Abstract
There is evidence for genetic overlap between cognitive abilities and schizophrenia (SCZ), and genome-wide association studies (GWAS) demonstrate that both SCZ and general cognitive abilities have a strong polygenic component with many single-nucleotide polymorphisms (SNPs) each with a small effect. Here we investigated the shared genetic architecture between SCZ and educational attainment, which is regarded as a “proxy phenotype” for cognitive abilities, but may also reflect other traits. We applied a conditional false discovery rate (condFDR) method to GWAS of SCZ (n = 82 315), college completion (“College,” n = 95 427), and years of education (“EduYears,” n = 101 069). Variants associated with College or EduYears showed enrichment of association with SCZ, demonstrating polygenic overlap. This was confirmed by an increased replication rate in SCZ. By applying a condFDR threshold <0.01, we identified 18 genomic loci associated with SCZ after conditioning on College and 15 loci associated with SCZ after conditioning on EduYears. Ten of these loci overlapped. Using conjunctional FDR, we identified 10 loci shared between SCZ and College, and 29 loci shared between SCZ and EduYears. The majority of these loci had effects in opposite directions. Our results provide evidence for polygenic overlap between SCZ and educational attainment, and identify novel pleiotropic loci. Other studies have reported genetic overlap between SCZ and cognition, or SCZ and educational attainment, with negative correlation. Importantly, our methods enable identification of bi-directional effects, which highlight the complex relationship between SCZ and educational attainment, and support polygenic mechanisms underlying both cognitive dysfunction and creativity in SCZ.
Keywords: pleiotropy, GWAS, conditional FDR
Introduction
Schizophrenia (SCZ) is characterized by psychotic symptoms but cognitive alterations are often seen,1 and cognitive impairment has been suggested to be a core feature of SCZ.2 However, in some studies or certain sub-samples of patients, increased cognitive functioning in persons with psychotic disorders has been reported,3,4 and there is an increase in creative professions among relatives of patients with SCZ. It has been argued that studying cognitive traits could help in understanding the etiology of SCZ.2,5
The estimated heritability of SCZ ranges from 60% in population-based studies to 75% in twin-based studies.6,7 Large genome-wide association studies (GWAS) by the Psychiatric Genomics Consortium (PGC) have identified 108 genomic loci associated with SCZ,8 which explain an estimated 18% of the heritability and confirm the polygenic architecture of SCZ. However, studies of the aggregated effect of common variants in SCZ suggest that additional single nucleotide polymorphisms (SNPs) may explain up to 40% of SCZ heritability.9,10 Epidemiological genetic studies in twins and GWAS show that general cognition is highly influenced by genetic factors and that common variants can explain 50% of inter-individual variance.11 Genetic overlap between general cognition (the g factor) and SCZ was recently reported,12 indicating that polygenic factors associated with general cognition are also implicated in SCZ. Further, the polygenic risk score of SCZ is associated with general cognition,13 and predicts creativity, measured by artistic society membership or creative profession.14 However, new statistical approaches are needed to identify the loci underlying these polygenic effects.
We have developed novel statistical tools for GWAS of polygenic traits based on a false discovery rate (FDR) approach.15–17 By leveraging additional information about the genetic variants, these methods increase the power to identify genomic loci in a GWAS with fewer type 2 errors and improved replication compared to standard P-value-based methods.16,18 Combining GWAS from 2 phenotypes that are relevant at the pathological or biological level provides additional insights into genetic overlap (defined as genetic variants being associated with more than one distinct phenotype) and may elucidate shared biology. The condFDR method permits identification of SNPs associated with both traits, and has been applied to phenotypes including psychiatric and neurological diseases,15–17,19 immune-related diseases,18 cardiovascular disease,20 and cancer.21 This approach can identify bi-directional overlap, unlike other methods used to investigate genetic correlations.22
Here we use data from GWAS on educational attainment23 (which is represented by 2 phenotypes: college completion, denoted “College,” n = 95 427; and years of education, denoted “EduYears,” n = 101 069) and SCZ (n = 82 315)8 to identify shared polygenic factors. Educational attainment is not a cognitive measure, but correlates with cognitive ability (r ~ .5) and is easily obtained in larger samples. It has thus been used as “proxy” for general cognition.23 It probably also represents other relevant traits, such as creativity.24 However, it remains to be determined whether educational attainment can be used to identify genetic overlap or shared genetic variants implicated in other phenotypes, like SCZ. The educational attainment GWAS identified association with several novel genomic loci, some of which were shared between College and EduYears and others that were trait-specific.23 Some of the variants associated with educational attainment subsequently showed association with cognitive performance.25,26 Here we investigate the polygenic overlap between SCZ and educational attainment using our FDR approach.
Methods
Participants
The relevant institutional review boards or ethics committees approved the research protocol of all the individual GWAS used here. All participants gave written informed consent. For the SCZ sample, we obtained GWAS results as summary statistics from the Schizophrenia Working Group of the PGC. This sample comprises 82 315 individuals from 49 non-overlapping case-control samples (58% male). Each cohort was tested separately under additive logistic regression and the results were merged by meta-analysis using an inverse-weighted fixed effects model. The inclusion criteria and phenotype characteristics of the different GWAS have been described previously.8
The GWAS for educational attainment comprised 2 measures, years of education and college completion, which were defined according to the UNESCO International Standard Classification of Education (ISCED). These measures were applied to 42 cohorts, all of Caucasian origin. 95% of the participants were older than 30. Years of education (EduYears), obtained from 101 069 individuals23 (59% female), is a quantitative variable defined as US-schooling-year equivalents after conversion. College completion (College), obtained from 95 427 individuals (59% female), is a binary measure which differentiates between individuals who do or do not hold a tertiary diploma according to ISCED standards. The correlation between the 2 measures is high (0.74–0.91), but EduYears reflects the mean distribution while College focuses on the upper tail of the phenotypic distribution. Each cohort was analyzed separately, including correction for population stratification, yielding gender-stratified summary results. After QC, the GWAS were merged in meta-analyses using genomic control and sample size weighting. For more details, see Rietveld et al.23
We utilized summary statistics (P-values, ORs, β-values and z-scores) for conditional and conjunctional FDR analyses. We corrected all P-values for inflation using a recent genomic control procedure.27 The analyses were performed on 2 283 442 markers which overlapped between the GWAS.
Statistical Analyses
A brief summary follows. For details, see supplementary methods and previous publications.15,16,18,19,21
Fold Enrichment Plots and Conditional Q–Q Plots
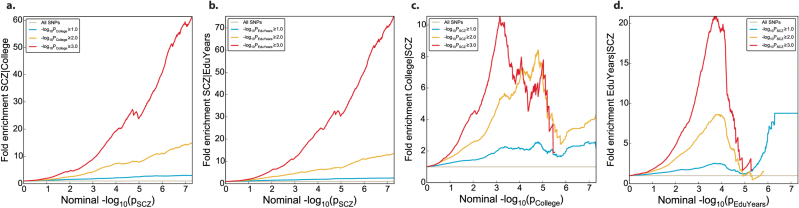
Genetic enrichment in one phenotype (eg, SCZ) is assessed using fold enrichment plots conditioned on the auxiliary phenotype (eg, College). Enrichment is present if the degree of deflection from the expected null line (horizontal line through 1) depends on the covariate stratum defined by the P-values of the corresponding markers in the phenotype used for conditioning (eg, −log10(P) > 1, >2, and >3 in College). We first compute the empirical cumulative distribution of −log10(P) values for SNP association with a given phenotype (eg, SCZ) for all SNPs, and then the cumulative −log10(P) values for each SNP stratum, which is determined by the P-value of these SNPs in the conditioning phenotype (eg, College). We then calculate the fold enrichment of each stratum as the ratio CDFstratum/CDFall between the −log10(P) cumulative distribution for that stratum and the cumulative distribution for all SNPs. The x-axis shows nominal P-values (−log10(P)); the y-axis shows fold enrichment. To assess polygenic effects below the standard GWAS significance threshold, we focused the fold enrichment plots on SNPs with nominal log10(P) < 7.3 (corresponding to P > 5×10−8).
Enrichment of statistical association is also visualized in Q–Q plots, which display nominal P-values from GWAS summary statistics (observed) as a function of empirical P-values expected under the global null hypothesis. Conditional Q–Q plots display the distribution of summary statistics for the primary trait conditioned on different P-value thresholds (−log10(P) > 1, >2, and >3) in the secondary trait. If enrichment of association with one trait is present among SNPs that are significantly associated with the other trait (pleiotropic enrichment), the conditional Q–Q plot will show successive leftward deflections.
Testing the Effect of Large Linkage Disequilibrium Blocks on Enrichment
To test whether the enrichment was driven by large blocks of linkage disequilibrium (LD), we performed the enrichment analyses after randomly pruning SNPs from each LD block (supplementary methods).
Verifying Enrichment Based on Conditional Replication Rates
For each of the 17 sub-studies contributing to the final PGC-SCZ meta-analysis, we independently adjusted the z-scores using intergenic inflation control. We sampled 1000 combinations of 8 and 9 sub-study groupings that were randomly assigned to discovery and replication sets to calculate a combined discovery z-score and a combined replication z-score for each SNP (average z-score across the sub-studies multiplied by the square root of the number of studies). For details, see supplementary methods.
Conditional FDR and Conjunctional FDR
We used conditional FDR to incorporate information from GWAS summary statistics of a second phenotype.15–17,19 The conditional FDR is the posterior probability of a SNP being null in the first phenotype given that the P-values in the first and second phenotype are as small as or smaller than the observed ones. Ranking SNPs by FDR or by P-values is equivalent, in that both give the same ordering of SNPs. In contrast, ranking SNPs according to conditional FDR will re-order the SNPs if the primary and secondary phenotypes are genetically related. To each SNP, we assigned a conditional FDR value for SCZ given the P-values for College or EduYears (denoted by condFDRSCZ|College and condFDRSCZ|EduYears) and vice versa (condFDRCollege|SCZ and condFDREduYears|SCZ) by computing condFDR estimates on a grid and interpolating these estimates into a 2-dimensional look-up table.
To identify SNPs significantly associated with both phenotypes, we used a genetic epidemiology framework based on the conjunctional FDR (conjFDR). ConjFDR is the posterior probability that a SNP is null for either phenotype or both simultaneously, given that the P-values for both traits are as small as or smaller than the observed P-value. A conservative estimate of conjFDR is given by the maximum of FDRtrait1|trait2 and FDRtrait2|trait1.28 While condFDR can be used to reorder association of SNPs to one trait based on additional information provided by the secondary trait, conjFDR pinpoints shared loci, since a low conjFDR occurs only if there is joint association with both traits.
Annotation of Genes to Genomic Loci
Genes were annotated to genomic loci by considering the entire region of association, ie, all SNPs without pruning. Genomic regions were defined as follows: each region must contain at least one SNP with condFDR < 0.01 before pruning; and the borders of the associated region are defined by all SNPs with condFDR < 0.01 without filtering for LD between the associated SNPs. Similar to the PGC-SCZ protocol, genomic loci less than 250kb apart were merged. For each interval, we calculated how many independent signals of association were present, based on performance in the pruned condFDR < 0.01 analysis with an LD threshold of r2 > .2 (ie, identifying clumps of associated SNPs). All refSeq genes located within the genomic interval were annotated to that interval. Each new genomic locus was searched for previously reported hits in the GWAS catalogue using UCSC browser tools (https://genome.ucsc.edu).29
We removed the Major Histocompatibility Complex (MHC) regions from the genomic loci associated with SCZ and submitted the coordinates of the other regions for protein-protein interaction analysis using DAPPLE v2.030 (http://www.broadinstitute.org/mpg/dapple/dappleTMP.php) with default parameters (1000 permutations, regulatory regions ±50kb).
Results
Polygenic Overlap Between Educational Attainment and SCZ
To investigate polygenic overlap we stratify the P-values from the SCZ GWAS conditioned on their P-values in the College or EduYears GWAS. Fold enrichment plots (figure 1) show the different enrichment of association between the traits. In SCZ, when the SNPs are selected for their association with College or EduYears, a marked enrichment of association was observed across different levels of significance (−log10(P) > 1, >2, and >3). This is also seen as a leftward deflection in the corresponding Q–Q plots of SCZ given association with College (supplementary figure 1). Clear enrichment remained after removing the MHC region and after random pruning of SNPs from each LD block (supplementary figure 2; supplementary methods). When we selected SNPs associated with SCZ and tested for enrichment of association with College or EduYears, the enrichment appeared to be weaker (figure 1; supplementary figure 1).
Fig. 1.
Fold enrichment of association across traits in pairwise comparisons. Fold enrichment plots of the nominal −log10(P) below the standard genome-wide association studies (GWAS) threshold of P < 5×10−8 in one phenotype as a function of the association level with the second phenotype, at the level of all single-nucleotide polymorphisms (SNPs) (grey), −log10(P) ≥ 1 (blue), −log10(P) ≥ 2 (yellow), −log10(P) ≥ 3 (red). Successive upward elevation in terms of all SNPs demonstrates polygenic enrichment of: (a) Schizophrenia (SCZ) association conditioned on college completion (College), (b) SCZ association conditioned on years of education (EduYears), (c) College conditioned on SCZ, (d) EduYears conditioned on SCZ.
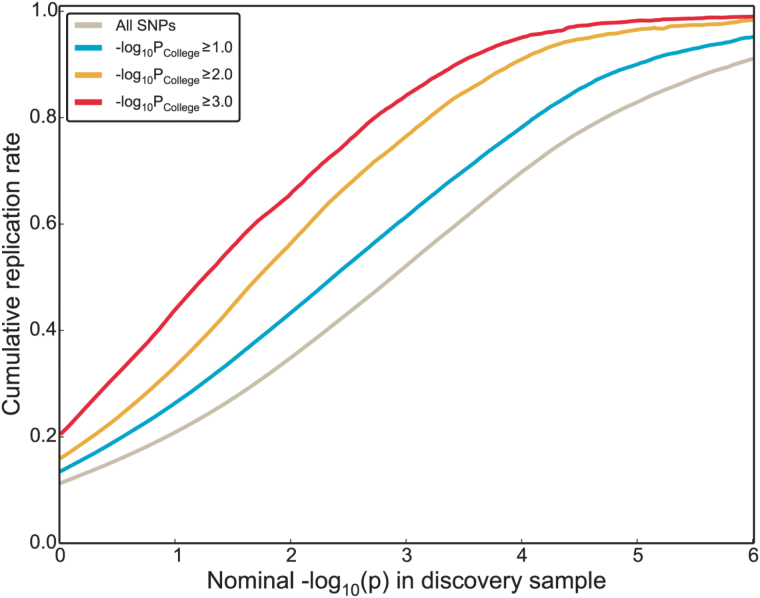
Increased Replication Rate for Shared Variants
We tested if the replication rate in SCZ samples would increase for SNPs with higher significance of association with College (supplementary methods). Figure 2 displays the average replication rate for each SNP within each −log10(P) stratum in the discovery sample. When the SNPs are stratified based on their association in the College GWAS, the replication rate in SCZ is increased compared to all SNPs. This stepwise increase in replication rate shows that the more significant the association with College, the higher the replication rate between SCZ discovery and replication samples, indicating higher likelihood of true findings.
Fig. 2.
Improvement in replication rate in schizophrenia (SCZ). Cumulative replication plot, showing the average replication rate (y-axis), defined as P-value < .05 in the replication samples and in the same direction as the discovery samples, for SCZ sub-studies for a range of single-nucleotide polymorphisms (SNPs) selected in the discovery sample based on their association P-value in the college completion (College) genome-wide association studies (GWAS).
Identification of SNPs and Genomic Loci Associated With SCZ Conditioned on Educational Attainment
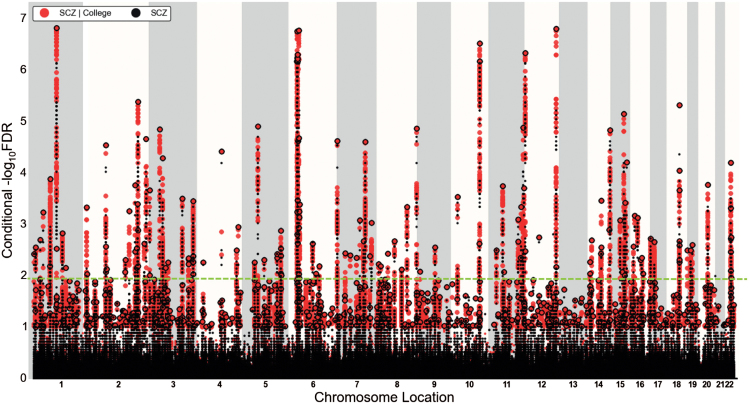
Using information from the genetic effects in College and EduYears, we leveraged the polygenic enrichment to identify specific SNPs associated with SCZ. For each SNP, we calculated the condFDR value in SCZ conditioned on the P-value of the SNP associations with College (denoted condFDRSCZ|College) or with EduYears (condFDRSCZ|EduYears). The condFDR values are visualized in 2-dimensional “look-up” tables (supplementary figure 3; supplementary methods). Using a significance threshold of condFDR<0.01 and after pruning the SNPs for LD at r2 > .2, we identified 153 independent SNPs associated with SCZ conditioned on College (supplementary table 1). The condFDRSCZ|College results are also visualized in a Manhattan plot in figure 3. Using the same significance threshold, 147 independent SNPs associated with SCZ conditioned on EduYears were identified with condFDRSCZ|EduYears (supplementary table 3). These SNPs were then clustered into loci (supplementary methods; supplementary tables 2 and 4).
Fig. 3.
Manhattan plot of Conditional FDRSCZ|College. Red data-points represent those single-nucleotide polymorphisms (SNPs) for which the FDR was improved by conditioning, whereas black points represent the SNPs that were not improved. All SNPs without pruning are shown, and the strongest signal in each linkage disequilibrium (LD) block is encircled in black. The strongest signal was identified after ranking all SNPs based on the condFDR and removing SNPs in LD r2 > .2 with any higher-ranked SNP. The green dashed line indicates the genome-wide significance threshold of condFDR < 0.01. SCZ; Schizophrenia, College; college completion.
Using condFDR, we identified 18 loci that become significant in SCZ when conditioned on College and 15 loci that became significant when conditioned on EduYears (table 1). Ten of the loci overlap. These loci were not tested for replication in the PGC-SCZ analysis because they did not pass the threshold for selection (P < 1×10−6). They should therefore be included in future replication studies.
Table 1.
Genomic Intervals Associated With SCZ|College or SCZ|EduYears but not SCZ Only
| Locus Positiona | Size (kb)b | SNP IDc | P value SCZd | condFDR SCZ|Collegee | condFDR SCZ|EduYearsf | Genesg |
|---|---|---|---|---|---|---|
| chr1:163734858-163734858 | 0 | rs4657304 | 9.43E-05 | 6.92E-03 | n.s. | NUF2* |
| chr1:244023666-244023666 | 0 | rs3008657 | 2.71E-05 | 9.11E-03 | n.s. | AKT3* |
| chr2:60713234-60713234 | 0 | rs10189857 | 6.60E-05 | 7.26E-03 | 2.55E-03 | BCL11A |
| chr2:145141540-145141540 | 0 | rs12991836 | 1.45E-04 | 8.41E-03 | n.s. | ZEB2* |
| chr3:71543757-71579021 | 35 | rs1499894 | 5.13E-05 | 5.47E-03 | n.s. | FOXP1 |
| chr3:161416295-161841017 | 425 | rs2175263 | 2.88E-05 | 4.50E-03 | 8.45E-03 | OTOL1* |
| chr5:88743218-88746330 | 3 | rs16867576 | 6.40E-06 | 4.87E-03 | 5.69E-03 | MEF2C* |
| chr6:43287892-43358361 | 70 | rs17209407 | 1.61E-04 | n.s. | 4.03E-03 | ZNF318 |
| chr6:56575670-56575670 | 0 | rs17684571 | 2.74E-04 | n.s. | 8.69E-03 | RNU6-71P, DST |
| chr6:108983526-108994825 | 11 | rs9398171 | 1.15E-05 | 6.47E-03 | 7.04E-03 | FOXO3 |
| chr7:41706131-41730930 | 25 | rs2237436 | 9.02E-05 | 3.93E-03 | n.s. | INHBA |
| chr7:71741796-71772928 | 31 | rs12670234 | 2.10E-04 | 4.46E-03 | 8.89E-03 | CALN1 |
| chr7:86403262-86459346 | 56 | rs13230421 | 5.36E-07 | n.s. | 8.83E-04 | GRM3 |
| chr7:104594252-105027645 | 433 | rs6466056 | 2.37E-06 | 2.09E-03 | 1.61E-03 | LINC01004, KMT2E, SRPK2 |
| chr8:8094869-8098037 | 3 | rs2945232 | 8.29E-06 | 6.41E-03 | 5.74E-03 | FAM86B3P |
| chr8:143736634-143749717 | 13 | rs6995314 | 1.42E-04 | n.s. | 7.93E-03 | JRK |
| chr9:7172497-7172497 | 0 | rs913587 | 1.23E-04 | 8.26E-03 | n.s. | KDM4C |
| chr10:3821560-3821560 | 0 | rs17731 | 1.66E-04 | n.s. | 9.20E-03 | KLF6 |
| chr14:35512994-35630376 | 117 | rs11156875 | 3.44E-05 | 2.01E-03 | 5.14E-03 | FAM177A1, LOC101927178, PPP2R3C, KIAA0391 |
| chr14:71361413-71605267 | 244 | rs17108804 | 3.95E-05 | 3.69E-03 | n.s. | PCNX |
| chr15:83254707-83254707 | 0 | rs783540 | 1.63E-05 | 9.77E-03 | n.s. | |
| chr16:63697133-63712718 | 16 | rs2018916 | 6.11E-06 | 4.44E-03 | 4.83E-03 | |
| chr18:77566534-77579811 | 13 | rs11663602 | 5.03E-05 | 3.15E-03 | 4.00E-03 | CPEB1 |
Note: SCZ, Schizophrenia; College, college completion; EduYears, years of education; SNP, single-nucleotide polymorphisms; condFDR, conditional false discovery rate; HGNC, HUGO Gene Nomenclature Committee. More details about the genomic loci can be found in supplementary tables 2 and 4.
aLocus position from hg 19 (chr:lower_boundary-upper_boundary).
bLocus size (kb)
crsID of the SNP with the lowest condFDR value.
d P-value in SCZ of this SNP.
econdFDR value of this SNP in SCZ|College (if <0.01). n.s., not significant.
fcondFDR value of this SNP in SCZ|EduYears (if <0.01). n.s., not significant.
gHGNC IDs of genes located in the interval. If no genes were located in the interval, the closest gene (within 100kb, if any) is indicated by *.
We used the same procedure to produce condFDRCollege|SCZ and condFDREduYears|SCZ. The results are visualized in 2-dimensional FDR “look-up” tables (supplementary figure 3), and Manhattan plots (supplementary figure 4). At the condFDR<0.01 significance threshold and after pruning, 3 independent SNPs were significant for condFDRCollege|SCZ (supplementary table 5) and 2 for condFDREduYears|SCZ (supplementary table 7). Each of these SNPs corresponded to a separate genomic locus (supplementary tables 6 and 8). For condFDRCollege|SCZ, 2 of the 3 loci were reported as being genome-wide significant in the original GWAS of educational attainment.23 In that study, the additional locus was not associated in the discovery sample only,23 but it became significant when the authors performed a combined analysis with their replication sample. For condFDREduYears|SCZ, 1 of the 2 loci was previously reported as being genome-wide significant.23
Identification of SNPs and Genomic Loci Associated With Both SCZ and Educational Attainment
To identify loci significantly associated with both phenotypes in each pairwise combination, we did conjFDR analysis. This procedure identifies loci with significant condFDR association in both SCZ conditioned on College (condFDRSCZ|College) and College conditioned on SCZ (condFDRCollege|SCZ). Thus, a conjFDR value for SCZ and College, denoted conjFDRSCZ&College, is assigned to each SNP. By interpolation into a bi-directional 2-dimensional FDR “look-up” table (supplementary figure 5), we identified 10 loci (shown in a Manhattan plot, supplementary figure 6) that were significantly associated with both phenotypes (conjFDR < 0.05; table 2). As denoted by the sign of the z-scores (table 2), the direction of effect of the loci which are associated with SCZ and College was the same in 6 of the 10 loci, and opposite in the remaining 4 loci. This suggests that the variants implicated in the genetic overlap between SCZ and college completion can have the same or opposite direction of effect.
Table 2.
Conjunctional FDR Between SCZ and Educational Attainment
| Locus Positiona | Size (kb)b | Phenotypesc | SNP IDd | SNP Positione | conjFDRf | Direction of Effect in SCZg | Direction of Effect in EduYears or Collegeh | Genesi |
|---|---|---|---|---|---|---|---|---|
| chr1:98187754-98651526 | 464 | SCZ & EduYears | rs4447033 | 98484291 | 1.43E-02 | + | + | DPYD, DPYD-AS2, MIR137HG, MIR2682, MIR137 |
| SCZ & College | rs2893376 | 98457303 | 4.46E-02 | + | + | |||
| chr1:176966823-177009393 | 43 | SCZ & EduYears | rs12724698 | 176979196 | 4.37E-02 | + | + | ASTN1, MIR488 |
| chr1:243376756-244025998 | 649 | SCZ & EduYears | rs2275155 | 243493907 | 1.43E-02 | + | + | CEP170, SDCCAG8, MIR4677, AKT3 |
| SCZ & College | rs3904683 | 243416525 | 3.98E-02 | + | + | |||
| chr2:7048216-7051170 | 3 | SCZ & EduYears | rs3922041 | 7050887 | 3.68E-02 | + | + | |
| chr2:57941184-58500140 | 559 | SCZ & EduYears | rs11885093 | 57941185 | 3.46E-02 | + | + | VRK2, FANCL |
| chr2:60704483-60727628 | 23 | SCZ & EduYears | rs7581162 | 60704484 | 1.46E-02 | − | − | BCL11A |
| chr2:162796516-162910222 | 114 | SCZ & EduYears | rs6707646 | 162808640 | 2.20E-02 | + | − | SLC4A10, DPP4 |
| chr2:174037346-174037346 | 0 | SCZ & EduYears | rs13004345 | 174037347 | 4.96E-02 | − | + | ZAK |
| chr2:193714080-194665571 | 951 | SCZ & EduYears | rs1913145 | 193753869 | 2.20E-02 | − | − | |
| chr3:16783340-16972210 | 189 | SCZ & EduYears | rs11928330 | 16958367 | 2.56E-02 | + | − | NMD3, SPTSSB |
| chr3:24104244-24152385 | 48 | SCZ & College | rs7612158 | 24109112 | 4.62E-02 | + | − | LINC00691 |
| chr3:160928708-161097267 | 169 | SCZ & EduYears | rs336572 | 161068014 | 3.66E-02 | + | + | |
| chr6:33647057-33791997 | 145 | SCZ & EduYears | rs943472 | 33738442 | 1.43E-02 | − | − | ITPR3, MNF1, IP6K3, LEMD2, MLN |
| SCZ & College | rs545787 | 33703230 | 1.49E-02 | − | − | |||
| chr6:56548358-56731703 | 183 | SCZ & EduYears | rs4415160 | 56686222 | 4.36E-02 | + | + | RNU6-71P, DST, LOC101930010 |
| chr6:104084383-104091971 | 8 | SCZ & College | rs9404453 | 104091972 | 4.48E-02 | + | + | |
| chr6:113387213-113442936 | 56 | SCZ & EduYears | rs2473938 | 113442937 | 3.74E-02 | + | + | |
| chr6:119113316-119113316 | 0 | SCZ & EduYears | rs9401090 | 119113317 | 4.02E-02 | − | − | |
| chr6:128305887-128328832 | 23 | SCZ & EduYears | rs9402011 | 128305888 | 4.96E-02 | − | − | PTPRK |
| chr7:24627941-24828054 | 200 | SCZ & EduYears | rs10486428 | 24627942 | 2.96E-02 | − | − | MPP6, DFNA5 |
| chr8:4815159-4818183 | 3 | SCZ & EduYears | rs11136811 | 4817716 | 2.20E-02 | + | − | CSMD1 |
| chr10:103562935-103720812 | 158 | SCZ & EduYears | rs17698831 | 103656466 | 2.11E-02 | + | − | MGEA5, KCNIP2-AS1, KCNIP2, C10orf76 |
| chr12:123447927-123829027 | 381 | SCZ & EduYears | rs941305 | 123715266 | 1.43E-02 | + | + | ABCB9, OGFOD2, ARL6IP4, PITPNM2, MIR4304, LOC100507091, MPHOSPH9, C12orf65, CDK2AP1, SBNO1 |
| SCZ & College | rs4275659 | 123447928 | 3.44E-02 | + | + | |||
| chr17:17654318-18029856 | 376 | SCZ & College | rs4925109 | 17661802 | 4.59E-02 | + | − | RAI1, SMCR5, SREBF1, MIR33B, TOM1L2, LRRC48, GID4, DRG2, MYO15A |
| chr18:44376848-44585954 | 209 | SCZ & EduYears | rs2246877 | 44577005 | 2.11E-02 | + | + | PIAS2, KATNAL2, TCEB3CL2, TCEB3CL, TCEB3CL, TCEB3C, TCEB3B |
| chr18:53207324-53463113 | 256 | SCZ & EduYears | rs590076 | 53260732 | 1.92E-02 | + | − | TCF4 |
| chr22:40063011-40091546 | 29 | SCZ & College | rs738315 | 40069245 | 4.77E-02 | − | + | CACNA1I |
Note: The following details are shown for each locus containing markers with conjFDR < 0.05.
aLocus position from hg 19 (chr:lower_boundary-upper_boundary).
bLocus size (kb).
cThe pair of phenotypes showing genetic overlap.
drsID of most significant SNP.
ePosition of most significant SNP.
fconjFDR of the most significant SNP.
gDichotomized direction of effect in SCZ obtained from the OR in the PGC-SCZ summary statistics, + effects are for OR > 1, - effects for OR < 1.
hDichotomized direction of effect in College or EduYears (depending on the phenotype pair in c) obtained from the summary statistics in Rietveld et al. (ref.23; + effects are for OR > 1 or positive beta values, − effects are for OR < 1 or negative beta values.
iThe genomic regions around the associated signals were defined by including all markers with conjFDR < 0.05; this column shows the HGNC IDs of genes located in the interval. Some regions have conjFDR significant markers for both SCZ&EduYears and SCZ&College; for these regions the best markers for each conjFDR pair are given.
The same procedure, when applied to SCZ and EduYears, identified 25 loci with significant conjFDRSZC&EduYears (table 2). Of the 25 loci, 16 had effects in the opposite direction, while 9 had effects in the same direction. Four of these 25 loci were also significant for conjFDRSCZ&College.
Gene Annotation
For all loci identified by condFDR < 0.01 (supplementary tables 2 and 4; table 1), we annotated all the genes located within each genomic region using the NCBI RefSeq Database.31 Of the regions that became associated with SCZ after condFDR analysis but were not associated in SCZ only (ie, the regions in table 1), 3 contained multiple genes, 13 contained one gene and the others were intergenic. Intergenic regions were annotated with the nearest gene within 100kb, if any. Further bioinformatics and fine mapping analyses will be required to identify likely causative genes or regulatory elements within these regions.
Of the genes identified, several are implicated in synaptic plasticity or transmission (MEF2C,32INHBA,33GRM334), brain development (AKT3,35BCL11A,36ZEB2,37FOXP1,38FOXO3,39SRPK2,40KLF641) or histone modifications (KMT2E, KDM4C). We screened all the genomic loci identified under condFDR for annotation in the GWAS catalogue. The region chr3:71543757-71579021 is also associated with ADHD.42 The locus chr7:86403262-86459346 was associated with SCZ in the previous PGC-SCZ GWAS but did not reach genome-wide significance in the latest PGC-SCZ GWAS.43
Genomic loci containing multiple genes cannot be studied at the pathway or gene set level by performing threshold-based pathway analysis. Therefore, we used DAPPLE30 to identify networks of interaction between the proteins encoded by the genes located in the genomic loci, after excluding the MHC (supplementary figure 7). DAPPLE analysis prioritized the following genes for follow-up studies: HSPA8, SFMBT1, ATXN7, FOXO3, GATAD2A, MYLPF, TSSK6, and KDM4A.
Discussion
We used a conditional FDR method to demonstrate polygenic overlap between SCZ and educational attainment (college completion and years of education), indicating shared polygenetic factors between SCZ and phenotypes that are influenced by cognitive abilities. Conditioning of the SCZ SNPs on College or EduYears allowed us to detect 23 SCZ-associated loci that were not identified earlier. Conjunctional FDR revealed 29 loci with bi-directional effects, ie, that are significantly associated with both SCZ and educational attainment (College or EduYears). Some loci had the same direction of effect in each pair of phenotypes while others had opposite effects, suggesting a bi-directional relationship between SCZ and educational attainment.
The present findings of 29 gene loci associated with both SCZ and College/EduYears are novel, as we are the first to apply the conjunctional FDR method to this topic. This methodology enables identification of the specific loci that are shared between 2 phenotypes, by leveraging the polygenic overlap.27 The loci identified in SCZ|College and SCZ|EduYears overlap extensively. This is mostly explained by the complete genetic correlation of the 2 phenotypes.44 The observed differences are probably due to the difference in power between the 2 phenotypes, since years of education is quantitative while college completion is binary. Within the genomic loci associated with SCZ conditioned on college attainment, we found additional genes implicated in synaptic plasticity or in neuronal plasticity, ie, axon guidance and neurite development and in histone modifications observed in the brain. Protein network analysis highlighted another gene involved in brain histone modification (KDM4A45), and 2 other genes implicated in brain development (ATXN746, FOXO339,47). The discovery of genes involved in histone modification and synaptic plasticity is in agreement with a report implicating these pathways across psychiatric disorders.48
Educational attainment seems to be a reasonably good proxy for general cognition.23 Our results are in agreement with 2 other studies indicating genetic overlap between SCZ and general cognition using polygenic risk scores for SCZ.12,13 Both studies identified polygenic effects in the opposite direction (ie, SCZ risk was associated with low cognition and vice versa) while we identified effects in both directions. In addition, several recent studies have used polygenic scores to look at the genetic overlap between educational attainment and SCZ, and the polygenic scores have often been derived from the same datasets that we used here.44,49,50 Most of these studies have shown a small genetic overlap, if any, between educational attainment and SCZ. In contrast, we show a clear enrichment. The main difference between these polygenic studies and ours is that they are limited to testing one direction of effect, while our method can identify shared genetic variants with effects in both directions.22 Successful completion of college education, or a greater number of years of education, is highly correlated with cognitive abilities but is also influenced by many other factors, like personality traits.23 A recent study showed that people with creative professions had an increased SCZ polygenic score,14 suggesting that polygenic variants associated with a higher risk of developing SCZ are also associated with higher scores on creativity scales. Interestingly, in their study on polygenic overlaps between cognitive traits, education attainment, and psychiatric disorders, Hill et al51 show that the polygenic correlation was negative between cognition and SCZ, while the correlation with educational attainment, while not significant, was positive. Their results support the bi-directionality that we observe in our study. This bi-directionality may reflect more complexity in the genetic overlap between SCZ and educational attainment than simply a potential detrimental effect of the genes associated with SCZ and cognitive abilities. This emphasizes the need to test for bi-directional effects between SCZ and cognition or educational attainment. As GWAS samples become more powerful (with greater numbers of participants phenotyped for more traits such as cognition, education, personality, etc.), it will be interesting to deconstruct the influence of different traits on SCZ, using polygenic tools that can identify genetic variants with bi-directional, as well as uni-directional effects.
We provide evidence for polygenic overlap between SCZ and educational attainment, and identify novel SCZ risk loci as well as overlapping loci associated with both SCZ and educational attainment. This suggests that polygenic factors may underlie some of the phenotypic overlap between SCZ and cognitive function, as well as other traits such as personality or creativity. Our results provide novel insight into the underlying pathophysiological mechanisms of SCZ.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
This work was supported by the Research Council of Norway (NFR; NORMENT-Centre of Excellence [#223273, #213837, #251134]); the South-East Norway Regional Health Authority (#2013-123); the KG Jebsen Foundation (SKGJ-MED-008); and the National Institutes of Health (U54EB020403, T32 EB005970, R01AG031224, R01EB000790, and RC2DA29475).
Supplementary Material
Acknowledgments
The authors thank all cohorts participating in the PGC and the Educational Attainment studies and the participants in these samples. The authors have declared that there are no conflicts of interest in relation to the subject of this study.
References
- 1. van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–645. [DOI] [PubMed] [Google Scholar]
- 2. Kahn RS, Keefe RS. Schizophrenia is a cognitive illness: time for a change in focus. JAMA Psychiatry. 2013;70:1107–1112. [DOI] [PubMed] [Google Scholar]
- 3. Carson SH. Creativity and psychopathology: a shared vulnerability model. Can J Psychiatry. 2011;56:144–153. [DOI] [PubMed] [Google Scholar]
- 4. Funaki T. Nash: genius with schizophrenia or vice versa? Pac Health Dialog. 2009;15:129–137. [PubMed] [Google Scholar]
- 5. Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. [DOI] [PubMed] [Google Scholar]
- 6. Lichtenstein P, Yip BH, Björk C, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Polderman TJ, Benyamin B, de Leeuw CA, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47:702–709. [DOI] [PubMed] [Google Scholar]
- 8. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee SH, DeCandia TR, Ripke S, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davies G, Tenesa A, Payton A, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16:996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lencz T, Knowles E, Davies G, et al. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the Cognitive Genomics consorTium (COGENT). Mol Psychiatry. 2014;19:168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McIntosh AM, Gow A, Luciano M, et al. Polygenic risk for schizophrenia is associated with cognitive change between childhood and old age. Biol Psychiatry. 2013;73:938–943. [DOI] [PubMed] [Google Scholar]
- 14. Power RA, Steinberg S, Bjornsdottir G, et al. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci. 2015;18:953–955. [DOI] [PubMed] [Google Scholar]
- 15. Andreassen OA, Djurovic S, Thompson WK, et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92:197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andreassen OA, Thompson WK, Dale AM. Boosting the power of schizophrenia genetics by leveraging new statistical tools. Schizophr Bull. 2014;40:13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andreassen OA, Thompson WK, Schork AJ, et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9:e1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu JZ, Hov JR, Folseraas T, et al. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat Genet. 2013;45:670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andreassen OA, Harbo HF, Wang Y, et al. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Mol Psychiatry. 2014;20:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andreassen OA, McEvoy LK, Thompson WK, et al. Identifying common genetic variants in blood pressure due to polygenic pleiotropy with associated phenotypes. Hypertension. 2014;63:819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Andreassen OA, Zuber V, Thompson WK, et al. Shared common variants in prostate cancer and blood lipids. Int J Epidemiol. 2014;43:1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schork AJ, Wang Y, Thompson WK, Dale AM, Andreassen OA. New statistical approaches exploit the polygenic architecture of schizophrenia–implications for the underlying neurobiology. Curr Opin Neurobiol. 2016;36:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rietveld CA, Medland SE, Derringer J, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parisi JM, Rebok GW, Xue QL, et al. The role of education and intellectual activity on cognition. J Aging Res. 2012;2012:416132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rietveld CA, Esko T, Davies G, et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci U S A. 2014;111:13790–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trampush JW, Lencz T, Knowles E, et al. Independent evidence for an association between general cognitive ability and a genetic locus for educational attainment. Am J Med Genet B Neuropsychiatr Genet. 2015;168B:363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schork AJ, Thompson WK, Pham P, et al. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Efron B. Large-Scale Inference: Empirical Bayes Methods for Estimation, Testing, and Prediction. Cambridge, NY: Cambridge University Press; 2010. [Google Scholar]
- 29. Karolchik D, Hinrichs AS, Furey TS, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rossin EJ, Lage K, Raychaudhuri S, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yuan Y, Singh D, Arikkath J. Mef2 promotes spine elimination in absence of delta-catenin. Neurosci Lett. 2013;536:10–13. [DOI] [PubMed] [Google Scholar]
- 33. Lau D, Bengtson CP, Buchthal B, Bading H. BDNF Reduces Toxic Extrasynaptic NMDA Receptor Signaling via Synaptic NMDA Receptors and Nuclear-Calcium-Induced Transcription of inhba/Activin A. Cell Rep. 2015;12:1353–1366. [DOI] [PubMed] [Google Scholar]
- 34. Walker AG, Wenthur CJ, Xiang Z, et al. Metabotropic glutamate receptor 3 activation is required for long-term depression in medial prefrontal cortex and fear extinction. Proc Natl Acad Sci U S A. 2015;112:1196–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baek ST, Copeland B, Yun EJ, et al. An AKT3-FOXG1-reelin network underlies defective migration in human focal malformations of cortical development. Nat Med. 2015;21:1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wiegreffe C, Simon R, Peschkes K, et al. Bcl11a (Ctip1) Controls Migration of Cortical Projection Neurons through Regulation of Sema3c. Neuron. 2015;87:311–325. [DOI] [PubMed] [Google Scholar]
- 37. van den Berghe V, Stappers E, Vandesande B, et al. Directed migration of cortical interneurons depends on the cell-autonomous action of Sip1. Neuron. 2013;77:70–82. [DOI] [PubMed] [Google Scholar]
- 38. Bacon C, Schneider M, Le Magueresse C, et al. Brain-specific Foxp1 deletion impairs neuronal development and causes autistic-like behaviour. Mol Psychiatry. 2015;20:632–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peng S, Zhao S, Yan F, et al. HDAC2 selectively regulates FOXO3a-mediated gene transcription during oxidative stress-induced neuronal cell death. J Neurosci. 2015;35:1250–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jang SW, Liu X, Fu H, et al. Interaction of Akt-phosphorylated SRPK2 with 14-3-3 mediates cell cycle and cell death in neurons. J Biol Chem. 2009;284:24512–24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salma J, McDermott JC. Suppression of a MEF2-KLF6 survival pathway by PKA signaling promotes apoptosis in embryonic hippocampal neurons. J Neurosci. 2012;32:2790–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lasky-Su J, Neale BM, Franke B, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1345–1354. [DOI] [PubMed] [Google Scholar]
- 43. Ripke S, Sanders AR, Kendler KS, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shen E, Shulha H, Weng Z, Akbarian S. Regulation of histone H3K4 methylation in brain development and disease. Philos Trans R Soc Lond B Biol Sci. 2014;369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumar G, Clark SL, McClay JL, et al. Refinement of schizophrenia GWAS loci using methylome-wide association data. Hum Genet. 2015;134:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pino E, Amamoto R, Zheng L, et al. FOXO3 determines the accumulation of α-synuclein and controls the fate of dopaminergic neurons in the substantia nigra. Hum Mol Genet. 2014;23:1435–1452. [DOI] [PubMed] [Google Scholar]
- 48. Network and Pathway Analysis Subgroup of the Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krapohl E, Euesden J, Zabaneh D, et al. Phenome-wide analysis of genome-wide polygenic scores [published online ahead of print August 25, 2015]. Mol Psychiatry. doi:10.1038/mp.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hubbard L, Tansey KE, Rai D, et al. Evidence of common genetic overlap between schizophrenia and cognition. Schizophr Bull. 2016;42:832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hill WD, Davies G, Group CCW, et al. Age-dependent pleiotropy between general cognitive function and major psychiatric disorders [published online ahead of print September 4, 2015]. Biol Psychiatry. doi:10.1016/j.biopsych.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.