Abstract
Background:
Drug resistance to targeted therapies occurs in lung cancer, and resistance mechanisms related to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) are continuously being discovered. We aimed to establish a novel method for highly parallel multiplexed detection of genetic mutations related to EGFR TKI-resistant lung cancer using Agena iPLEX chemistry and matrix-assisted laser desorption ionization time-of-flight analysis on the MassARRAY mass spectrometry platform.
Methods:
A review of the literature revealed 60 mutation hotspots in seven target genes (EGFR, KRAS, PIK3CA, BRAF, ERBB2, NRAS, and BIM) that are closely related to EGFR TKI resistance to lung cancer. A total of 183 primers comprised 61 paired forward and reverse amplification primers, and 61 matched extension primers were designed using Assay Design Software. The detection method was established by analyzing nine cell lines, and by comparison with LungCarta™ kit in ten lung cancer specimens. EGFR, KRAS, and BIM genes in all cell lines and clinical samples were subjected to Sanger sequencing for confirming reproducibility.
Results:
Our data showed that designed panel was a high-throughput and robust tool, allowing genotyping for sixty hotspots in the same run. Moreover, it made efficient use of patient diagnostic samples for a more accurate EGFR TKIs resistance analysis. The proposed method could accurately detect mutations in lung cancer cell lines and clinical specimens, consistent with those obtained by the LungCarta™ kit and Sanger sequencing. We also established a method for detection of large-fragment deletions based on single-base extension technology of MassARRAY platform.
Conclusions:
We established an effective method for high-throughput detection of genetic mutations related to EGFR TKI resistance based on the MassARRAY platform, which could provide more accurate information for overcoming cancers with de novo or acquired resistance to EGFR-targeted therapies.
Keywords: Drug Resistance, Epidermal Growth Factor Receptor, Lung Cancer, MassARRAY, Targeted Molecular Therapy
Introduction
Most patients with epidermal growth factor receptor (EGFR)mutant lung cancers develop acquired resistance to EGFR tyrosine kinase inhibitors (TKIs). This resistance to treatment with EGFR TKIs often involves both pharmacological and biological mechanisms. The biological mechanisms involve three main categories of molecular features: alterations in the drug target, activation of alternative signaling pathways, and phenotypic changes.[1] Because many resistance alterations have been defined, the screening of multigene resistance mutations associated with EGFR TKIs will become the preferred approach for routine clinical practice.[2]
The detection of genetic mutations can be implemented based on single-base extension technology and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) using the MassARRAY iPLEX platform, which utilizes multiplex polymerase chain reaction (PCR). Target sequences are amplified by amplification primers, and extension primers located one base before the mutation site that are complementary to the amplification products are used to perform single-base extension reactions. Single-nucleotide polymorphisms can be distinguished using MALDI-TOF MS according to the molecular weight of the different extension bases of the screening site.[3] Moreover, the MassARRAY platform is also ideal for the screening of multiple mutations, as its design is both accurate and flexible.[2] Therefore, it is of great significance to establish a multigene detection method that is especially suitable for detecting EGFR TKI-resistant mutations.
Methods
Ethical approval
Informed consent was obtained from each patient, and the study was conducted in accordance with the Declaration of Helsinki and was approved by the Local Ethics Committee of Guangdong General Hospital (No. GDREC2013013(R2)).
Materials
Patient specimens and cell lines
We randomly selected a cohort of ten lung cancer specimens from the Guangdong Lung Cancer Institute of Guangdong General Hospital in 2016. All samples, which were stored at −80°C after being frozen in liquid nitrogen, were assessed by two pathologists to ensure that more than 50% of the sample consisted of tumor tissue. We used nine nonsmall cell lung cancer cell lines (H460, PC9, H1650, H1975, A549, GLC82, L78, HCC827, and H2228), which were purchased from the cell bank of the Chinese Academy of Sciences in Shanghai.
Reagents and instruments
QIAsymphony DNA Mini Kit (Qiagen, Valencia, Germany); LungCarta ™ kit, PCR Accessory Set, iPLEX Pro Reagent Kit and SpectroCHIP® (Agena Bioscience, San Diego, CA, USA); H2O (Sigma-Aldrich, St. Louis, MO, USA); QIAsymphony SP (Qiagen, Valencia, Germany); Ex Taq ™ Hot Start Version Kit (Takara Biotechnology, Dalian, China); Thermo NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA, USA); MassARRAY® Nanodispenser and MassARRAY® Analyzer (Agena Bioscience, San Diego, CA, USA); ABI 3730xl Sequencing Machine; and PCR Machine (Life Technologies, Carlsbad, CA, USA) were used.
Methods
Preparation of polygenic primer panel
Determination of the driver genes of lung cancer
Based on our review of the literature and data on EGFR-targeted resistance to lung cancer, as well as on previous findings from our institution, seven target oncogenes (EGFR, KRAS, PIK3CA, BRAF, ERBB2, NRAS, and BIM) that are closely related to the targeted therapy resistance of the EGFR gene were used in the polygenic primer panel.
Determination of the hotspots of driver genes
Our review of the Catalogue of Somatic Mutations in Cancer (COSMIC) database identified the COSMIC identifier numbers of the following seven genes: EGFR (ENST00000275493), KRAS (ENST00000256078), PIK3CA (ENST00000263967), BRAF (ENST00000288602), ERBB2 (ENST00000269571), NRAS (ENST00000369535), and BIM (ENST_00000393256). According to the mutation frequencies of the seven oncogenes in lung cancer, 60 resistance mutations related to EGFR gene targeted therapy were added to the polygenic primer panel [Table 1].
Table 1.
The newly designed panel contains protein mutation loci of 7 genes
| Gene | Exon | Protein mutation locus |
|---|---|---|
| EGFR | Exon 18 | p.G719SCDA |
| Exon 19 | p.E746_A750delELREA p.E746_T751>A p.E746_S752>V p.L747_T751delLREAT p.L747_P753>S p.L747_S752delLREATS p.L747_A750>P | |
| Exon 20 | p.T790M p.S768I p.G796SDA p.C797YSA p.L747S p.D761YN p.T854APS p.A763_Y764insFQEA p.V769_D770insASV p.D770_N771insSVD p.H773_V774insNPH | |
| Exon 21 | p.L858R p.L861Q | |
| KRAS | / | p.G12CRSDVA p.G13CRSDAV p.Q61H |
| PIK3CA | / | p.E542KQVAG p.E545KQAGVD p.R1023Q p.H1047RLY |
| BRAF | / | p.G469AVER p.D594NHGVA p.L597RQPSV p.V600MLEAGKRD |
| BIM | / | Del2903 |
| ERBB2 | / | p.S310FY p.R678Q p.L755S p.D769YHN p.A775_G776insYVMA p.V777LM p.V842I |
| NRAS | / | p.Q61K |
EGFR: Epidermal growth factor receptor. “/” indicates unclassfied Exon.
Design of the polygenic primer panel
The genome sequence numbers of the following seven target genes were identified in GenBank: EGFR (NG_007726.3), KRAS (NG_007524.2), PIK3CA (NG_012113.2), BRAF (NG_007873.3), ERBB2 (NG_007503.1), NRAS (NG_007572.1), and BIM (NG_029006.1). According to the mutation label and format requirements of the MassARRAY platform, we marked 60 mutant loci in the genomic DNA (gDNA) sequences. It is important to note that the BIM gene contains a large-fragment deletion of 2903 bp, making it difficult to design the large-fragment deletion in one assay; therefore, we designed the BIM wild-type and BIM deletion in two separate assays. To include all 61 assays in the polygenic primer panel, the relevant parameters of Assay Design Software (ADS) were adjusted. We set the maximum number of loci capable of being detected simultaneously to 10 mutant loci. Sixty-one loci were randomly distributed in 12 wells using ADS according to the primer design (avoidance of the formation of dimers/mismatches, etc.). In total, 183 primers comprising 61 paired forward and reverse amplification primers and 61 matched single-base extension primers were designed. Target sequences were amplified using amplification primers, and extension primers were located one base before the mutation site and were complementary to the amplification products. Single-base extension reactions were then performed.
Configuration of the polygenic primer panel
Primers were synthesized by Shanghai Sangon Biological Engineering Technology and Service Co., Ltd. The polygenic primer panel was configured as follows: (1) Amplification primers were first diluted to 10 μmol/L. The working liquid was a mixture of all primers in each well, including 0.5 μmol/L of each primer. According to the ADS parameters, the forward and reverse amplification primers were distributed into 12 pipes. The forward primer P1-F and reverse amplification primer P1-R were designated as Group 1; the forward primer P2-F and reverse amplification primer P2-R were designed as Group 2, etc. (2) Extension primers were first diluted to 500 μmol/L, and the primers were mixed according to molecular weight. Extension primers E1 were Group 1, extension primers E2 were Group 2, and so on (E1-E12). The 12 amplification primers corresponded to 12 extension primers (e.g., P1 corresponded to E1, etc.).
Establishment of a detection method
The detection method was verified by analyzing nine cell lines (H460, PC9, H1650, H1975, A549, GLC82, HCC827, H1299, and H2228) and ten lung cancer specimens. The proposed method was then validated by comparison with the LungCarta ™ kit or previously reported results. The gDNA of a healthy person, a sample of foreskin tissue was obtained from individuals after we obtained their informed consent, was used as a negative control, and H2O was used as a blank control. Each sample required a total of 120 ng of gDNA for 10 ng/well × 12 wells.
This procedure was carried out using the MassARRAY system platform. Experiments using the LungCarta ™ kit were performed according to the manufacturer's protocol. The method of detection used by the polygenic primer kit was as follows:
Polymerase chain reaction
gDNA was extracted from cell lines and patient specimens according to the manufacturer's protocol and was quantified on a NanoDrop ND-1000 spectrophotometer. gDNA was amplified using a PCR Accessory Set. The thermocycling cocktail was composed of 0.5 μl PCR buffer (10×), 0.4 μl MgCl2 (25 mmol/L), 0.1 μl dNTPs (25 mmol/L), 0.2 μl PCR enzyme (5 U/μl), 1 μl amplification primer mix (P1-P12), 1 μl gDNA (10 ng/μl), and H2O (final volume 5 μl). The thermocycling conditions were: 94°C for 2 min; this was followed by 45 cycles at 94°C for 30 s, 56°C for 30 s, 72°C for 1 min, and a final extension step at 72°C for 5 min.
Shrimp Alkaline Phosphatase reaction
dNTPs in the PCR products were removed using Shrimp Alkaline Phosphatase (SAP). For this reaction, 0.3 μl SAP (1.7 U/μl) and 0.17 μl SAP buffer (10×) were added to step 1 PCR products, and H2O was added to a final volume of 7 μl. Reaction conditions were 37°C for 40 min and 85°C for 5 min.
Extension reaction
The single-base extension reaction was performed using the iPLEX Pro Reagent Kit to hybridize and elongate the extension primers at the nucleotide position of interest. For the single-base extension, 0.2 μl Typeplex buffer (10×), 0.2 μl Typeplex Termination Mix (10×), 0.041 μl Typeplex Thermosequenase (33 U/μl), and 0.804 μl extension primers (E1–E12) were mixed with step 2 PCR products, and H2O was added to a final volume of 9 μl. Reaction conditions were: 94°C for 30 s, followed by 35 cycles of (94°C for 5 s, [52°C for 5 s, 80°C for 5 s], 5 cycles), and a final extension at 72°C for 3 min.
Desalination
For desalination, 41 μl H2O and 15 mg clean resin (96-well microplates) were added to step 3 extension products. The plate was rotated for 30–60 min and then centrifuged at 3200 ×g for 5 min.
Spotter and analysis
The supernatant from step 4 extension products was spotted onto a matrix-precoated SpectroCHIP® using the MassARRAY® Nanodispenser and scanned using the MassARRAY® Analyzer. The results were analyzed by MassArray® Workstation software (version 4.0, Agena Bioscience, San Diego, CA, USA). Mutations were distinguished using MALDI-TOF MS according to molecular weight. Peaks in the mass spectra were identified as mutations.
Direct sequencing
The newly established methods were evaluated by Sanger sequencing of EGFR, KRAS, and BIM genes in nine cell lines and the lung cancer specimens. EGFR and KRAS mutations were detected by Sanger sequencing using a previously published protocol.[4] Primers used for sequencing analysis of the BIM gene were as follows: forward (F), CCTCATGATGAAGGCTAACTCAA; reverse wild-type (R-wt), TGGTGGTCACTTGTCAGAGGTT; and reverse mutation (R-mut), TGTTCTCCATA GAGGCTGTGCC. For this reaction, 5 μl PCR Ex Taq ™ buffer (10×), 4 μl dNTPs (10 mmol/L), 0.5 μl Ex Taq ™ HS enzyme (5 U/μl), 0.5 μl each primer (12.5 μmol/L), 1 μl DNA, and 38 μl H2O were added to a final volume of 50 μl. Reaction conditions were: 94°C for 7 min, followed by 35 cycles at 94°C for 30 s, 60°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 10 min. Sequencing data were analyzed using Sequencing Analysis Software version 5.2 (Applied Biosystems, Foster City, CA, USA).
Results
The polygenic primer panel contains mutation sites
We selected 60 mutation hotspots in seven target genes (EGFR, KRAS, PIK3CA, BRAF, ERBB2, NRAS, and BIM) that are closely related to EGFR TKI-targeted therapy resistance. The proposed panel included primarily the acquired resistance mutations to EGFR TKI treatment that often occur in exon 20 of the EGFR gene, including EGFR T790M, T797N, and exon 20 insertion mutations, as well as de novo resistance variants such as a 2903 bp large-fragment deletion of the BIM gene. It is important to note that EGFR-sensitive mutations, such as the exon 19 deletion and exon 21 L858R, were also included in the designed panel. Synchronous screening-sensitive and -resistance mutations will provide more detailed information for analyzing the resistance mechanism of EGFR TKI-targeted therapies. The detailed protein mutation sites of genes are shown in Table 1.
Establishment of a detection method in cell lines
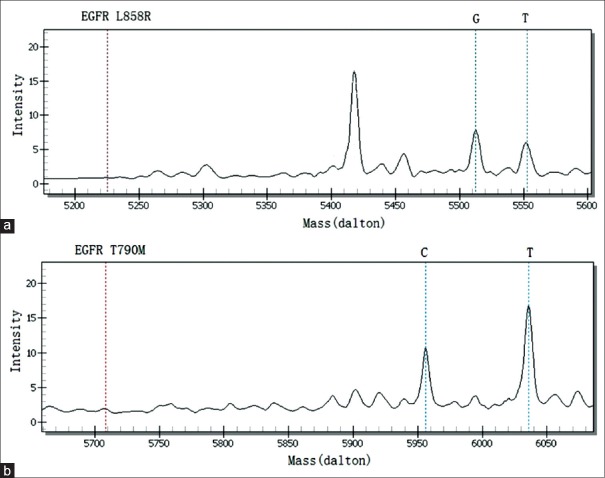
The detection method that utilizes the polygenic primer panel was established by analyzing nine lung cancer cell lines. All cell lines were confirmed using Sanger sequencing. The resistance panel did not include PTEN loss and ALK fusion mutations, rendering it unable to detect the corresponding mutations in H1650 and H2228 cell lines. Other results from the newly established method were consistent with previously reported mutations in cell lines. Detailed mutation sites within genes and proteins are shown in Table 2. The detection results of H1975, an EGFR TKI-resistant cell line harboring L858R and T790M mutations, are shown in Figure 1. No mutations were detected in either the negative control or blank control.
Table 2.
Validation results of designed resistance panel in lung cancer cell lines
| Cell line | Previously reported ATCC | Designed resistance panel |
|---|---|---|
| H460 | KRAS mutation | KRAS_Q61H (c.183A>T) |
| PIK3CA mutation | PIK3CA_E545K (c.1633G>A) | |
| PC9 | EGFR_Exon 19 deletion | p.E746_A750delELREA (c.2235-2249del15) |
| H1650 | EGFR_Exon 19 deletion | p.E746_A750delELREA (c.2235-2249del15) |
| PTEN loss | The new panel does not include PTEN gene | |
| H1975 | EGFR_L858R | EGFR_L858R (c.2573T>G) |
| EGFR_T790M | EGFR_T790M (c.2369C>T) | |
| A549 | KRAS mutation | KRAS_G12S (c.34G>A) |
| GLC82 | EGFR_L858R | EGFR_L858R (c.2573_2574TG>GT) |
| HCC827 | EGFR_Exon19 deletion | p.E746_A750delELREA (c.2236_2250del15) |
| H2228 | EML4-ALK fusion | No mutation |
| The new panel does not include ALK fusion | ||
| H1299 | EGFR/ALK/KRAS negative | No mutation |
ATCC: American Type Culture Collection; EGFR: Epidermal growth factor receptor.
Figure 1.
EGFR_L858R and EGFR_T790M were detected by the newly designed panel in H1975 lung cancer cells. (a) EGFR_L858R was detected by the newly designed panel (G: 5512.60); (b) EGFR_T790M was detected by the newly designed panel (T: 6035.80). EGFR: Epidermal growth factor receptor.
Establishment of a detection method in lung cancer specimens
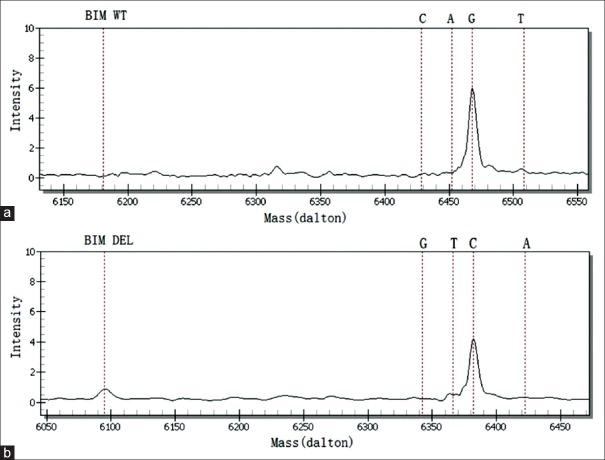
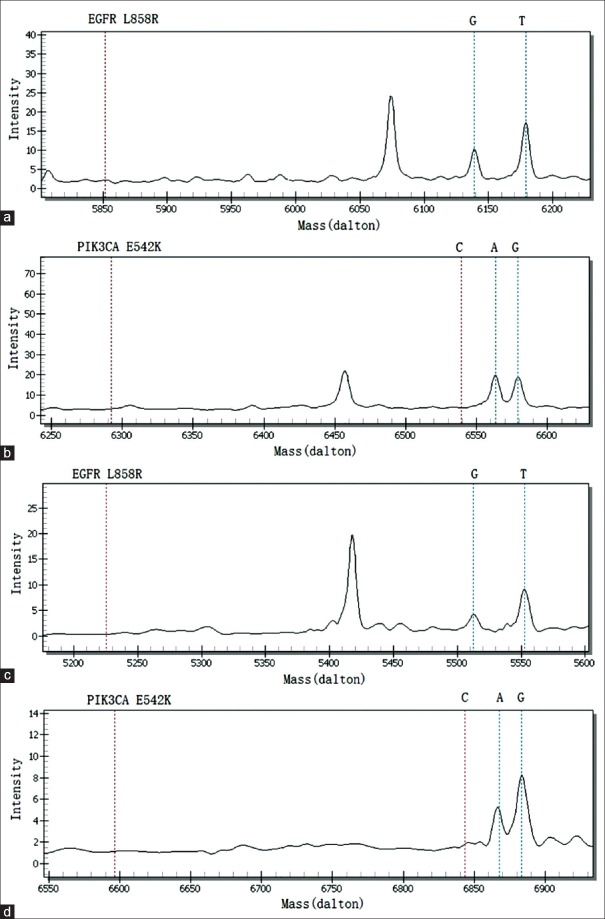
The detection method that utilizes the polygenic primer panel was established by analyzing ten lung cancer tissue specimens and was validated by comparison with the LungCarta ™ kit [Table 3]. A 2903 bp large-fragment deletion of the BIM gene was found in lung cancer tissue sample No. 20455 using the newly established method [Figure 2]; however, the LungCarta ™ kit was unable to detect the mutation. In addition, the resistance panel did not include a P53 mutation, rendering it unable to detect the P53_Y220C in sample No. 33070. With respect to the other clinical lung cancer specimens, the findings were consistent with observations using the LungCarta ™ kit [the result of tissue sample No. 22840 shown in Figure 3]. We also found that two of the ten specimens harbor resistance variations; sample No. 20455 harbored EGFR_L858R and BIM_DEL, and sample No. 22840 harbored EGFR_L858R and PIK3CA_E542K.
Table 3.
Comparison results between newly designed resistance panel and LungCarta™ kit in lung cancer tissue samples
| Sample number | LungCarta™ results | Designed resistance panel |
|---|---|---|
| 20455 | EGFR_L858R | EGFR_L858R (c.2573T>G) |
| BIM_DEL | ||
| 20483 | EGFR_L858R | EGFR_L858R (c.2573T>G) |
| 22840 | EGFR_L858R | EGFR_L858R (c.2573T>G) |
| PIK3CA_E542K | PIK3CA_E542K (c.1624G>A) | |
| 27001 | EGFR_L858R | EGFR_L858R (c.2573T>G) |
| 33030 | EGFR_Exon 19 deletion | p.E746_A750delELREA (c.2235_2249del15) |
| 33032 | No mutation | No mutation |
| 33040 | No mutation | No mutation |
| 33052 | EGFR_Exon 19 deletion | EGFR_p.L747_P753>S (c.2240_2257del18) |
| 33070 | EGFR_Exon 19 deletion | p.E746_A750delELREA (c.2235_2249del15) |
| P53_Y220C | ||
| 33071 | No mutation | No mutation |
EGFR: Epidermal growth factor receptor.
Figure 2.
BIM_DEL and BIM_WT were detected by the newly designed panel in lung cancer tissue sample No. 20455. (a) BIM_WT was detected by the newly designed panel (G: 6468.20); (b) BIM_DEL was detected by the newly designed panel (C: 6382.20).
Figure 3.
Coexistence of EGFR_L858R and PIK3CA_E542K was detected in the newly designed panel and in the LungCarta™ kit in lung cancer tissue sample No. 22840. (a) EGFR_L858R was detected by LungCarta™ (G: 6139.00); (b) PIK3CA_E542K was detected by LungCarta™ (A: 6563.30); (c) EGFR_L858R was detected by the newly designed panel (G: 5512.60); (d) PIK3CA_E542K was detected by the newly designed panel (A: 6867.50). EGFR: Epidermal growth factor receptor.
Design of a detection method for large-fragment deletions
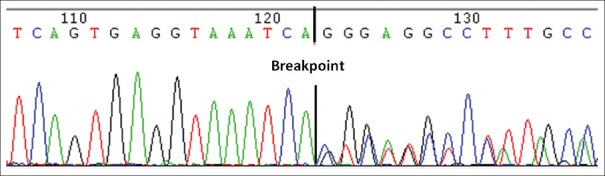
We designed the respective BIM wild-type and BIM deletion using two separate assays (the sequence after the extended single base differs). Both the wild-type and deletion assays contained peaks in the mass spectra of the BIM gene, indicating a large-fragment deletion; only the wild-type peak was observed in the no-deletion specimen. According to the newly designed panel, one of the specimens harbored a BIM gene variation. This result was further confirmed by direct sequencing [Figure 4]. We established a detection method for large-fragment deletions based on single-base extension technology using the MassARRAY platform.
Figure 4.
The BIM gene was detected in sample No. 20455 using Sanger sequencing.
Discussion
The genes and mutations included in the newly designed panel were chosen according to three types of EGFR TKI resistance mechanisms. The first mechanism is the acquired resistance to EGFR TKI treatment, which often occurs in exon 20 of the EGFR gene. The most common alteration of acquired resistance involves the secondary T790M mutation, which accounts for approximately 50% of EGFR TKI resistance,[5] and the C797S mutation, which is related to resistance to the third-generation EGFR TKIs.[6] EGFR exon 20 insertion mutations, which account for approximately 10% of all EGFR mutations, are generally associated with an insensitivity to available TKIs.[7,8,9] Other rare secondary mutations in EGFR, including L747S, D761Y, and T854A, have been described in patients with acquired resistance.[10,11,12] The second mechanism is the activation of alternative pathways. For example, variations in the BIM gene are associated with intrinsic EGFR TKI resistance.[13] Several other types of acquired resistance to EGFR TKIs have also been identified, including mutations in PIK3CA, KRAS, NRAS, ERBB2, and BRAF.[14,15,16,17,18] The third mechanism is the loss of activating EGFR mutations, which is believed to contribute to the acquired resistance to EGFR TKIs in lung cancer cells.[19] Our panel includes sensitive mutations such as G719X in exon 18, the exon 19 deletion, and L858R and L861Q mutations in exon 21. Synchronous screening of sensitive and resistance mutations will provide more detailed information for the analysis of the resistance of EGFR TKI-targeted therapies.
The LungCarta ™ kit[20,21] was used to analyze 214 mutations of 26 oncogenes that include therapeutic targets of lung cancer. We compared the newly designed resistance panel with the LungCarta ™ kit, and the results were highly consistent in lung cancer specimens. However, our panel detected a large-fragment deletion in the BIM gene that the LungCarta ™ kit did not. In addition, our panel did not detect the P53 gene mutation that was detected in the LungCarta ™ kit, which may be because the genes and mutations included in our panel were mostly intentional, selected resistance mutations related to EGFR TKI therapy. It is important to note that the BIM gene has a large-fragment deletion of 2903 bp, rendering it difficult to design one assay using the MassARRAY platform; therefore, we designed the BIM wild-type and BIM deletion in two separate assays, which differs from the routine design. Herein, we established a new method to detect large-fragment deletions using the single-base extension platform, which has obvious advantages for synchronously screening multiple genes.
The proposed MALDI-TOF multiplex detection method of EGFR TKI-resistant mutations demonstrates the following advantages: (1) the proposed method can be used to detect EGFR TKI therapy-related resistance mutations that are rarely reported. The screening results provide detailed information for EGFR TKI-targeted resistance. (2) The proposed method is a high-throughput technique and can synchronously detect 60 loci of 7 genes related to EGFR TKI resistance in 12 wells, which provides a sensitive panel-based approach to make efficient use of patient diagnostic samples for a more accurate resistance analysis. (3) The polygenic primer panel can reduce routine testing costs. After clinical and translational applications, it will produce direct economic benefits and optimize clinical resource configurations. (4) Large-fragment deletions can be detected via the single-base extension reaction method on the MALDI-TOF platform.
However, the established resistance panel also presents some limitations, the most important of which is not including all the drug resistance loci related to EGFR-targeted therapies. First, there are too many genes and mutations involved in EGFR-targeted drug resistance; therefore, some low-frequency mutations could not be included in the panel due to throughput restrictions. For example, we included only five types of high-frequency insertion mutation in exon 20 based on the COSMIC database; other low-frequency insertion mutations were not included in our panel.[8] Second, our panel could not synchronously screen for gene amplifications due to limitations of the single-base extension technology (i.e., MET amplification accounts for 20% of all EGFR TKI resistance cases,[22] and ERBB2 and FGFR1 amplification also contributes to EGFR TKI resistance).[23,24]
Collectively, we provide a relatively comprehensive detection method for understanding the complexity of EGFR TKI resistance and choosing the appropriate treatment in tumors resistant to EGFR TKIs. It is necessary to develop more efficient approaches that would synchronously detect more resistance variants to EGFR TKIs.
Financial support and sponsorship
This work was supported by the Special Fund for Research in the Public Interest from the National Health and Family Planning Commission of PRC (No. 201402031) and the Special Fund for Research in the Public Interest and Capacity Building from the Guangdong Science and Technology Department (No. 2014A020212225).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Yuan-Yuan Ji
References
- 1.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: Learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–81. doi: 10.1038/nrclinonc.2014.104. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 2.Mondolfi AP, Singh RR. Genotyping of frequent mutations in solid tumors by PCR-based single-base extension and MassARRAY analysis. Methods Mol Biol. 2016;1392:83–101. doi: 10.1007/978-1-4939-3360-0_9. doi: 10.1007/978-1-4939-3360-0_9. [DOI] [PubMed] [Google Scholar]
- 3.Li P, Li Y, Zhou AH, Chen S, Li J, Wen XT, et al. Association Study of a proliferation-inducing ligand, spermatogenesis associated 8, platelet-derived growth factor receptor-alpha, and POLB polymorphisms with systemic lupus erythematosus in Chinese Han Population. Chin Med J. 2016;129:2085–90. doi: 10.4103/0366-6999.189055. doi: 10.4103/0366-6999.189055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen YS, Cai L, Zhang XW, Zhu JF, Zhang ZC, Shao JY, et al. Concurrent oncogene mutation profile in Chinese patients with stage IB lung adenocarcinoma. Medicine (Baltimore) 2014;93:e296. doi: 10.1097/MD.0000000000000296. doi: 10.1097/MD.0000000000000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–2. doi: 10.1038/nm.3854. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang JC, Sequist LV, Geater SL, Tsai CM, Mok TS, Schuler M, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: A combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015;16:830–8. doi: 10.1016/S1470-2045(15)00026-1. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 8.Yasuda H, Park E, Yun CH, Sng NJ, Lucena-Araujo AR, Yeo WL, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med. 2013;5:216ra177. doi: 10.1126/scitranslmed.3007205. doi: 10.1126/scitranslmed.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: Prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–9. doi: 10.1158/1535-7163.MCT-12-0620. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–79. doi: 10.1371/journal.pmed.0040315. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–501. doi: 10.1158/1078-0432.CCR-06-1570. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 12.Bean J, Riely GJ, Balak M, Marks JL, Ladanyi M, Miller VA, et al. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin Cancer Res. 2008;14:7519–25. doi: 10.1158/1078-0432.CCR-08-0151. doi: 10.1158/1078-0432.CCR-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng KP, Hillmer AM, Chuah CT, Juan WC, Ko TK, Teo AS, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–8. doi: 10.1038/nm.2713. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 14.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan JL, Zhong WZ, An SJ, Yang JJ, Su J, Chen ZH, et al. KRAS mutation in patients with lung cancer: A predictor for poor prognosis but not for EGFR-TKIs or chemotherapy. Ann Surg Oncol. 2013;20:1381–8. doi: 10.1245/s10434-012-2754-z. doi: 10.1245/s10434-012-2754-z. [DOI] [PubMed] [Google Scholar]
- 16.Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin YL, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–33. doi: 10.1073/pnas.1203530109. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minami Y, Shimamura T, Shah K, LaFramboise T, Glatt KA, Liniker E, et al. The major lung cancer-derived mutants of ERBB2 are oncogenic and are associated with sensitivity to the irreversible EGFR/ERBB2 inhibitor HKI-272. Oncogene. 2007;26:5023–7. doi: 10.1038/sj.onc.1210292. doi: 10.1038/sj.onc.1210292. [DOI] [PubMed] [Google Scholar]
- 18.Greulich H, Kaplan B, Mertins P, Chen TH, Tanaka KE, Yun CH, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012;109:14476–81. doi: 10.1073/pnas.1203201109. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabara K, Kanda R, Sonoda K, Kubo T, Murakami Y, Kawahara A, et al. Loss of activating EGFR mutant gene contributes to acquired resistance to EGFR tyrosine kinase inhibitors in lung cancer cells. PLoS One. 2012;7:e41017. doi: 10.1371/journal.pone.0041017. doi: 10.1371/journal.pone.0041017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamoto I, Sakai K, Morita S, Yoshioka H, Kaneda H, Takeda K, et al. Multiplex genomic profiling of non-small cell lung cancers from the LETS phase III trial of first-line S-1/carboplatin versus paclitaxel/carboplatin: Results of a West Japan Oncology Group Study. Oncotarget. 2014;5:2293–304. doi: 10.18632/oncotarget.1906. doi: 10.18632/oncotarget.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn AM, Hickson N, Adaway M, Priest L, Jaeger E, Udar N, et al. Diagnostic mutation profiling and validation of non-small-cell lung cancer small biopsy samples using a high throughput platform. J Thorac Oncol. 2015;10:784–92. doi: 10.1097/JTO.0000000000000473. doi: 10.1097/JTO.0000000000000473. [DOI] [PubMed] [Google Scholar]
- 22.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 23.Takezawa K, Pirazzoli V, Arcila ME, Nebhan CA, Song X, de Stanchina E, et al. HER2 amplification: A potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–33. doi: 10.1158/2159-8290.CD-12-0108. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao JL, Liu RJ, Zhou JH, Meng SH. Fibroblast growth factor receptor 1 gene amplification in nonsmall cell lung cancer. Chin Med J. 2016;129:2868–72. doi: 10.4103/0366-6999.194649. doi: 10.4103/0366-6999. [DOI] [PMC free article] [PubMed] [Google Scholar]