Abstract
The aim of this study was to review empirical studies examining associations between candidate genes and adverse events (AEs) from methylphenidate (MPH) use in children and adolescents. The PubMed, EMBASE, CINAHL, and Web of Science databases were searched from their inception until March 2017. We included empirically based articles on pharmacogenetic studies in 0–17-year-old patients that investigated associations between specific candidate genes, their polymorphisms, and reported AEs. We extracted information about study design, setting, type of AE reporter, studied genes and their polymorphisms, age and gender, administered doses, method of genotyping, outcome measures, and main findings. A total of nine articles reporting information about four double-blind, placebo-controlled, cross-over studies and five open-label cohort studies were eligible for inclusion. Studies were published from 2006 onward and included a total of 998 patients (3–17-year-olds) diagnosed with attention-deficit hyperactivity disorder (ADHD). Studies predominantly involved males and lasted from 1 to 12 weeks. Studies used polymerase chain reaction and single nucleotide polymorphism genotyping methodology. Reported AEs were significantly associated with the following genes: appetite reduction (CES1*G); buccal-lingual movements (T1065G); diastolic blood pressure (ADRA2A Mspl C/C-GC); emotionality (DAT1*9/9); irritability (SNAP25 T1065G); picking (DRD4*7/DRD4*4); social withdrawal (DRD4*7/DRD4*4); somatic complaints (DAT1*10/10); tics (5-HTTLRP*S/L*L/L; SNAP25 T1065G); sadness (CES1*rsl12443580); and vegetative symptoms (5-HTTLPR). In conclusion, only few MPH pediatric pharmacogenetic studies were located, and large between-study heterogeneity was found. Studies were of naturalistic design and of short duration. They included small patient samples, poorly standardized treatment regimens, and limited outcome assessments. In the future, more pharmacogenomic studies in ADHD are needed, preferably using randomized, controlled study designs and of longer duration (more than 6 months).
KEYWORDS: Adverse drug reaction, adverse event, attention-deficit hyperactivity disorder, Methylphenidate, pediatric, pharmacogenetics
INTRODUCTION
Attention-deficit hyperactivity disorder (ADHD) is a behavioral disorder with an estimated prevalence of 5.3%–7.1% in children and adolescents.[1,2] ADHD is characterized by inattentiveness, hyperactivity, and impulsivity and affects boys more frequently than girls.[2,3] The ADHD disorder is generally diagnosed in childhood but in many cases persists into adulthood. Pharmacological treatment for ADHD is currently determined empirically, as there are few predictors of response to stimulants, type of stimulant, or optimal dose.[4,5] Methylphenidate (MPH) is recommended as the first-line treatment of ADHD symptoms.[6] Common reported adverse events (AEs) to MPH use are headache, abdominal pain, decreased appetite, and delayed onset of sleep,[7] but also serious and rarely occurring AEs such as aggression, depression, and mania have been reported.[8] There is little knowledge about long-term safety aspects from MPH use in the treatment of ADHD.[9] The etiology of ADHD is not yet fully understood, but it is believed that genetic factors play an important role and that the disorder is polygenic.[10] Several studies have shown a high heritability in ADHD.[11,12,13] Serious AEs are one of the primary reasons why ADHD patients discontinue medical treatment with MPH.[9] Since knowledge about rare and serious AEs from MPH use in children and adolescents is limited, we need to explore this topic further. The strong heritability of ADHD and wide individual variability in dosing and tolerability have increased the interest in pharmacogenomic studies of stimulant response as well as identification of patients who are responsive or unresponsive to a medication, and individuals who are at risk of experiencing a serious AE. Pharmacogenomic studies may help to elucidate the associations between specific genes and occurrence of AEs, which may lead to safer treatment of ADHD and improve medication adherence.[11-13] Therefore, we aimed to study possible associations between genes and AEs reported in the pediatric population from MPH use to detect information about unknown, rare, and serious AEs.
The aim of this study is to review published articles examining associations between specific candidate genes and AEs reported for MPH in 0–17-year-old patients diagnosed with ADHD.
METHODS
Literature search
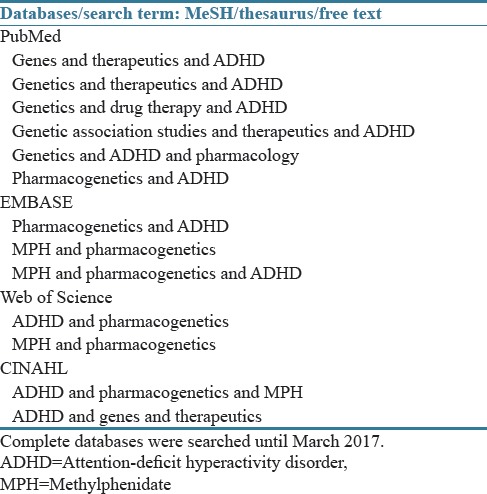
We systematically searched the references in the databases PubMed, EMBASE, CINAHL, and Web of Science from inception until July 2016. Literature searches were updated in March 2017 and no new relevant articles were found. The complete databases were searched without language restrictions for the combination of the search terms: “genes,” “genetics,” “genetic association studies,” “pharmacogenetics,” “pharmacogenomics,” “therapeutics,” “drug therapy,” “pharmacology,” “ADHD,” and “methylphenidate.” Each search contained two or three of the listed terms. The search was performed both with MeSH terms on PubMed and EMTREE thesaurus on EMBASE and also free-text searches in all the four databases. The full search strategy is shown in Table 1. The reference lists of the identified articles were manually checked for additional potential relevant articles.
Table 1.
Search strategy
Inclusion and exclusion criteria
The authors retrieved and assessed articles identified during the literature searches based on information provided in the title, abstract, and descriptor/search terms. Randomized controlled trials and all the other types of clinical and naturalistic trials, both blinded and open label, were included in this review. Only pharmacological interventions using MPH and with the purpose of examining the pharmacogenetics of ADHD were included. Studies reporting data on children and adolescents with a diagnosis of ADHD, either combined type or one of the two subtypes, were included. Articles were excluded if any of the following applied: the population was based on adults or healthy volunteers, the sample was not treated with MPH, or the study did not take into account specific AEs of ADHD treatment.
Extraction of data
Information about authors, study design, location of the study, candidate genes, polymorphisms, population size, age of the participants, administered doses of MPH, treatment period, method of genotyping, outcome measures used in the studies, and main findings were extracted from all full-text articles. The identification of relevant articles followed the QUOROM guidelines.[14] The first author extracted data from the included articles, and the second and third authors checked all data extractions. In the event of disagreement, consensus was reached through discussion.
Analysis
For all the included articles, we analyzed time of publication, characteristics of explored patient populations, reported AEs, investigated candidate genes and their polymorphisms, and the genetic profile of the included children.
Classification of reported adverse events
For each included article, any AEs reported were classified according to system organ class (SOC) and preferred term (PT) in keeping with the Medicinal Dictionary for Regulatory Activities terminology, which is an international standard terminology for classifying medicinal information.[15] AEs that could be related to more than one SOC were classified according to their primary SOC. AEs that could not be linked to a specific term (PT) were classified according to their high-level term or high-level group term.
Seriousness of reported adverse events
It was not possible to apply a common definition of AEs across studies, as the included papers used a range of definitions. Severity of reported AEs was classified according to the criteria defined in Volume 9 of the rules governing medicinal products in the European Union guidelines.[16] Here, serious AEs are divided into: resulting in death, life-threatening, requiring hospitalization or prolongation of existing hospitalization, resulting in persistent or significant disability/incapacity in the reporter's opinion, a congenital anomaly/birth defect, and other medically important conditions. Others were classified as nonserious.[16]
RESULTS
The literature search identified 948 potentially relevant articles, but after the initial screening, the number was narrowed down to 67 relevant records. After the full-text assessment, studies including healthy volunteers and adults were excluded (n = 7), as well as studies investigating stimulant treatment other than MPH (n = 7) were excluded. Studies examining associations between genes and reported AEs from MPH use, but did not report the specific types of studied AEs on PT level, were also excluded (n = 51). A flow chart of the selection and review process, including reasons for exclusion, is shown in Figure 1. There were no cases of disagreement and consequently, nine studies were included in the review.
Figure 1.
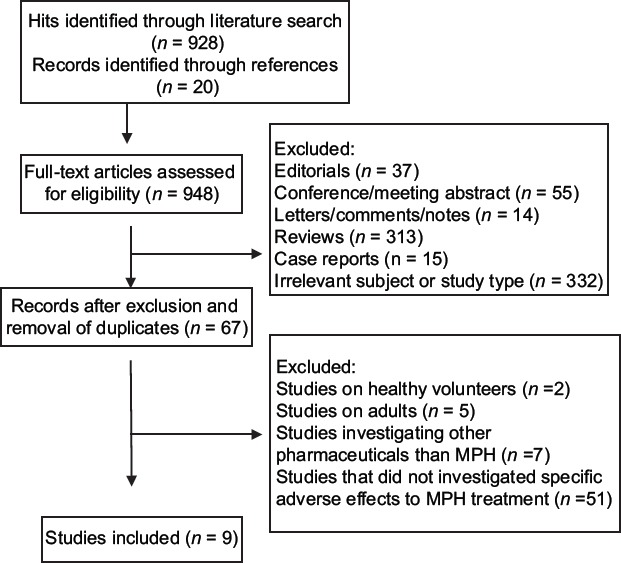
Flow chart of selection and review process for included/excluded studies
Study characteristics
A summary of study characteristics is displayed in Table 2. Articles were published from 2006 onward. In four studies, a double-blind, placebo-controlled, cross-over study design was used,[17,19,20,21] whereas in five studies, an open-label, naturalistic study design was applied.[18,19,22,23,24] Across studies, the treatment period varied from 1 to 12 weeks. Different types of participants reported the AE data. Reported AEs were noted by parents and teachers in two studies,[17,22] by parent and child in three studies,[18,19,20] by physicians in two studies,[21,24] parents in one study,[23] and by parents, children, and physicians in one study.[25] A total of 1128 children aged 3–17 years were included in the studies, and of these, 998 children completed the studies. The number of male patients was higher than female patients, ranging from 77% to 85% across studies. In all studies, the included patients were diagnosed with ADHD, but only in five of these, the patients were MPH treatment naïve before entering the study,[17,21,22,23,25] and in two studies, no information about this topic was found.[19,24] DNA samples for gene sequencing in the form of blood or buccal cell samples were requested before the study participants agreed to participate in the study. In two studies, DNA from both blood and saliva was examined,[17,22] whereas in six studies, only DNA from blood was used.[18,20,21,23,24,25] Three studies used single nucleotide polymorphism (SNP) genotyping in the measurement of genetic variations of SNPs,[21,22,23] and six studies used polymerase chain reaction techniques.[17,18,19,20,24,25]
Table 2.
Summary of study characteristics
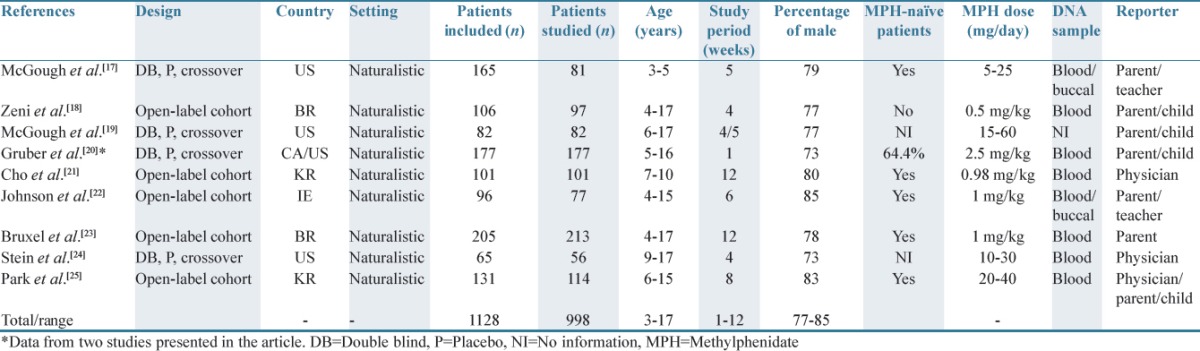
Table 3 displays the characteristics of reported AEs (PT), possibly related to MPH use. The studied AEs were of the following types (SOCs): general disorders and administration site conditions (appetite loss/reduction, weight loss); gastrointestinal disorders (abdominal pain/stomach ache); investigations (blood pressure, heart rate); nervous system disorders (buccal-lingual movements, headache, and tics/abnormal movements), and psychiatric disorders (anxiety, emotionality, insomnia, irritability, over-focused, picking/nail biting, sleeping problems, sadness, staring, social withdrawal, and tendency to cry). Only in two studies, the reported AEs were rated as being serious.[23,25] Across studies, the following candidate genes and their polymorphisms were examined: adrenergic genes: adrenergic α2A-receptor gene (ADRA2A); dopaminergic genes: dopamine transporter gene (DAT1/SLC6A3), norepinephrine transporter gene (SLC6A2), dopamine receptor D4 gene (DRD4), and serotonin transporter gene (5-HTT/SLC6A4). Other studied genes were synaptosomal-associated protein 25 kDa gene (SNAP-25), carboxylesterase 1 gene (CES1), and catechol-O-methyltransferase gene (COMT).
Table 3.
Pharmacogenetic studies investigating possible gene–adverse event associations from methylphenidate use in 0-17-year-old patients

Detected gene–adverse event associations
McGough et al., 2006, showed that the SNAP25 variant T1069C was associated with the AEs: tics (P < 0.02) and buccal (oral)-lingual movements (P < 0.01).[17] The SNAP25 variant T1065G was found to be associated with irritability (P = 0.04).[17] DRD4 variants were associated with picking (DRD4*4 repeat) (P = 0.03), increasing dose-predicted irritability (DRD4 promoter) (P = 0.05), and social withdrawal (DRD4*7/DRD4*4) (P = 0.03).[17] The study by McGough et al., 2009, showed that the AE “irritability” was predicted by COMT Val/Val (P = 0.02), and that vegetative symptoms were predicted by 5HTTLPR (P = 0.03).[19] In the study by Gruber et al., 2009, it was found that children with the DAT1*9/9 genotype displayed higher score on the emotionality scores which increased further during MPH treatment (P < 0.05).[20] Children with the DAT1*10/10 genotype displayed a significant increase in somatic complaint factor scores during MPH treatment relative to the other genotypes (P < 0.05).[20] In the study by Cho et al., 2012, children with ADHD and having the ADRA2 MspI*C/C genotype showed a 12.5% increase in diastolic blood pressure compared to baseline, whereas children with the G/C genotype showed a 2.3% increase in DPB after MPH treatment.[21] In the study by Johnson et al., 2013, an association between the AE sadness and the two CES1 SNP markers rs12443580 and rs224613 was found.[22] Bruxel et al., 2013, showed that the AE “appetite reduction” was associated with the gene CES1–75 T>G polymorphism (P = 0.05).[23] In studies by Zeni et al., 2007,[18] and Stein et al., 2014,[24] no significant association was found between the studied AEs and candidate genes. Park et al., 2015, documented a significant association between the gene 5-HTTLPR*S/L and nail biting (P < 0.001).[25]
DISCUSSION
To our knowledge, this is the first study that has systematically reviewed the empirical literature on pharmacogenetic studies investigating associations between specific candidate genes and AEs reported from MPH use in children and adolescents. Only few significant associations between the studied genes and reported AEs were found. The included studies were of naturalistic setting without a control/comparator group, which made it impossible to distinguish whether the reported AEs were related to MPH use and/or etiology of the ADHD disease. ADHD medications are usually prescribed for long-term treatment, but the included studies only investigated short-term treatment with MPH. The study duration was short (<3 months) and the included patient populations were of small size which made it difficult to obtain significant statistical power to detect significant gene–AE associations. Most studies focused on potential genes of susceptibility for ADHD, mainly noradrenergic and dopaminergic genes, as the suspected effect of MPH on ADHD symptoms is on dopaminergic and noradrenergic pathways.[4,5] Empirical studies applying full genome sequencing techniques of the included patients were not located. The lack of significant study results can question whether the most appropriate candidate genes and polymorphisms have been selected for the study.
In the recent years, there has been an increasing interest in the field of personalized medicine as it is assumed that pharmacogenomic studies can open the door to new treatment protocols.[26] This is believed to improve the effect of medical treatment, decrease the risk of serious AEs, and improve medication adherence.[26] The included studies found only few significant associations between AEs and studied genes. Lack of significant associations may be explained by the short study duration, nonserious AEs being studied, low number of patients, presence of comorbidities, and drug–drug interactions that were not studied/adjusted for. Limited information about the patients’ genetic profiles, MPH blood concentration, medicine use, comorbidities, ethnicities, etc., is also a major limitation for evaluating whether the detected gene–AE associations were of clinical relevance.
The objective of this study was to review studies reporting AEs from MPH use in children and adolescents that have examined possible associations with candidate genes and their polymorphism. Since we were only able to locate nine relevant articles, a systematic review on this topic seems premature, but for researchers within interests in the diagnosis and treatment of ADHD, this review is important. Almost all the included studies were of observational character and therefore the studies only provide limited information about possible gene–AE associations. The findings of the review are determined by the quality and number of included studies. Limitations of this study are (1) the scarcity of pharmacogenomic studies assessing AEs to MPH, (2) the heterogeneity in methodology used in the included studies, (3) the low number of participants, and (4) the short study duration. The differences between the included studies encompassed study design, study duration, outcome measures, control for comorbidities, applied inclusion/exclusion criteria, and the use of patients, parents, teachers, and/or physicians for reporting of AEs. The included articles predominantly analyzed candidate genes and their genetic polymorphisms based on prior pharmacogenomic studies and known risk alleles for ADHD. Studies were also biased in terms of gender, as the majority of included patients were males. The studies were conducted over a period of approximately 10 years in six different countries, with a great deal of inconsistency in reporting and classifying AEs and limited data on the children's comorbidities, medicine use, and medication compliance. The included articles provided only limited information about the applied definitions of AEs, prevalence of AEs, selection of studied AEs, classification and evaluation of AE cases with respect to causality, criteria of seriousness, and type of AE. The findings of this review are further limited by the quality and content of the included studies but illustrate that information about AEs and genetic associations is limited and more knowledge is needed. In some studies, dropout of patients was observed due to serious AEs and problems with storage of blood, and therefore the final sample sizes were probably not large enough to detect significant gene–AE associations.[27] In some studies, higher rates of amplification failure for DNA derived from buccal cells versus blood were found, mostly due to poor techniques, but this limitation does not bias the usefulness of the method.[17] Across studies, DNA information was collected by either blood or buccal sampling; thus, authors were not able to compare genotype results from one source versus the other, and therefore future pharmacogenomic investigations should rely on blood collection or establishment of suitable sampling protocols to ensure adequate collection for accurate genotyping of buccal cell samples. Another limitation of the included studies involves the use of saliva as a source of DNA. Using saliva for DNA is common in pediatric studies and has resulted in a number of valuable pharmacogenetic studies.
Only few MPH pediatric pharmacogenomic studies were located in the literature, and large between-study heterogeneity was found. The majority of studies were of naturalistic design and of short duration. They included small patient samples, poorly standardized treatment regimens, and limited outcome assessments. Therefore, more pharmacogenomic studies in ADHD are needed, preferably using randomized controlled study designs and of longer duration (>6 months).
AUTHORS’ CONTRIBUTION
The original idea for this paper was devised by BJ, MM, and LA. BJ conducted the literature searches and appraised the papers with support from MM and LA. All authors wrote the first draft of the manuscript, and all authors contributed to and critically revised subsequent manuscripts.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Polanczyk G, de Lima MS, Horta BL, Biederman J, Rohde LA. The worldwide prevalence of ADHD: A systematic review and metaregression analysis. Am J Psychiatry. 2007;164:942–8. doi: 10.1176/ajp.2007.164.6.942. [DOI] [PubMed] [Google Scholar]
- 2.Willcutt EG. The prevalence of DSM-IV attention-deficit/hyperactivity disorder: A meta-analytic review. Neurotherapeutics. 2012;9:490–9. doi: 10.1007/s13311-012-0135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshimasu K, Barbaresi WJ, Colligan RC, Killian JM, Voigt RG, Weaver AL, et al. Gender, attention-deficit/hyperactivity disorder, and reading disability in a population-based birth cohort. Pediatrics. 2010;126:e788–95. doi: 10.1542/peds.2010-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelham WE, Jr, Greenslade KE, Vodde-Hamilton M, Murphy DA, Greenstein JJ, Gnagy EM, et al. Relative efficacy of long-acting stimulants on children with attention deficit-hyperactivity disorder: A comparison of standard methylphenidate, sustained-release methylphenidate, sustained-release dextroamphetamine, and pemoline. Pediatrics. 1990;86:226–37. [PubMed] [Google Scholar]
- 5.Barbaresi WJ, Katusic SK, Colligan RC, Weaver AL, Leibson CL, Jacobsen SJ. Long-term stimulant medication treatment of attention-deficit/hyperactivity disorder: Results from a population-based study. J Dev Behav Pediatr. 2006;27:1–10. doi: 10.1097/00004703-200602000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Bolea-Alamañac B, Nutt DJ, Adamou M, Asherson P, Bazire S, Coghill D, et al. Evidence-based guidelines for the pharmacological management of attention deficit hyperactivity disorder: Update on recommendations from the British association for psychopharmacology. J Psychopharmacol. 2014;28:179–203. doi: 10.1177/0269881113519509. [DOI] [PubMed] [Google Scholar]
- 7.Feldman HM, Reiff MI. Clinical practice. Attention deficit-hyperactivity disorder in children and adolescents. N Engl J Med. 2014;370:838–46. doi: 10.1056/NEJMcp1307215. [DOI] [PubMed] [Google Scholar]
- 8.Aagaard L, Hansen EH. The occurrence of adverse drug reactions reported for attention deficit hyperactivity disorder (ADHD) medications in the pediatric population: A qualitative review of empirical studies. Neuropsychiatr Dis Treat. 2011;7:729–44. doi: 10.2147/NDT.S26403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gajria K, Lu M, Sikirica V, Greven P, Zhong Y, Qin P, et al. Adherence, persistence, and medication discontinuation in patients with attention-deficit/hyperactivity disorder - A systematic literature review. Neuropsychiatr Dis Treat. 2014;10:1543–69. doi: 10.2147/NDT.S65721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabriela ML, John DG, Magdalena BV, Ariadna GS, Francisco de LP, Liz SM, et al. Genetic interaction analysis for DRD4 and DAT1 genes in a group of Mexican ADHD patients. Neurosci Lett. 2009;451:257–60. doi: 10.1016/j.neulet.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Larsson H, Chang Z, D’Onofrio BM, Lichtenstein P. The heritability of clinically diagnosed attention deficit hyperactivity disorder across the lifespan. Psychol Med. 2014;44:2223–9. doi: 10.1017/S0033291713002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lichtenstein P, Carlström E, Råstam M, Gillberg C, Anckarsäter H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psychiatry. 2010;167:1357–63. doi: 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- 13.Silberg JL, Gillespie N, Moore AA, Eaves LJ, Bates J, Aggen S, et al. Shared genetic and environmental influences on early temperament and preschool psychiatric disorders in Hispanic twins. Twin Res Hum Genet. 2015;18:171–8. doi: 10.1017/thg.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.QUOROM Guidelines for Systematic Reviews and Meta-Analysis. [Last accessed on 2016 Oct 07]. Available from: http://www.consort-statement.org/downloads .
- 15.MedDRA. [Last accessed on 2016 Oct 07]. Available from: http://www.meddramsso.com .
- 16.European Commission. Volume 9. Pharmacovigilance 2012. Medicinal Products of Human Use and Veterinary Products. [Last accessed on 2016 Oct 03]. Available from: http://www.ec.europa.eu/enterprise/pharmaceuticals/eudralex/homev9.htm .
- 17.McGough J, McCracken J, Swanson J, Riddle M, Kollins S, Greenhill L, et al. Pharmacogenetics of methylphenidate response in preschoolers with ADHD. J Am Acad Child Adolesc Psychiatry. 2006;45:1314–22. doi: 10.1097/01.chi.0000235083.40285.08. [DOI] [PubMed] [Google Scholar]
- 18.Zeni CP, Guimarães AP, Polanczyk GV, Genro JP, Roman T, Hutz MH, et al. No significant association between response to methylphenidate and genes of the dopaminergic and serotonergic systems in a sample of Brazilian children with attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:391–4. doi: 10.1002/ajmg.b.30474. [DOI] [PubMed] [Google Scholar]
- 19.McGough JJ, McCracken JT, Loo SK, Manganiello M, Leung MC, Tietjens JR, et al. A candidate gene analysis of methylphenidate response in attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2009;48:1155–64. doi: 10.1097/CHI.0b013e3181bc72e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruber R, Joober R, Grizenko N, Leventhal BL, Cook EH, Jr, Stein MA. Dopamine transporter genotype and stimulant side effect factors in youth diagnosed with attention-deficit/hyperactivity disorder. J Child Adolesc Psychopharmacol. 2009;19:233–9. doi: 10.1089/cap.2008.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho SC, Kim BN, Cummins TD, Kim JW, Bellgrove MA. Norepinephrine transporter -3081(A/T) and alpha-2A-adrenergic receptor MspI polymorphisms are associated with cardiovascular side effects of OROS-methylphenidate treatment. J Psychopharmacol. 2012;26:380–9. doi: 10.1177/0269881111405356. [DOI] [PubMed] [Google Scholar]
- 22.Johnson KA, Barry E, Lambert D, Fitzgerald M, McNicholas F, Kirley A, et al. Methylphenidate side effect profile is influenced by genetic variation in the attention-deficit/hyperactivity disorder-associated CES1 gene. J Child Adolesc Psychopharmacol. 2013;23:655–64. doi: 10.1089/cap.2013.0032. [DOI] [PubMed] [Google Scholar]
- 23.Bruxel EM, Salatino-Oliveira A, Genro JP, Zeni CP, Polanczyk GV, Chazan R, et al. Association of a carboxylesterase 1 polymorphism with appetite reduction in children and adolescents with attention-deficit/hyperactivity disorder treated with methylphenidate. Pharmacogenomics J. 2013;13:476–80. doi: 10.1038/tpj.2012.25. [DOI] [PubMed] [Google Scholar]
- 24.Stein MA, Waldman I, Newcorn J, Bishop J, Kittles R, Cook EH., Jr Dopamine transporter genotype and stimulant dose-response in youth with attention-deficit/hyperactivity disorder. J Child Adolesc Psychopharmacol. 2014;24:238–44. doi: 10.1089/cap.2013.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SY, Kim EJ, Cheon KA. Association between 5-HTTLPR polymorphism and tics after treatment with methylphenidate in Korean children with attention-deficit/hyperactivity disorder. J Child Adolesc Psychopharmacol. 2015;25:633–40. doi: 10.1089/cap.2014.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beger RD, Dunn W, Schmidt MA, Gross SS, Kirwan JA, Cascante M, et al. Metabolomics enables precision medicine: “A white paper, community perspective”. Metabolomics. 2016;12:149. doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong EP, Park JW. Sample size and statistical power calculation in genetic association studies. Genomics Inform. 2012;10:117–22. doi: 10.5808/GI.2012.10.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]


