Abstract
Background.
Glioblastoma (GBM) is the most common primary brain tumor, with dismal prognosis. The failure of drug–radiation combinations with promising preclinical data to translate into effective clinical treatments may relate to the use of simplified 2-dimensional in vitro GBM cultures.
Methods.
We developed a customized 3D GBM culture system based on a polystyrene scaffold (Alvetex) that recapitulates key histological features of GBM and compared it with conventional 2D cultures with respect to their response to radiation and to molecular targeted agents for which clinical data are available.
Results.
In 3 patient-derived GBM lines, no difference in radiation sensitivity was observed between 2D and 3D cultures, as measured by clonogenic survival. Three different molecular targeted agents, for which robust clinical data are available were evaluated in 2D and 3D conditions: (i) temozolomide, which improves overall survival and is standard of care for GBM, exhibited statistically significant effects on clonogenic survival in both patient-derived cell lines when evaluated in the 3D model compared with only one cell line in 2D cells; (ii) bevacizumab, which has been shown to increase progression-free survival when added to standard chemoradiation in phase III clinical trials, exhibited marked radiosensitizing activity in our 3D model but had no effect on 2D cells; and (iii) erlotinib, which had no efficacy in clinical trials, displayed no activity in our 3D GBM model, but radiosensitized 2D cells.
Conclusions.
Our 3D model reliably predicted clinical efficacy, strongly supporting its clinical relevance and potential value in preclinical evaluation of drug–radiation combinations for GBM.
Keywords: erlotinib, glioblastoma, glioma stemlike cells, ionizing radiation, three-dimensional, VEGF
Glioblastoma (GBM) is the most common primary brain tumor and is associated with dismal prognosis.1 Tumors exhibit inherent chemo- and radioresistance, which has been attributed to a subpopulation of cancer cells termed ‘GBM stem-like cells’ (GSC).2 Comprehensive genomic and molecular characterization of GBM has identified a number of promising targets, and preclinical studies have shown activity of several molecularly targeted agents against GBM cell lines.3 To date, however, these agents have failed to improve clinical outcomes for GBM patients.4 One explanation for the discrepancy between preclinical and clinical data may be the lack of preclinical models that faithfully recapitulate the clinical scenario. In particular, established cancer cell lines cultured in simplified 2D in vitro systems undergo profound phenotypical changes and have been reported to exhibit markedly different responses to cytotoxic treatments than those observed in patients.5 In the context of radiation therapy 3D culture of lung and head and neck cancer cells embedded in laminin-rich extracellular matrix (ECM) has been shown to promote radiation resistance compared with 2D culture.6–8 Colorectal cancer cell lines cultured in similar laminin-rich ECM 3D conditions also exhibited changes in cellular morphology, phenotype, and gene expression and were resistant to treatment with epidermal growth factor receptor (EGFR) inhibitors compared with 2D cells.5,9 These observations provide a potential explanation for the frequent failure of results derived from conventional 2D cell culture systems to predict clinical efficacy.10
To examine the hypothesis in the context of GBM, in which all molecular targeted agents so far tested in patients have been ineffective, we developed a customized 3D cell culture system that recapitulates key histological features of GBM such as high cellularity, a relative paucity of stromal components, and the presence of radioresistant GSC. Patient-derived GBM cells were cultured in stem cell promoting medium in order to preserve the GSC phenotype. Based on accumulating evidence that GSC localize preferentially to hypoxic and perivascular regions associated with high levels of vascular endothelial growth factor (VEGF),11,12 and because GBM generally exhibit high levels of hypoxia and VEGF,13,14 the culture medium was supplemented with VEGF. Using this 3D model, we evaluated cellular responses to radiotherapy alone and in combination with molecular targeted agents previously assessed in the clinic and compared survival data with results derived from 2D simplified cultures and with data from clinical trials in GBM. Our data indicate that this novel 3D in vitro model may have broad utility as a clinically representative system in which to evaluate potential new therapies for this cancer of unmet need.
Materials and Methods
Cell Culture and Radiation Treatment
E2, R10, and G7 GBM cell lines were obtained from Colin Watts (Cambridge) and are derived from anonymized patient resection specimens as previously described.15,16 Cell lines were routinely cultured on Matrigel-coated plates (0.2347mg/mL in Adv/Dulbecco’s modified Eagle’s medium) in serum-free “stem cell” medium (Supplementary materials). For laminin-rich 3D cultures, cells were seeded in suspension in 1:2 diluted Matrigel (4.75mg/mL in stem cell media). For Alvetex 3D cultures, Alvetex scaffolds were permeabilized according to manufacturer’s instructions and coated with diluted Matrigel as for 2D cultures.
Clonogenic Survival Assays
Seeded were 250 cells per well in Matrigel-coated plates or scaffolds or embedded in Matrigel plugs for 3D-E experiments. Eighteen hours after seeding, cells were either sham irradiated or irradiated (1–9 Gy) and incubated for 2 to 3 weeks prior to methanol fixation and crystal violet staining. Visible colonies were counted manually. For 3D cultures, cells were incubated with MTT reagent and colonies counted manually. Curves represent mean surviving fraction ± SD of 3 independent experiments (in triplicate) fitted to a linear quadratic model.
Mouse Experiments
Female CD1 nude mice were orthotopically injected with 1×105 cells grown for 7 days in 3D-Alvetex scaffolds or in 2D conditions as previously performed.16 Mice were monitored for the duration of experiment and were sacrificed when they became symptomatic (isolated, hunched, reduced mobility, and/or weight loss greater than 20%).
Ethical Approval
Animal experiments were in compliance with all regulatory guidelines, as described in the Animals Act 1986 Scientific Procedures on living animals regulated by the Home Office in the United Kingdom.
Flow Cytometry
For 2D cultures, cells were detached from plastic by incubation with trypsin (1mL) (Gibco) and fixed with 70% ice-cold ethanol/phosphate buffered saline (PBS). For 3D cultures, cells were detached with trypsin (1mL per scaffold) and incubated for 15min in a rotating platform at 30rpm. Fixed cells were stained with 1 µg/mL propidium iodide/PBS in the presence of RNase A (10 µg/mL). Cell cycle distribution was determined according to manufacturer’s instructions (BD Biosciences). Data were analyzed using FlowJo software (Tristar).
Protein Extraction
For 2D and 3D cultures, cells were incubated in 1% sodium dodecyl sulfate–Tris buffer for 20min on ice; 3D cultures were transferred to a rotating platform at 100rpm and incubated for 10min. Recovered lysates were clarified using Qiagen columns.
Gene Expression Array
Four days after plating cells in either 2D or 3D conditions, RNA was extracted with TRIzol reagent. RNA sequencing (RNA-seq) analysis was performed using the IlluminaNextSeq500 for a PolyA selection RNA library, with a paired-end sequencing model and 33M depth for triplicate experimental repeats of 2D and 3D G7 or E2 cultures. Differential transcript expression analysis was performed using TopHat and Cufflinks. Expression array data were analyzed according to the following criteria: statistical significance was set at q value of <0.1, where P value was set at <.05.
Quantitative Real-Time PCR
Cells were grown as for gene expression array assays, RNA extraction was performed using TRIzol reagent (Life Technologies), and cDNA was prepared using the Quantitech Reverse Transcription Kit (Qiagen, cat. no. 205311), both as per manufacturer’s instructions. Quantitative PCR was conducted using SYBR green (Quanta, cat. no. 95072-012) and the Bio-Rad CFX platform, with a 60ºC annealing temperature and the primer pairs that are tabulated in the Supplementary materials.
Fluorescence In situ Hybridization Assay
Cells were grown in either 2D or 3D conditions for 4 days and fixed with 2% paraformaldehyde for 15min. Fluorescence in situ hybridization (FISH) analysis was performed using a histology FISH Accessory Kit (Dako, cat. no. K5799) and an EGFR/CEN-7 FISH probe mix (Dako, cat. no. Y5500) according to manufacturer’s instructions. Z-stack images were acquired using a Zeiss 780 confocal microscope with a 63× magnification objective and analyzed with Zen black software.
Immunofluorescence
Cells were plated at identical seeding densities either on Matrigel-coated coverslips or on Matrigel-coated Alvetex scaffolds. Fixed cells (2% paraformaldehyde/PBS) were permeabilized with 1% Triton/PBS, blocked with 2% bovine serum albumin/Tris-buffered saline/0.5% Tween-20, and incubated with phalloidin for actin detection (Abcam) or with the respective primary antibodies (Supplementary materials). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) present in mounting medium (VectaShield).
Statistical Analyses
All experiments were performed in triplicate and data points reported as ±SEM. P values were obtained using the unpaired 2-tailed t-test in GraphPad Prism software. Kaplan–Meier mouse survival curves were analyzed using Minitab 17 Statistical software with pairwise comparisons and log rank (Mantel–Cox) analysis.
Results
Characterization of Primary GSC Cultured in the Novel 3D-Alvetex System
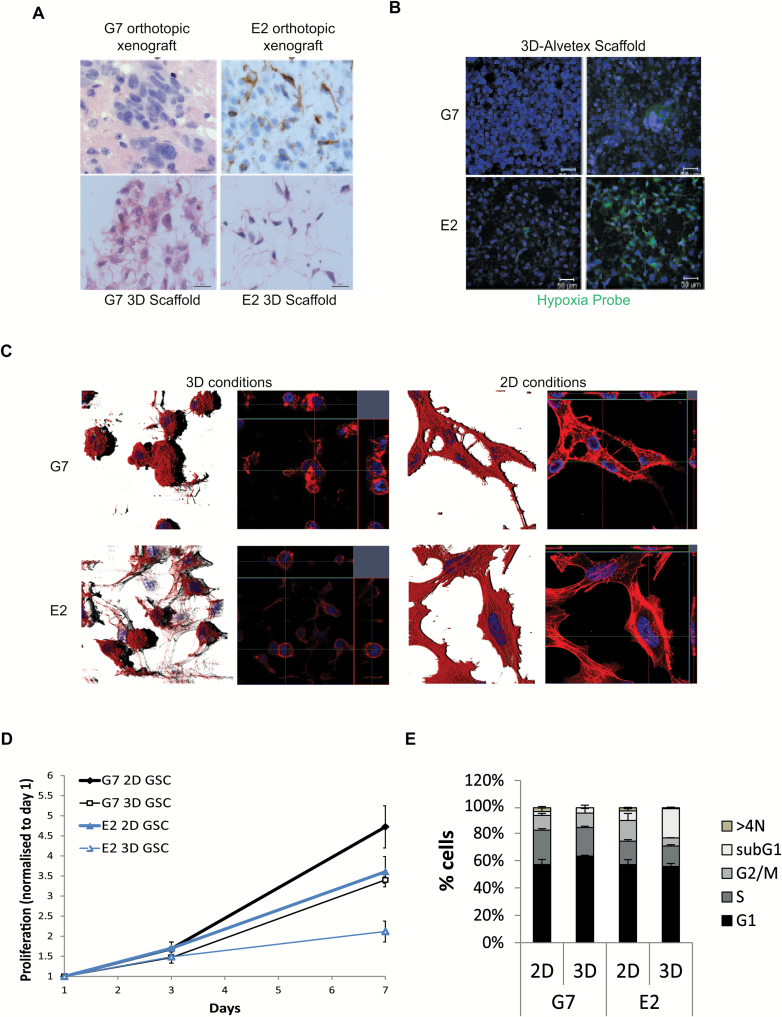
Previously published 3D cancer models have generally reproduced the characteristics of carcinomas, in which cells are embedded in a dense, laminin-rich microenvironment and are thus inappropriate for GBM studies. In GBM, laminin expression is sparse and generally restricted to blood vessels (Supplementary Fig. 1A 17). Using Alvetex technology,18 we established a novel in vitro GBM model which promotes 3D growth of patient-derived GSC on a polystyrene scaffold. To replicate the perivascular stem cell niche, cells were cultured in serum-free stem cell medium supplemented with VEGF as well as EGF and basic fibroblast growth factor, which are routinely used in GSC cultures,19 and both 2D and 3D surfaces were coated with diluted (1:40) laminin-rich ECM (Supplementary Fig. 1B). Three-dimensional cultures recapitulated the histopathololgical features of GBM in general and of orthotopic xenografts derived from the corresponding cell cultures in particular. Key features include high cellularity (G7 cell line: Fig. 1A, left panels), invasion (E2 cell line: Fig. 1A, right panels), and hypoxic regions (Fig. 1B). The specific hypoxia marker pimonidazole revealed cell line dependent effects of the 3D model on oxygen concentrations: pimonidazole staining was barely detectable in 3D G7 cultures but was observed in approximately 20% of sections obtained from 3D E2 cultures (Fig. 1B). These observations were supported by direct measurements made with an oxygen probe (OxyLite, Oxford Optronics) applied to the surface of scaffolds: oxygen levels in G7 3D cultures were generally higher and more uniform (mean value 68.9±4.03 mmHg, 9.06% O2) than those in E2 cultures, which ranged from 20.6±2.07 mmHg (2.6% O2) to 63.6±6.48 mmHg (8.48 O2) depending on the region measured. GSC cultured in 3D conditions acquired a spherical morphology that contrasted with the flattened, elongated morphology typical of adherent cells in 2D culture (Fig. 1C). By promoting highly cellular and invasive 3D growth of GSC in reduced ECM conditions, the model recapitulates essential features of the GBM microenvironment
Fig. 1.
Characterization of GSC grown in 3D conditions. (A) Representative hematoxylin and eosin and human leukocyte antigen images of orthotopic tumors derived from intracranially injected G7 or E2 cells, respectively, and G7 or E2 cells grown on 3D-Alvetex scaffolds. (B) GSCs were grown in Alvetex scaffolds and incubated with pimonidazole one hour prior to fixation (2% paraformaldehyde). Scaffolds were immunostained using an anti-pimonidazole antibody (green) and counterstained with DAPI (blue). Two representative images of 3D cultures are shown for each cell line. (C) Immunofluorescent images of F-actin (Alexa Fluor 568 Phalloidin, red) and nuclear staining (DAPI; blue) of cells in 3D and 2D GSC conditions. (D) G7 and E2 GSC (5x104) were seeded in 12-well Alvetex scaffolds or in T25 flasks and proliferation of 2D and 3D cultured cells measured according to Alvetex scaffolds MTT viability assay instructions (http://reinnervate.com/using-alvetex/workflow-2-culturing-monitoring-3d-cell-growth/). Graph of mean±SD (n = 3). Statistical significance is observed in both cell lines at day 7 (t-test, P<.005). (E) Cell cycle distribution of GSC cultured in 2D or 3D conditions for 24 hours and analyzed by flow cytometry after propidium iodide staining (n = 3). Graph of mean±SEM (n = 3).
Cells in 3D cultures displayed reduced proliferation rates compared with 2D (Fig. 1D, day 7), consistent with the non-exponential growth profiles observed in vivo and with other published 3D models in vitro.20 VEGF supplementation increased 3D cell proliferation and had no effect on 2D conditions (Supplementary Fig. 2A). Three-dimensional culture had no effect on cell cycle distribution in G7 cells; however, reduced G2/M populations and increased sub-G1 populations were observed in 3D cultured E2 cells (Fig. 1E), which can be likened to the high prevalence of cell loss and necrosis observed in GBM, and these effects were not affected by VEGF addition (Supplementary Fig. 2B and 2C, respectively).
Primary 3D Culture Preserves the GBM Stem-like Phenotype
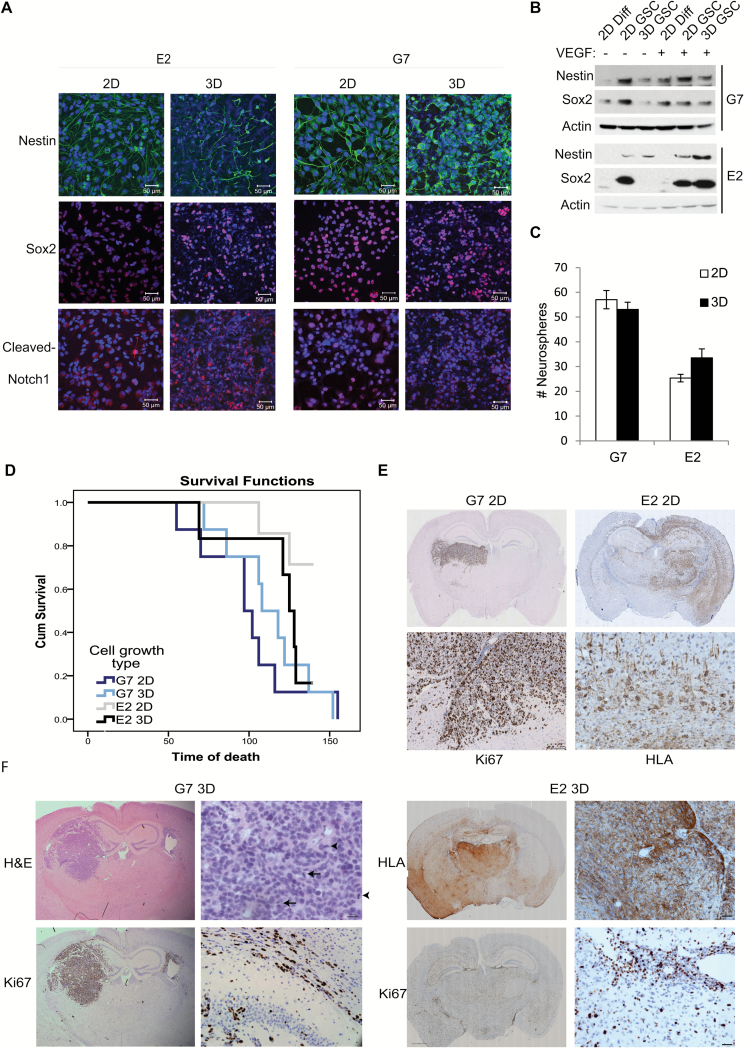
G7 and E2 cells cultured in both 2D and 3D conditions retained expression of the putative stem cell markers nestin, Sox2, and cleaved Notch1 as assessed by immunofluorescence (Fig. 2A) and western blot (Fig. 2B). Removal of VEGF from 3D culture medium was associated with increased expression of the differentiation marker glial fibrillary acidic protein and a decrease in stem cell marker expression but had no effect on these parameters in 2D cultures (Supplementary Fig. 3). Indeed, the negative effects of VEGF deprivation on 3D culture “stemness” were equivalent to the well-established differentiating effects of fetal calf serum on 2D cultures (Supplementary Fig. 3). These data corroborate VEGF’s role in maintaining a cancer stem cell phenotype in 3D conditions,21,22 and the absence of effect in 2D models emphasizes the critical influence of the 3D microenvironment on growth factor signaling. Consistent with the stem cell marker data, neurosphere formation capacity, a well-established surrogate of the GSC phenotype, was preserved and equivalent in cells cultured in 2D or 3D conditions (Fig. 2C); and in vivo tumorigenicity, the gold standard assay of the cancer stem cell phenotype, was also maintained. Tumors were generated in 100% of immunocompromised mice (Crl:NU(NCr)-Foxn1nu) subjected to intracranial injection of G7 GSC that had been cultured in either 2D or 3D conditions, and no statistical significant difference in mouse survival was observed (Fig. 2D). In the case of E2 cells, 8/8 mice injected with 2D cultured GSC generated tumors detectable on histology compared with 7/8 mice injected with 3D GSC. However, whereas most mice from the 3D GSC cohort had become symptomatic by the end of the study (140 days after injection), only 2 mice from the 2D cohort developed symptoms (Fig. 2D). Accelerated tumor progression was also observed in mice injected with CD133+ E2 cells sorted by flow cytometry (data not shown), indicating that the 3D microenvironment recapitulates the tumorigenic attributes of the stem cell niche. Similarly, G7 tumors derived from 2D and 3D GSC displayed key histopathological features of GBM, including high rates of cellular proliferation and mitosis, and infiltrative growth along white matter tracts (Fig. 2E and 2F). Tumors derived from 3D and 2D cultured E2 cells expressed high levels of the proliferation marker Ki67 and were remarkably invasive: in these mice, human leukocyte antigen‒1ABC (HLA) positive tumor cells were detected in both hemispheres and frequently localized around blood vessels (Fig. 2E and 2F). Together these results demonstrate that GSC cultured in the novel 3D system retain their tumorigenic potential and generate orthotopic tumors that recapitulate the GBM phenotype.
Fig. 2.
GSCs grown on 3D conditions retain their stemness and tumorigenicity. (A, B) Immunofluorescent images (A, scale bar 50 µm) and western blot analysis (B) of E2 and G7 GSCs grown on 2D or 3D conditions of stem cell marker expression. (C) Neurosphere formation assays for G7 and E2 GSC previously grown for 7 days in 2D or 3D stem cell conditions. Mean values ±SEM (n = 3). (D) Kaplan–Meier survival curves showing overall survival of individual cohorts of mice orthotopically injected with G7 and E2 cells grown on 2D or 3D conditions. Pairwise comparisons using log rank (Mantel–Cox) analysis: E2 2D versus E2 3D cells P = .082; G7 2D versus 3D cells P = .519. (E and F) Hematoxylin and eosin, Ki67, and human leukocyte antigen (HLA) immunohistochemistry of paraffin-embedded brain sections containing orthotopic tumors derived from intracranially injected G7 cells or E2 cells grown in 2D conditions (E) or 3D conditions (F) for 7 days prior to injection (n = 8). → indicates mitotic bodies.
Radiation Responses of GSC in 2D and 3D Cultures
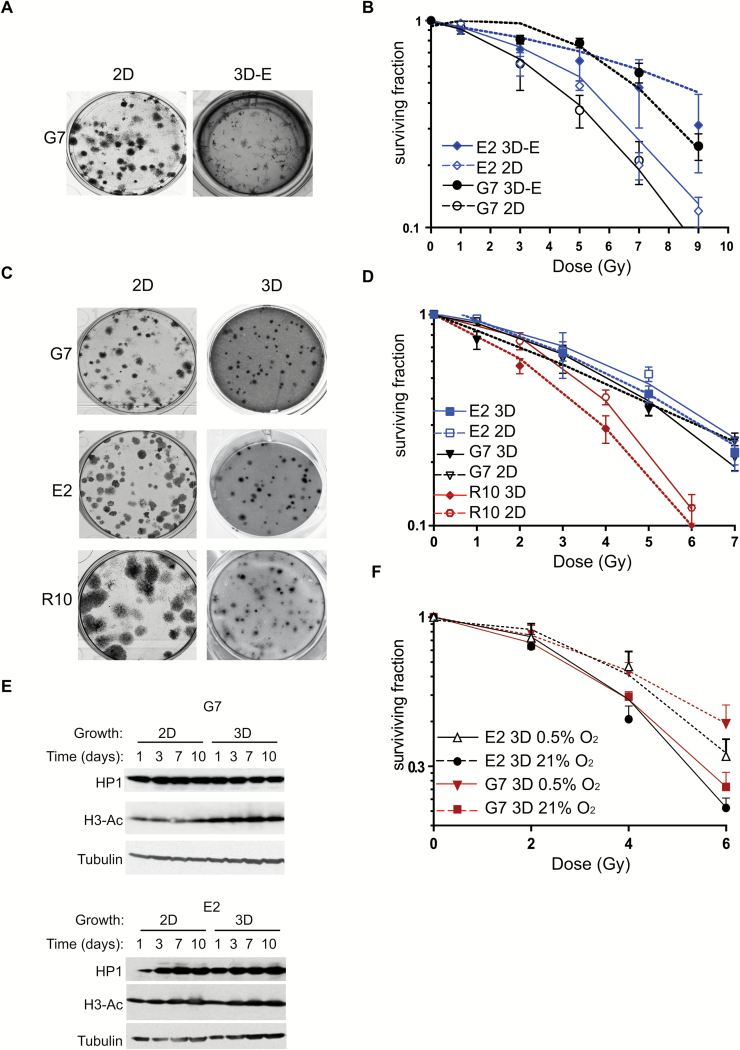
To evaluate the impact of 2D and 3D culture conditions on radiation responses of GSC, clonogenic survival assays were conducted in the relevant models, with cell irradiation and colony formation taking place in situ, and similar plating efficiencies were observed in the 2D and 3D models (data not shown). To distinguish between 3D growth and ECM effects, clonogenic survival experiments were initially conducted using GSC cultured either as monolayers on ECM-coated plates (2D) or embedded in laminin-rich ECM (3D-E), which has previously been shown to increase radiation resistance in lung and head and neck carcinoma cell lines.6 GSC cultured in 3D-E conditions were significantly more radioresistant than corresponding 2D cultures, consistent with previous studies in other cell types (Fig. 3A, B). In contrast, the radiation sensitivity of E2 and G7 cells cultured in the 3D-Alvetex scaffold (3D) model was equivalent to that observed in 2D cultures, except for the R10 primary cell line, in which a modest increase in radiosensitivity was observed in 3D (Fig. 3C, 3D). This novel finding indicates firstly that ECM concentrations can affect ligand binding and effects of ligands on signaling and cell behavior, which might have a greater influence on radiation resistance than 3D growth per se and secondly that 3D in vitro models must be tailored to reflect the microenvironmental features of the relevant tumor type.
Fig. 3.
Radiation responses of GSC in 2D and 3D conditions. (A, C) Representative images of GSC colonies after 21 days growing on plastic (2D), embedded in Matrigel (3D-E, A) or in 3D-Alvetex scaffolds (3D, C). (B, D) Clonogenic survival curves of G7 and E2 cells grown in 2D and 3D-E (B) or 2D and 3D-Alvetex scaffold conditions (D) and irradiated with single doses of X-rays (0–9 Gy; n = 3). Curves are significantly different in (B) for both cell lines by 2-way ANOVA (2D vs 3D-E P < .0001, calculated by ANOVA general linear model. No statistical significance was observed for 2D versus 3D G7 (P = .1) or E2 (P = .1) data in (D). R10 3D cultures were significantly more radiosensitive than 2D cultures (P = .01). (E) Western blot analysis of G7 and E2 cell lysates extracted from cells grown in 2D or 3D. (F) Clonogenic survival curves as (D). Hypoxic cultures of both G7 and E2 GSCs are significantly more resistant than normoxic cultures (2-way ANOVA analysis; P = .0021 and P = .0004, respectively).
Changes in cellular morphology and an associated increase in heterochromatin have previously been reported to account for the radioprotection observed in 3D-E conditions.6 In our 3D-Alvetex cultures, GSC acquired a spherical morphology that was distinct from the flattened, elongated morphology of the 2D populations (Fig. 1C). In contrast, cells grown in 3D-E conditions, while less flattened than 2D cells, acquired a more elongated morphology than cells cultured on the scaffold. Furthermore, western blot analysis revealed no effect of 3D-Alvetex culture on levels of heterochromatin protein 1; indeed, a slight increase in histone H3 acetylation (a marker of euchromatin) was observed (Fig. 3E). These observations are consistent with RNA-seq data (see below), which showed decreased expression of histone deacetylase genes in 3D GSC populations (Table 1). Taken together, our results indicate no association among chromatin condensation, cellular morphology, and radiation sensitivity in GSC.
Table 1.
Reduced expression of histone deacetylases in 3D GSC compared with 2D GSC
| Gene | Locus |
Log2_fold
2D v 3D |
Cell Line |
Significance
(P value) |
|---|---|---|---|---|
| HDAC5 | chr17:42154120-42201014 | −0.28206 | G7 | .02705 |
| HDAC6 | chrX:48660486-48683380 | −0.24577 | G7 | .0224 |
| HDAC7 | chr12:48166966-48213763 | −0.61262 | G7 | 5.00E-05 |
| −0.53918 | E2 | .03 | ||
| HDAC9 | chr7:18126571-19036992 | −0.84383 | G7 | .00065 |
| −0.8634 | E2 | 5.00E-05 |
Source: RNA sequencing (RNA-Seq) analysis of gene expression performed on G7 and E2 cells grown on plastic (2D) or 3D- Alvetex scaffolds (3D).
Because the high cellularity and low stromal component of the 3D-Alvetex model more closely resembled the GBM microenvironment, we subsequently focused on characterizing radiation responses of GSC in this model, with the primary aim of validating it as a clinically relevant model of GBM.
Regions of low oxygen concentration (hypoxia) are a cardinal feature of GBM, and it is widely recognized that hypoxic cells are radioresistant.23 To document the impact of hypoxia on radiation responses of 3D GSC and to more closely mimic the hypoxic cancer stem cell niche, clonogenic survival assays were performed under oxic (21% O2) and clinically relevant hypoxic conditions (0.5% O2). As expected, cells were significantly more radioresistant at 0.5% O2 than at 21% O2 (Fig. 3F). These results demonstrate that hypoxia confers further radioprotection to the already radioresistant 3D GSC populations, and add further weight to our hypothesis that the 3D model can recapitulate key features of GBM.
The EGFR Tyrosine Kinase Inhibitor Erlotinib Enhances Radiosensitivity of 2D GSC but Has No Effect in 3D Cultures
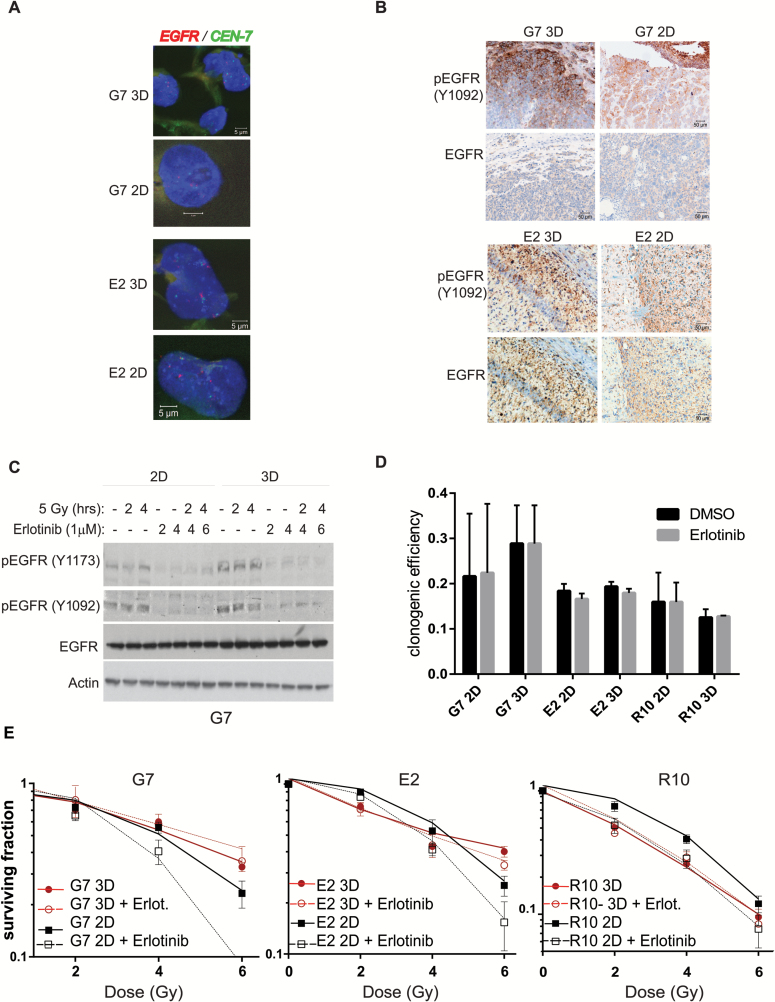
In the past decade, a series of international, multicenter, phase III randomized studies have failed to improve outcomes for patients with GBM, either through intensification of chemotherapy or addition of molecular targeted therapies such as cilengitide or EGFR inhibitors such as erlotinib to standard treatment. Results of phase II trials studying the addition of molecular targeted agents including erlotinib to standard therapy have been equally discouraging.24–26 EGFR gene amplification is a common feature of GBM that is associated with poor prognosis,27 and a number of preclinical studies have demonstrated radiopotentiating efficacy of erlotinib in conventional 2D GBM cultures.28 A modest amplification of the EGFR gene in our patient-derived cell lines was observed, with no difference between 2D and 3D populations (Fig. 4A). G7 and E2 cells also exhibited chromosome 7 gain (centromere to chromosome 7 staining by FISH, red, Fig. 4A), a typical cytogenetic feature of GBM. Motivated by the large disparity between the preclinical and clinical effects of erlotinib, we selected this drug to investigate whether the 3D model provides a more clinically relevant system than 2D cell culture. EGFR expression and activation in E2 and G7 GSC grown in 2D and 3D conditions expressed similar levels of total and activated (phospho-Y1092) EGFR protein both in orthotopic xenografts derived from cells cultured in 2D and 3D conditions and in vitro (Fig. 4B, Supplementary Fig. 4A). Erlotinib inhibited EGFR activity in both 2D and 3D G7 cultures as demonstrated by decreased phosphorylation of its active sites (Y1173 and Y1092) at baseline and following radiation treatment (Fig. 4C); however, no single agent activity of erlotinib was observed in either 2D or 3D GSC in 3 different primary GBM cell lines, as assessed by clonogenic formation efficiency (Fig. 4D). In contrast, the effects of erlotinib on radiation sensitivity were profoundly affected by the microenvironment. While erlotinib significantly enhanced the radiosensitivity of 2D cultures of all 3 cell lines, it had no effect on radiation sensitivity in the 3D model (Fig. 4E). No radiosensitization by erlotinib was confirmed in neurosphere formation assays (Supplementary Fig. 4C), a clinically relevant measure of GSC survival and self-renewal in which single cells generate 3D sphere structures devoid of a scaffold or extrinsic ECM, although a reduction in neurosphere formation efficiency was observed following erlotinib treatment alone (Supplementary Fig. 4B). Hence while EGFR activation might be required for GBM tumorigenicity, our data demonstrate that EGFR inhibition has no therapeutic effect in 3D models in combination with radiation.
Fig. 4.
Radiosensitization of GSCs by EGFR inhibition is determined by growth conditions. (A) Representative immunofluorescence images of EGFR gene (green) and chromosome 7 centromere (red) staining by FISH assay in G7 and E2 2D and 3D GSC. Nonnuclear staining reflects background autofluorescence. (B) Representative immunohistochemistry images of phosphorylated and total EGFR in G7 and E2 orthotopic tumors from cells grown on 3D or 2D conditions for 7 days. (C) Protein extracts of G7 GSCs grown in 2D or 3D conditions obtained at different time points after treatment with erlotinib (1 µM) and/or ionizing radiation (5 Gy) were analyzed for total and phosphorylated EGFR by western blot. Actin served as loading control. (D) Clonogenic survival efficiency of G7, E2, and R10 cells treated with either vehicle (dimethyl sulfoxide [DMSO]) or erlotinib (1 µM) 20 hours following seeding and left for the duration of the experiment (18 days). Graph depicts mean±SD. (E) Clonogenic survival of G7, E2, and R10 cells grown in 2D and 3D conditions and irradiated with single doses of X-rays (0–6 Gy; n = 3) 2 hours after treatment with erlotinib (1 µM) or DMSO. Erlotinib treatment significantly increased the radiosensitivity of G7, E2, and R10 GSCs under 2D conditions (ANOVA; P < .0001, P = .0006, and P = .0016, respectively). No effect of erlotinib was observed in 3D conditions compared with DMSO (G7 P = .1; E2 P = .1007 and R10 P = .842).
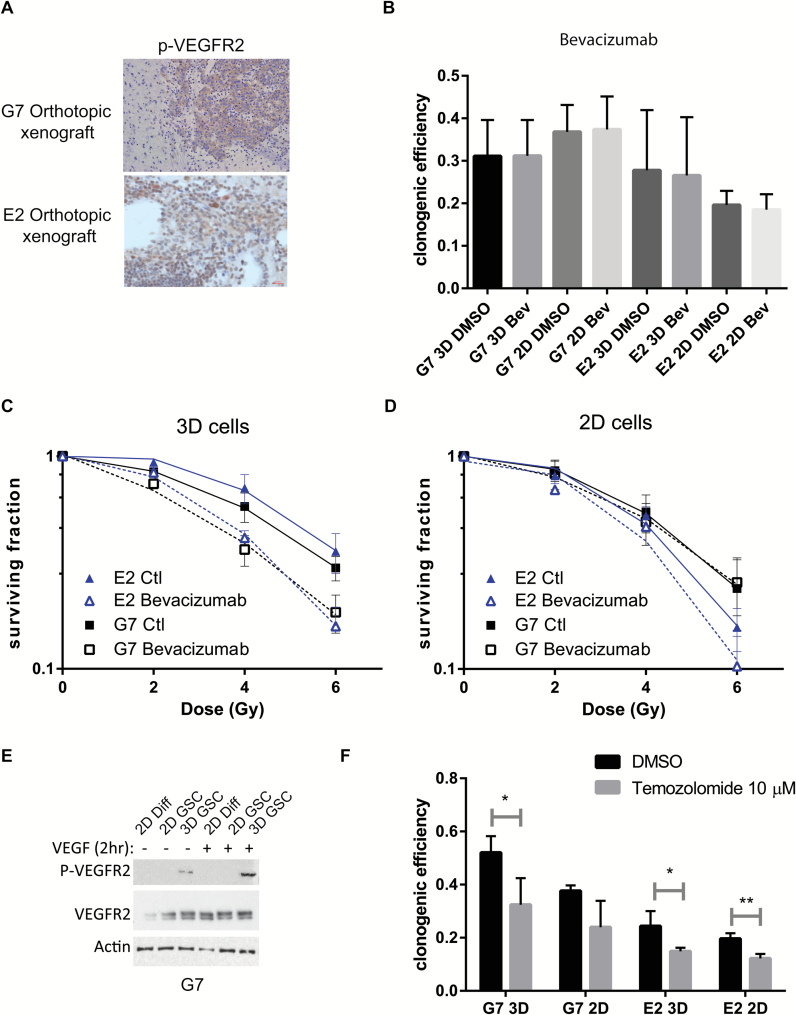
The Novel 3D GSC Model Recapitulates the Activity of Temozolomide and Bevacizumab Observed in the Clinic
Next, we evaluated 2 agents that have documented clinical activity against GBM: temozolomide, which is part of standard of care1,29; and the anti-VEGF monoclonal antibody bevacizumab, which is the only molecular targeted agent to have shown evidence of radiopotentiating activity against GBM in the form of increased progression-free survival in 2 randomized phase III trials.30,31 Constitutive activation of the VEGF receptor VEGFR2 was observed in G7 and E2 orthotopic xenografts (Fig. 5A). Bevacizumab treatment alone had no effect on clonogenic efficiency of GSC in 2D or 3D conditions (Fig. 5B). In contrast, whereas bevacizumab treatment significantly radiosensitized 3D GSCs, but had no radiosensitizing activity in 2D cultures (Fig. 5C and 5 D, respectively). Bevacizumab also significantly potentiated the effects of radiation on neurosphere formation of both cell lines (Supplementary Fig. 4E), as well as presenting activity as a single agent in the E2 cell line (Supplementary Fig. 4D). Radiosensitization in this alternative 3D model confirmed our findings in the 3D-Alvetex system and reproduced the effects observed in clinical trials.
Fig. 5.
Radiosensitization of GSCs by bevacizumab and temozolomide is determined by growth conditions. (A) Representative immunohistochemistry images of phospho-VEGFR2 in G7 and E2 orthotopic tumors grown from 3D GSC. (B) Clonogenic survival efficiency of G7 and E2 cells treated with either vehicle (PBS) or bevacizumab (0.1 µg/mL) 20 hours following seeding and left for the duration of the experiment (18 days). Graph depicts mean±SD. (C and D) Clonogenic survival of G7, E2, and R10 cells grown in 3D (C) and 2D (D) conditions and irradiated with single doses of X-rays (0–6 Gy; n = 3) 2 hours after treatment with bevacizumab (0.1 µg/ mL) or vehicle (PBS). Bevacizumab treatment significantly increased the radiosensitivity of G7 and E2 GSCs under 3D conditions (ANOVA; control vs bevacizumab P < .01 and P < .05, respectively). (E) Protein extracts of G7 GSCs grown in 2D or 3D conditions in the absence or presence of VEGF-A (30ng/mL) were analyzed for total and phospho-VEGFR2 by western blot. (F) Clonogenic survival efficiency of G7 and E2 cells treated with either vehicle (dimethyl sulfoxide; DMSO) or temozolomide (10 µM) as in (B). Statistical significance (t-test) *P < .05, **P < 0.05.
Consistent with this 3D-specific effect, VEGFR2 protein levels were higher in 3D cells than 2D cells in the absence of VEGF, and the active, phosphorylated form of VEGFR2 was only detected in 3D G7 cells (Fig. 5E). Although addition of VEGF induced expression of VEGFR2 in 2D GSC and differentiated cells, no detectable levels of phosphorylated VEGFR2 were observed.
Finally, activity of the clinically active agent temozolomide on 2D and 3D GSC cultures was assessed. Treatment with temozolomide significantly reduced the clonogenic efficiency of 3D GSC in both G7 and E2 GSCs, whereas in 2D conditions this cytotoxic effect only reached statistical significance in the E2 cell line (Fig. 5F). Taken together, these findings corroborate our hypothesis that the novel 3D GBM culture system replicates effects observed in the clinic, both negative and positive clinical outcomes, and will facilitate meaningful preclinical assessment of novel molecular targets.
Genome-wide Gene Expression Analysis in 2D and 3D GSC
Expression array analysis was performed to identify specific gene expression patterns associated with 2D and 3D culture conditions which might explain differential responses to treatment. Comparison of 2D and 3D datasets demonstrated a higher number of genes to be differentially expressed in G7 (4027 genes) than E2 (361 genes) GSCs (full list provided in Table S1). Found to be upregulated were 2108 transcripts and 1916 downregulated in G7 GSC grown on 3D conditions compared with 2D, while 222 transcripts were upregulated and 137 downregulated in E2 3D cells. When the 2 cell lines were matched together, 160 transcripts were differentially expressed in 2D versus 3D in both cell lines (80 upregulated, 35 downregulated, and 45 exhibiting opposite expression patterns).
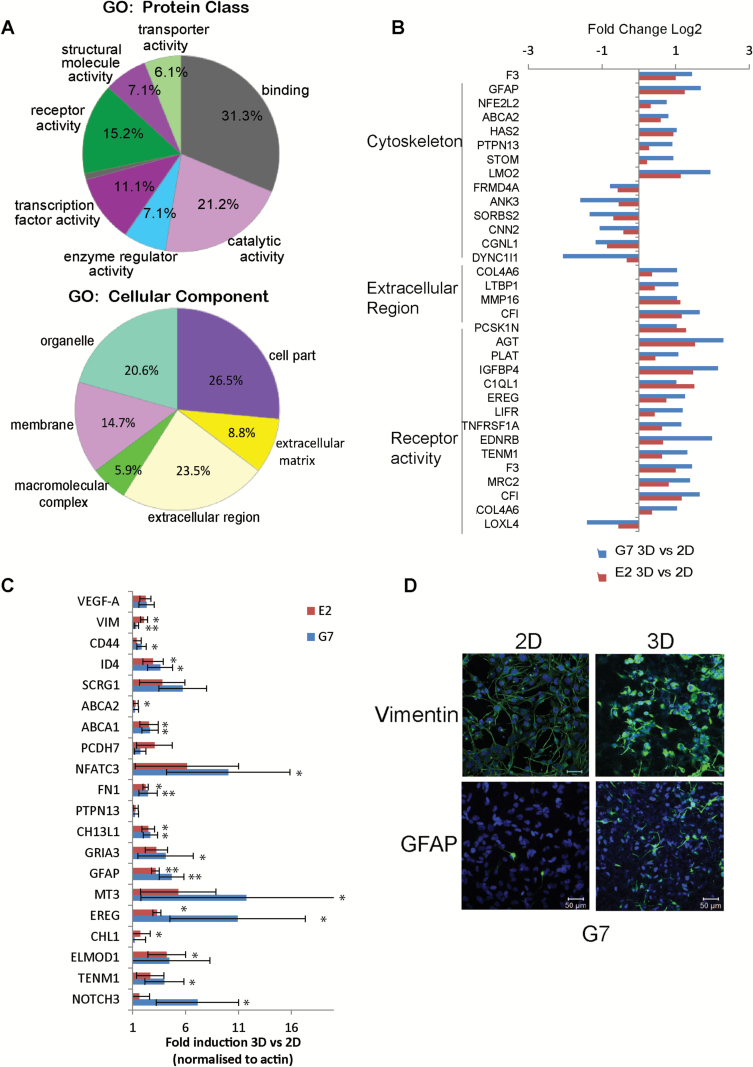
Gene ontology analysis of upregulated transcripts in 3D conditions by protein class using Panther32 identified genes associated with receptor, transporter, and transcription factor activity (Fig. 6A). These include genes associated with the EGFR pathway such as EREG,33ABCA1 and ABCA2,34IGFBP4,35PTPN13,36 and NOTCH337; and with VEGFR activity such as VEGFA and EDNRB. Cellular component analysis revealed an increase in expression of genes belonging to the organelle, membrane (100% belonging to cytoskeleton), extracellular region, and ECM categories (Fig. 6A). Of note, increased expression of genes associated with “stemness” (eg, CEBPD, NOTCH3, ID; and Wnt signaling) was observed in 3D cultures (Supplementary Fig. 6B). Several genes from the “mesenchymal” GBM signature were upregulated in 3D cultures of both E2 and G7 cells, including CHI3L1, RELB, FN1, VIM, and CEBPD (Supplementary Fig. 6A); however, our transcriptome data revealed no clear segregation of our cultures with the proposed GBM subtypes previously reported,38 exemplifying the intratumoral transcriptional heterogeneity observed in GBM.39
Fig. 6.
RNA-seq analysis of 3D versus 2D GSC cultures. (A) Gene ontology (GO) analysis of transcripts significantly upregulated in the 3D model in both G7 and E2 cultures. GO of molecular function (upper chart) and cellular components (lower chart) are represented. (B) Bar charts representing differentially expressed genes in 3D compared with 2D G7 and E2 cells within the cytoskeleton, extracellular matrix region, and receptor activity categories. (C) Real-time PCR validation of representative genes upregulated in the 3D GBM model. Bars represent mean±SD of cDNA expression from 3 independent experiments performed in duplicate. t-Test *P < .05; **P < .005. (D) Immunofluorescent images of vimentin (upper panel) and glial fibrillary acidic protein (lower panel) of G7 cells grown in either 2D or 3D conditions.
Quantitative real-time PCR validation of the transcriptome data regarding the 3D versus 2D comparison of 20 genes confirmed upregulation of the selected target genes with 100% success rate (Fig. 6C). Immunofluorescence analysis of G7 3D cells confirmed upregulation of the cytoskeleton proteins vimentin and glial fibrillary acidic protein in 3D compared with 2D G7 cells, recapitulating human GBM immunohistochemistry (Fig. 6D).
Although different patterns were observed in the 2 cell lines studied, the marked changes in cytoskeleton, transcriptional activity, and receptor activity associated with the transition to 3D culture are consistent with the observed morphological changes and might explain the different effects of molecular targeted therapies against EGFR or VEGFR observed in the 2D and 3D models.
Discussion
Here we describe a customized, 3D cell culture system that recapitulates key histological features of GBM. Our data indicate that (i) responses of GBM cells to growth factor receptor therapies in combination with radiation are profoundly affected by 2D versus 3D culture; (ii) meaningful preclinical assessment of these and other molecular targeted agents requires experimental models that recapitulate key features of tumors in vivo; and (iii) the 3D GBM model developed in our laboratory has potential value in these analyses. The new 3D model reproduced the clinical results of 3 molecular targeted therapies, demonstrating its reliability predicting clinical outcomes and indicating superiority over conventional 2D models which have failed to predict clinical efficacy.
In the context of radiation therapy, 2D and 3D GSC exhibited similar sensitivity in our models, which might be explained by the stem cell culture conditions that enrich for the radioresistant GSC population. Further characterization of our 3D GBM model in terms of proteomics, metabolomics, and biophysical properties such as stiffness will further our understanding of the potential value of our model in evaluating radiation, drug, and radiation–drug responses.
One important feature of our 3D model was the requirement for VEGF supplementation for stem cell phenotype preservation, as previously identified for EGF and basic fibroblast growth factor.24 VEGF promotes neurosphere formation in GSC cultures21 and proliferation of neural stem cells,40 which share common features with GSCs. Our 3D model not only corroborates VEGF’s role in promoting GSC proliferation (see Supplementary Fig. 4B), but also implicates VEGF in cell fate determination. This “stemness” function of VEGF is not novel but importantly has only been observed in vitro in 3D cancer stem cell conditions such as neurospheres,21 or when studied in vivo.22 This mechanistic observation is of biological interest and corroborates the clinical significance of our 3D model. Understanding how VEGF enhances the cancer stem cell phenotype will be critical for developing effective therapeutic strategies to target GSC populations. Our data indicate that there may yet be potential in targeting the VEGF signaling pathway in GBM but that greater understanding of this complex pathway is required in order to identify the best therapeutic approach. Because both endothelial and tumor cells secrete VEGF, blocking paracrine and autocrine VEGF/VEGFR2 autocrine loops may be required to achieve remission and long-term cure.
Overall, the results obtained from our 3D GSC model replicate the therapeutic responses to molecular targeted therapies observed in GBM clinical trials. As well as increasing our understanding of the clinical effects and limitations of radiation therapy in the management of patients with GBM, our data support the concept that new potential treatments for GBM should be evaluated in clinically meaningful 3D models before proceeding to in vivo and clinical testing. Our findings support the 3D-Alvetex model as a platform for modeling drug and radiation responses in this cancer of unmet need.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
Funding for this work was provided by the Medical Research Council [64420] to A.C., a Chief Scientist Office (CSO) project ETM/405 to A.C. and N.G.R., and Wellcome Trust grant 097821/Z/11/Z to N.G.R..
Conflict of interest statement. The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
Cell lines were kindly donated by Dr Colin Watts, University of Cambridge.
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. [DOI] [PubMed] [Google Scholar]
- 3. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rich JN, Reardon DA, Peery T, et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol. 2004;22(1):133–142. [DOI] [PubMed] [Google Scholar]
- 5. Luca AC, Mersch S, Deenen R, et al. Impact of the 3D Microenvironment on Phenotype, Gene Expression, and EGFR Inhibition of Colorectal Cancer Cell Lines. PloS one. 2013;8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Storch K, Eke I, Borgmann K, et al. Three-dimensional cell growth confers radioresistance by chromatin density modification. Cancer Res. 2010;70(10):3925–3934. [DOI] [PubMed] [Google Scholar]
- 7. Zschenker O, Streichert T, Hehlgans S, et al. Genome-wide gene expression analysis in cancer cells reveals 3D growth to affect ECM and processes associated with cell adhesion but not DNA repair. PloS one. 2012;7(4):e34279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hehlgans S, Lange I, Eke I, et al. 3D cell cultures of human head and neck squamous cell carcinoma cells are radiosensitized by the focal adhesion kinase inhibitor TAE226. Radiotherapy and oncology: journal of the European Society for Therapeutic Radiology and Oncology. 2009;92(3):371–378. [DOI] [PubMed] [Google Scholar]
- 9. Poschau M, Dickreuter E, Singh-Muller J, et al. EGFR and beta1-integrin targeting differentially affect colorectal carcinoma cell radiosensitivity and invasion. Radiotherapy and oncology: journal of the European Society for Therapeutic Radiology and Oncology. 2015;116(3):510–516. [DOI] [PubMed] [Google Scholar]
- 10. Prados MD, Schold SC, Sc Jr, Fine HA, et al. A randomized, double-blind, placebo-controlled, phase 2 study of RMP-7 in combination with carboplatin administered intravenously for the treatment of recurrent malignant glioma. Neuro Oncol. 2003;5(2):96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takano S, Yoshii Y, Kondo S, et al. Concentration of vascular endothelial growth factor in the serum and tumor tissue of brain tumor patients. Cancer Res. 1996;56(9):2185–2190. [PubMed] [Google Scholar]
- 12. Plate KH, Rizau W. Angiogenesis in malignant gliomas. Glia. 1995;15(3):339–347. [DOI] [PubMed] [Google Scholar]
- 13. Flynn JR, Wang L, Gillespie DL, et al. Hypoxia-regulated protein expression, patient characteristics, and preoperative imaging as predictors of survival in adults with glioblastoma multiforme. Cancer. 2008;113(5):1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shibahara I, Kumabe T, Kanamori M, et al. Imaging of hypoxic lesions in patients with gliomas by using positron emission tomography with 1-(2-[18F] fluoro-1-[hydroxymethyl]ethoxy)methyl-2-nitroimidazole, a new 18F-labeled 2-nitroimidazole analog. J Neurosurg. 2010;113(2):358–368. [DOI] [PubMed] [Google Scholar]
- 15. Fael Al-Mayhani TM, Ball SLR, Zhao J-W, et al. An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumors. J Neurosci Methods. 2009;176(2):192–199. [DOI] [PubMed] [Google Scholar]
- 16. Ahmed SU, Carruthers R, Gilmour L, et al. Selective Inhibition of Parallel DNA Damage Response Pathways Optimizes Radiosensitization of Glioblastoma Stem-like Cells. Cancer Res. 2015;75(20):4416–4428. [DOI] [PubMed] [Google Scholar]
- 17. Kawataki T, Yamane T, Naganuma H, et al. Laminin isoforms and their integrin receptors in glioma cell migration and invasiveness: Evidence for a role of alpha5-laminin(s) and alpha3beta1 integrin. Exp Cell Res. 2007;313(18):3819–3831. [DOI] [PubMed] [Google Scholar]
- 18. Bokhari M, Carnachan RJ, Cameron NR, et al. Novel cell culture device enabling three-dimensional cell growth and improved cell function. Biochem Biophys Res Commun. 2007;354(4):1095–1100. [DOI] [PubMed] [Google Scholar]
- 19. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 20. Fischbach C, Chen R, Matsumoto T, et al. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4(10):855–860. [DOI] [PubMed] [Google Scholar]
- 21. Hamerlik P, Lathia JD, Rasmussen R, et al. Autocrine VEGF-VEGFR2-Neuropilin-1 signaling promotes glioma stem-like cell viability and tumor growth. J Exp Med. 2012;209(3):507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beck B, Driessens G, Goossens S, et al. A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumors. Nature. 2011;478(7369):399–403. [DOI] [PubMed] [Google Scholar]
- 23. Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev. Cancer. 2008;8(3):180–192. [DOI] [PubMed] [Google Scholar]
- 24. Brown PD, Krishnan S, Sarkaria JN, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J Clin Oncol. 2008;26(34):5603–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. dos Santos MA, Pignon JP, Blanchard P, et al. Systematic review and meta-analysis of phase I/II targeted therapy combined with radiotherapy in patients with glioblastoma multiforme: quality of report, toxicity, and survival. J Neurooncol. 2015;123(2):307–314. [DOI] [PubMed] [Google Scholar]
- 26. Prados MD, Chang SM, Butowski N, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27(4):579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shinojima N, Tada K, Shiraishi S, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63(20):6962–6970. [PubMed] [Google Scholar]
- 28. Chinnaiyan P, Huang S, Vallabhaneni G, et al. Mechanisms of enhanced radiation response following epidermal growth factor receptor signaling inhibition by erlotinib (Tarceva). Cancer Res. 2005;65(8):3328–3335. [DOI] [PubMed] [Google Scholar]
- 29. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomized phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 30. Lai A, Tran A, Nghiemphu PL, et al. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J Clin Oncol. 2011;29(2):142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Narayana A, Gruber D, Kunnakkat S, et al. A clinical trial of bevacizumab, temozolomide, and radiation for newly diagnosed glioblastoma. J Neurosurg. 2012;116(2):341–345. [DOI] [PubMed] [Google Scholar]
- 32. Mi H, Muruganujan A, Casagrande JT, et al. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8(8):1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oliveras-Ferraros C, Cufi S, Queralt B, et al. Cross-suppression of EGFR ligands amphiregulin and epiregulin and de-repression of FGFR3 signalling contribute to cetuximab resistance in wild-type KRAS tumor cells. Br J Cancer. 2012;106(8):1406–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hopper-Borge EA, Nasto RE, Ratushny V, et al. Mechanisms of tumor resistance to EGFR-targeted therapies. Expert Opin Ther Targets. 2009;13(3):339–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guix M, Faber AC, Wang SE, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118(7):2609–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scrima M, De Marco C, De Vita F, et al. The nonreceptor-type tyrosine phosphatase PTPN13 is a tumor suppressor gene in non-small cell lung cancer. Am J Pathol. 2012;180(3):1202–1214. [DOI] [PubMed] [Google Scholar]
- 37. Alqudah MA, Agarwal S, Al-Keilani MS, et al. NOTCH3 is a prognostic factor that promotes glioma cell proliferation, migration and invasion via activation of CCND1 and EGFR. PloS one. 2013;8(10):e77299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao Z, Kong Y, Yang S, et al. Upregulation of Flk-1 by bFGF via the ERK pathway is essential for VEGF-mediated promotion of neural stem cell proliferation. Cell Res. 2007;17(1):73–79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.