J., Golic K.G. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci USA. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Moon S.J., Montell C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc Natl Acad Sci USA. 2007;104:14110–14115. doi: 10.1073/pnas.0702421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Moon S.J., Wang X., Ren Q., Montell C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr Biol. 2008;18:1797–1801. doi: 10.1016/j.cub.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Choi M.S., Kang K., Kwon J.Y. Behavioral analysis of bitter taste perception in Drosophila larvae. Chem Senses. 2016;41:85–94. doi: 10.1093/chemse/bjv061. [DOI] [PubMed] [Google Scholar]
- Kwon J.Y., Dahanukar A., Weiss L.A., Carlson J.R. Molecular and cellular organization of the taste system in the Drosophila larva. J Neurosci. 2011;31:15300–15309. doi: 10.1523/JNEUROSCI.3363-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDue E.E., Chen Y.C., Jung A.Y., Dahanukar A., Gordon M.D. Pharyngeal sense organs drive robust sugar consumption in Drosophila. Nat Commun. 2015;6:6667. doi: 10.1038/ncomms7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Moon S.J., Montell C. Multiple gustatory receptors required for the caffeine response in Drosophila. Proc Natl Acad Sci USA. 2009;106:4495–4500. doi: 10.1073/pnas.0811744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Moon S.J., Wang Y., Montell C. A Drosophila gustatory receptor required for strychnine sensation. Chem Senses. 2015;40:525–533. doi: 10.1093/chemse/bjv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra D., Miyamoto T., Rezenom Y.H., Broussard A., Yavuz A., Slone J., Russell D.H., Amrein H. The molecular basis of sugar sensing in Drosophila larvae. Curr Biol. 2013;23:1466–1471. doi: 10.1016/j.cub.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T., Slone J., Song X., Amrein H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell. 2012;151:1113–1125. doi: 10.1016/j.cell.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S.J., Lee Y., Jiao Y., Montell C. A Drosophila gustatory receptor essential for aversive taste and inhibiting male-tomale courtship. Curr Biol. 2009;19:1623–1627. doi: 10.1016/j.cub.2009.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Kwon J.Y. Heterogeneous expression of Drosophila gustatory receptors in enteroendocrine cells. PLoS ONE. 2011;6:e29022. doi: 10.1371/journal.pone.0029022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H.M., Warr C.G., Carlson J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci USA. 2003;100(Suppl 2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K., Brady R., Jr, Cravchik A., Morozov P., Rzhetsky A., Zuker C., Axel R. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- Shim J., Lee Y., Jeong Y.T., Kim Y., Lee M.G., Montell C., Moon S.J. The full repertoire of Drosophila gustatory receptors for detecting an aversive compound. Nat Commun. 2015;6:8867. doi: 10.1038/ncomms9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford J.W., Lynd K.M., Jung A.Y., Gordon M.D. Integration of taste and calorie sensing in Drosophila. J Neurosci. 2012;32:14767–14774. doi: 10.1523/JNEUROSCI.1887-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R.F. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- Stocker R.F. Taste perception: Drosophila - a model of good taste. Curr Biol. 2004;14:R560–561. doi: 10.1016/j.cub.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Sweeney S.T., Broadie K., Keane J., Niemann H., O’Kane C.J. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- Tissot M., Stocker R.F. Metamorphosis in Drosophila and other insects: the fate of neurons throughout the stages. Prog Neurobiol. 2000;62:89–111. doi: 10.1016/s0301-0082(99)00069-6. [DOI] [PubMed] [Google Scholar]
- Vosshall L.B., Stocker R.F. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Weiss L.A., Dahanukar A., Kwon J.Y., Banerjee D., Carlson J.R. The molecular and cellular basis of bitter taste in Drosophila. Neuron. 2011;69:258–272. doi: 10.1016/j.neuron.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.V., Ni J., Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340:1334–1338. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]