Abstract
Background.
Glioblastoma (GBM) is the deadliest primary brain cancer in adults. Emerging innovative therapies hold promise for personalized cancer treatment. Improving therapeutic options depends on research relying on relevant preclinical models. In this line we have established in the setting of the GlioTex project (GBM and Experimental Therapeutics) a GBM patient-derived cell line (GBM-PDCL) library. A multi-omic approach was used to determine the molecular landscape of PDCL and the extent to which they represent GBM tumors.
Methods.
Single nucleotide polymorphism array, expression arrays, exome sequencing, and RNA sequencing were used to measure and compare the molecular landscapes of 20 samples representing 10 human GBM tumors and paired GBM-PDCLs.
Results.
Copy number variations were similar for a median of 85% of the genome and for 59% of the major focal events. Somatic point mutations were similar in a median of 41%. Mutations in GBM driver and “druggable” genes were maintained in 67% of events. Mutations that were not conserved in the PDCL were mainly low allelic fraction and/or non-driver mutations. Based on RNA expression profiling, PDCLs cluster closely to their parental tumor with overexpression of pathways associated with cancer progression in PDCL.
Conclusions.
Overall, PDCLs recapitulate pivotal molecular alterations of paired-parental tumors supporting their use as a preclinical model of GBM. However, some driver aberrations are lost or gained in the passage from tumor to PDCL. Our results support using PDCL as a relevant preclinical model of GBM. Further investigations of changes between PDCLs and their parental tumor may provide insights into GBM biology.
Keywords: cancer, cell lines, genome, glioblastoma
Glioblastoma (GBM) is the most common and the most devastating primary brain cancer in adults. Despite intensive treatments (ie, surgery, radiation therapy, and/or chemotherapy), prognosis of GBM patients remains dismal, with a median overall survival between 12 and 18 months appealing for new treatments.1
Most of the innovative therapeutic strategies for cancer treatment developed in the last decade incorporate drugs targeting specific oncogenic proteins or signaling pathways. Such promising approaches are already used in non-CNS tumors.23 The potential of these approaches is being enhanced by the comprehensive molecular mapping of thousands of tumors, thus identifying novel oncogenic targets.4 In addition, the growing number of targeted drugs5 should enable the application of tumors’ molecular information to offer a growing number of therapeutic options, personalized to each patient’s disease state.
Development of novel treatments is highly dependent on relevant preclinical models recapitulating biology of human tumors. Indeed, before reaching clinical routine, innovative treatments are tested most often in cancer cell lines (CCLs) and animal models. Accordingly, efforts to create large molecularly comprehensively annotated CCL libraries were carried out.6
The use of CCLs as models to investigate potential efficacy of novel drugs is built on the assumption that important parts of tumor biology are represented in these cellular models. Hence, it seems important to examine in a quantitative manner whether indeed CCL libraries represent the molecular landscape of parental tumors.
We have established a cell line library of GBM patient-derived cell lines (GBM-PDCLs). In this study, we measured and compared the molecular profiles, obtained using high-throughput technologies, of a set of parental tumors and paired GBM-PDCLs. Multi-omics is the study of those biological factors that end with “-omic,” such as genomic, proteomic, transcriptomic, and metabolomic. Indeed, we hypothesize that the measurement of multi-omics molecular profile changes in this cohort of paired tumors/GBM-PDCLs can address and shed light on potential molecular aberrations and biological processes that are being lost or gained during the transition from human GBM to GBM-PDCL.
Materials and Methods
Human GBM Samples
Fresh tumor samples from 10 patients with newly diagnosed de novo GBM were collected. The patient characteristics are given in Table 1. Blood samples were available for 7 patients. Samples come from the tissue bank OncoNeuroThèque and were accrued over 6 years. The PDCL derivation success rate was 32%. Collection of tumor and blood samples, clinicopathological information, and molecular analysis were undertaken with informed consent and with the relevant ethical board approval in accord with the tenets of the Declaration of Helsinki.
Table 1.
Patient characteristics
| Patient | PDCL | Blood | Age | Sex |
|---|---|---|---|---|
| 2197T | 4339 | NA | 59 | F |
| 2211T | 4371 | NA | 76 | F |
| 3716T | 5706 | 3716_S | 67 | M |
| 3718T | 6190 | NA | 78 | F |
| 3427T | 6240 | 3427_S | 72 | F |
| 3719T | 7015 | 3719_S | 74 | M |
| 3722T | 7060 | 3722_S | 59 | M |
| 3523T | 7097 | 3523_S | 70 | F |
| 3724T | 7142 | 3724_S | 65 | M |
| 4724T | N13-1520 | 4724_S | 53 | M |
GBM–PDCL Preparation
Within 3 hours post-resection, tumors collected in Hank’s Buffered Salt Solution were mechanically dissociated and then maintained in neurosphere growth conditions using Dulbecco’s modified Eagle’s medium/F12 supplemented with 1% penicillin/streptomycin, B27 (Life Technologies), epidermal growth factor (EGF; 20ng/mL), and basic fibroblast growth factor (20ng/mL) (Peprotech). Tumor cells in culture were amplified for at least 8 passages, after which the cell line was considered established.
To standardize cell material preparation, 1×106 cells, from established PDCLs (ie, ≥8 passages) were plated in a T75 flask. Three days later, culture medium was renewed, and after 24 hours, cells were collected, centrifuged, and snap frozen.
Copy Number Variation Analysis
Copy number variation (CNV) analysis was performed using the iSelect Infinium HumanOmniExpress v1.0 Illumina chip platform and the GPHMM (Global Parameters Hidden Markov Model) algorithm.7
Whole Exome Sequencing
Exome capture was performed using the Capture Agilent SureSelect All Exon V5+UTR kit according to manufacturer’s protocol and for 5 samples using the Nextera Rapid Capture Exome Kit. A paired-end 2×75 base sequencing was performed by HiSeq 2000. Data analysis used Genome Analysis Toolkit (GATK) best practices pipeline8 as detailed in the Supplementary materials. Somatic mutation analysis used Mutect9 for tumor samples with blood-paired DNA data and the GATK HaplotypeCaller8 for tumor samples without blood-paired DNA data. For the tumor samples without blood-paired DNA data, only mutations that were not described in the Single Nucleotide Polymorphism Database (dbSNP) were considered.
Point Mutations Annotation and Interpretation
Point mutations were annotated by Oncotator.10 IntOGen software11 was used for functional impact prediction in the cancer biology context. All mutations of the exome were analyzed. We also examined mutations in several gene subsets: (i) the Catalogue of Somatic Mutations in Cancer (COSMIC) Gene Census: 547 general cancer-related genes,12 (ii) GBM driver genes: 23 frequently mutated GBM driver genes,13 and (iii) “druggable genes”: 69 genes that can be targeted by FDA-approved drugs.14
RNA Chip Analysis
Expression array analysis was performed by Affymetrix Human Genome U133 Plus 2.0 array. Data analysis methods are given in Supplementary materials. The Limma package15 was used for differential expression analysis (P < .05 with false discovery rate [FDR] correction). GBM subtype16 classification was performed using single-sample gene set enrichment analysis (ssGSEA) in GenePattern,17 as reported by Brennan et al.13
RNA Sequencing (RNA-Seq) Analysis
Libraries were generated from total RNA and constructed according to manufacturer protocols. Paired-end sequencing (2×150bp) was performed by a Nextseq 500 machine using the High Output Kit (300 cycles). Data analysis methods are given in the Supplementary materials.
Pathway Analysis
Qiagen’s Ingenuity Pathway Analysis (IPA)18 was used to assess pathways involving genes that were differentially expressed between parental tumors and PDCLs. Two statistical measures were used for pathway assessment: (i) P-value for the enrichment of each pathway’s genes in the set of differentially expressed genes, FDR correction applied (P < .05), and (ii) the activation/inhibition measure for each pathway, with threshold Z=1.
GSEA19 was performed for the complete transcriptome using GenePattern17 implementation. Gene lists representing the pathways of Biocarta, the Kyoto Encyclopedia of Genes and Genomes (KEGG), and the Pathway Interaction Database (PID) were extracted from MsigDB (the Molecular Signatures Database).20
Clonal Analysis
Clonal analysis was performed by Absolute algorithm.21 This algorithm requires input of basic CNV segmentation profiles and somatic point mutations. For the CNV segmentation, we used the CBS (circular binary segmentation) algorithm22 and the somatic point mutation data as described above.
Statistical Analysis
Statistical analysis was performed using the R programming language.
Results
Copy Number Variations
The frequencies of CNV, in parental tumors and paired PDCLs, are reported in Fig. 1. As expected in the parental GBM, the most common chromosomal alterations are: (i) chromosome 7 gain 100%, (ii) chromosome 10 loss 90%, (iii) chromosome 6 loss 50%, and (iv) chromosome 9 loss 20%. Classical focal genomic alterations targeting GBM driver genes are also detected: (i) EGFR amplification 50%, (ii) CDKN2A homozygous deletion 60%, (iii) MDM2 amplification 10%, (iv) PIK3CA amplification 10%, and (v) CDKN2C homozygous deletion 10%.
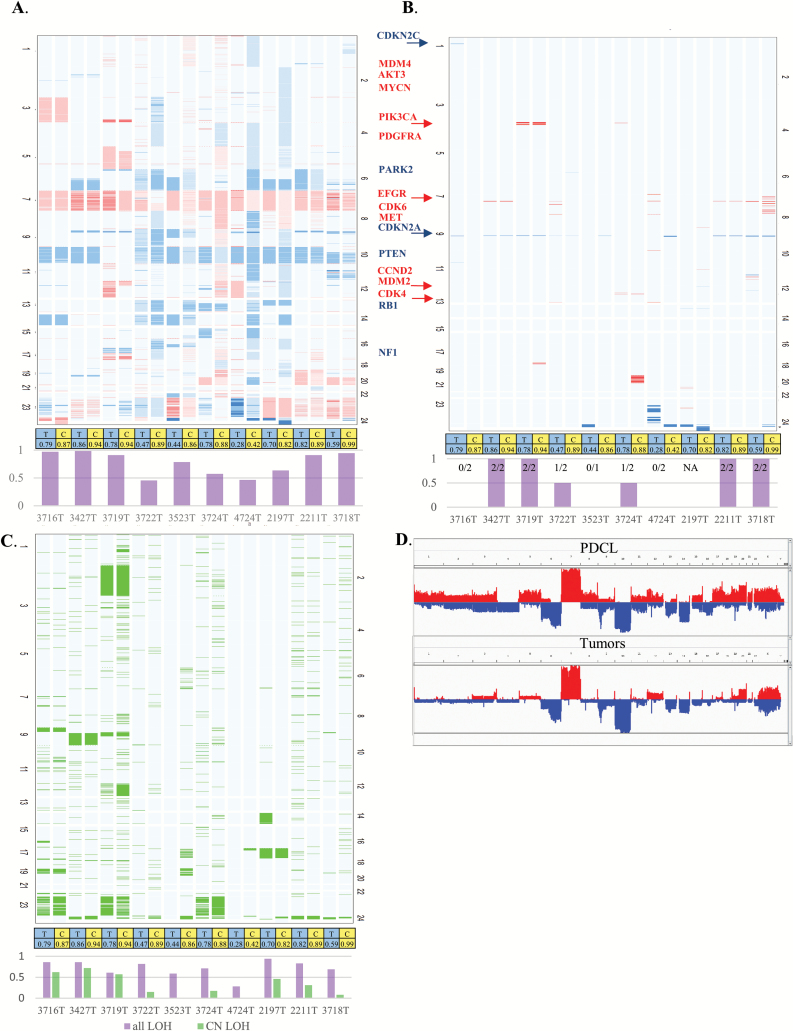
Fig. 1.
Copy number variation landscape. (A). Tumor and PDCL heatmap. Tumors (T) and their paired PDCL (C) are adjacent to each other. Red denotes gain and blue denotes loss in relation to estimated ploidy. Darker color stands for higher gain or deeper deletion. The bar chart at the bottom gives the level of agreement for each tumor–PDCL pair. (B) Heatmap filtered for high-level amplification (CN≥ploidy+3) is denoted in red, and deeper deletions (CN=0 or CN≤1 if ploidy is 4) are colored blue. The bar chart at the bottom gives the level of agreement for each tumor–PDCL pair. (C) Heatmap describing estimated CN-LOH. The bar chart at the bottom gives the level of agreement for each tumor–PDCL pair with purple for general LOH and light green for CN-LOH. (D). Genomic landscape of the group of PDCLs (top) and group of tumors (bottom).
Copy number state (gain, normal, or loss) was compared between parental GBM and paired PDCLs. The median level of agreement, compared at bins of 1000 base pairs, was 85% (range: 46%−99%) (Fig. 1A). The level of agreement for the most common large alterations (chromosome 7 gain, chromosome 10 loss, chromosome 6 loss, and chromosome 9 loss) was 90%. The 10% difference consisted of chromosome 6 loss in one PDCL and chromosome 9 loss in 2 PDCLs that were not detected in their paired parental tumor.
Across the parental tumor samples, 15 focal genomic alterations (high-level amplifications and homozygous deletions) of known GBM driver genes were detected (Fig. 1B): (i) 10/15 were maintained in their paired PDCLs and (ii) 5/15—including 2 EGFR amplifications, one CDKN2C homozygous deletion, one CDKN2A homozygous deletion, and one CDK4 amplification—were not detected in paired PDCLs. Of note, 2 CDKN2A homozygous deletions measured in the PDCL were not found in the parental tumor.
The frequencies of copy-neutral loss of heterozygosity (CN-LOH), in the parental tumors and PDCLs, are reported in Fig. 1C. The most common large chromosomal level CN-LOH was in chromosome 9 (30%), and the level of agreement between tumors and PDCLs was 100% for large chromosome CN-LOH. Comparison of all areas defined as CN-LOH showed poor agreement between tumor and PDCL (range 0–72%) with median of 24%. For all types of LOH (CN-LOH, deletion-LOH, and gain-LOH), the level of agreement range is 28%–94% with a median of 77%.
Point Mutations
Exome sequencing analysis was performed for the 10 tumors, 10 paired PDCLs, and 7 corresponding available blood samples. Mean coverage was 76X ±13. Point mutation analysis was carried out to identify somatic mutations for the 14 samples with corresponding blood DNA and to identify non-dbSNP mutations (termed “novel”) for the 6 samples without blood DNA (Fig. 2). Overall 1988 somatic mutations (including intronic and silent mutations; see Supplementary file 1) were detected across the samples: (i) median 41% mutations existed in both tumor and PDCL, (ii) median 19% mutations were present in the parental tumors only, and (iii) median 36% were present in the PDCL only. Of note, the 2 samples presenting the lowest frequency of maintained mutations (PDCLs 7015 and N13-1520: 14% and 17 %, respectively) contained 2 different missense TP53 mutations (C176F and R248Q, respectively) that appeared only in the PDCL and not in the parental tumor. These 2 mutations are described as frequent somatic mutations in the COSMIC database and are predicted to have high impact on protein activity. Interestingly, a minority of cells in parental tumors corresponding to these PDCLs stained positive for tumor protein (TP)53—suggesting the existence of these mutations in minor subclones that were positively selected when cultured as PDCLs (Fig. 2D). When considering mutations in the subset of COSMIC genes, a median of 44% of mutations existed in both tumors and PDCLs. For the subset of GBM driver genes, the corresponding number was 50%.
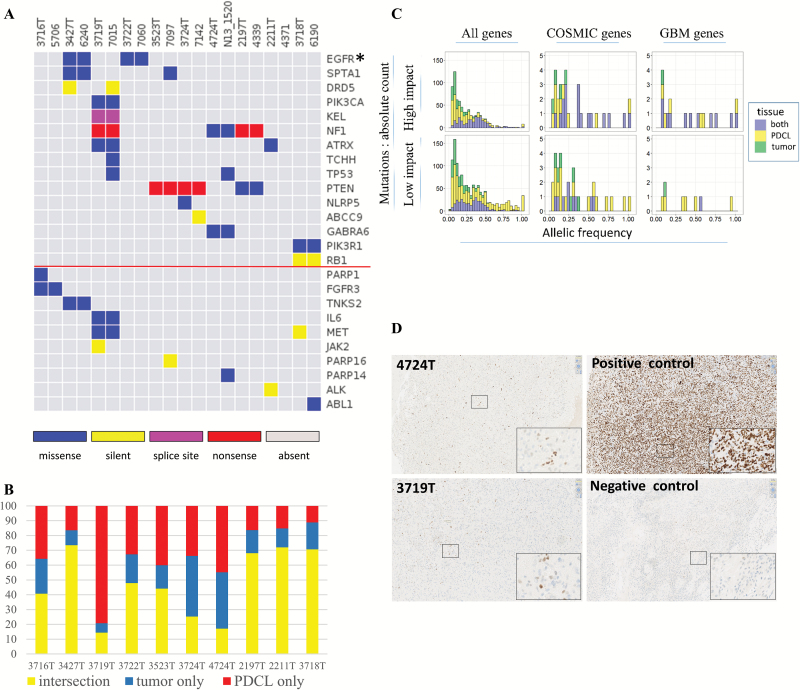
Fig. 2.
Point mutations landscape. (A) Point mutations for GBM driver genes (top) and druggable genes (bottom, below the red line). Tumors and their paired PDCL are adjacent to each other. Different colors are given for the mutation types. For the left 7 pairs, germ line information was used for somatic mutations inference. For the right 3 pairs, germ line information was unavailable and mutations defined as “novel” are shown (see “Methods” section). (B) Frequency of mutations that appeared in both tumor and PDCL (yellow), tumor only (blue), PDCL only (red). (C) Mutation characteristics for the combined set of somatic mutations. In each histogram, the distribution of allelic fraction (x-axis) of mutations is given. The y-axis denotes mutation count. Each histogram bar is divided for the tissues in which the mutations were detected: (i) both tumor and PDCLs, (ii) PDCL only, and (iii) tumor only. The 6 histograms are ordered in columns and rows. The columns define gene set groups: (i) all genes, (ii) COSMIC genes, (iii) GBM driver genes. The rows define the predicted functional impact class (“impact,” “no impact”). (D) TP53 staining for parental tumors 4724T and 3719T.
Detailed description, focused on the non-silent mutations, in the subsets of GBM driver genes and “druggable” genes is of special interest due to the genes’ biological and therapeutic relevance. Mutations in GBM driver genes were maintained in 11/16 events across the samples. Only 1/16 mutations was detected in parental tumors and disappeared in their paired PDCLs (NLRP5 mutation). Four of 20 mutations were not detected in the parental tumors but were found in PDCL (one SPTA1 mutation, one TCHH mutation, and 2 TP53 mutations). For the subset of “druggable” genes (including EGFR), 6/8 events were maintained. One of 8 mutations was detected in a parental tumor but was not detected in its paired GBM-PDCL (PARP1 mutation). One of 8 mutations was found in GBM-PDCL, while it was not detected in the paired parental tumor (PARP14 mutation). As expected, silent mutations in both gene subsets were significantly less preserved compared with non-silent mutations (0/5 preserved events, P = .003, binomial test).
In order to further characterize the mutations that were different between parental tumors and paired PDCLs, we estimated allelic fraction of the mutations (ie, how many cells carry this mutation). As shown in Fig. 2C, mutations that appear in parental tumor only or PDCL only have lower allelic fraction compared with mutations that appear in both parental tumor and PDCL (P < 2.2e-16, chi-square test).
We used IntOGen functional prediction for each mutation to classify into putatively functional mutations versus nonfunctional mutations (Fig. 2C). Forty-six percent of mutations detected in both tumors and PDCLs were putatively functional versus 33% of the mutations detected in tumors or PDCL exclusively (P = 7.8E-8, chi-square test). For the subset of COSMIC genes, 65% of mutations detected in both tumors and PDCL were putatively functional versus 38% of the mutations detected in tumors or PDCL exclusively (P = .057). Finally, for GBM driver genes, 92% of mutations detected in both tumors and PDCLs were putatively functional versus 50% of the mutations detected in tumors or PDCLs exclusively (P = .06).
Interestingly, when considering only mutations with allelic fraction >10% and putatively functional biological impact, the median agreement between parental tumors and PDCL raised and reached 48%, 67%, and 79% for all genes, COSMIC genes, and GBM driver genes, respectively.
The subgroup of mutations that were detected in tumor only and not in PDCL are of special interest. Overall, a median of 19% of mutations belong to this group. These mutations have lower allelic fraction compared with mutations detected in both tissues (0.15 vs 0.32, P < 2.2e-16, chi-square test). Fewer of these mutations are predicted to have functional impact (38% vs 46%, P = .02, chi-square test). A minority (6%) of functional COSMIC gene mutations belong to this group (CAMTA1, SLC45A3, PTPRB, KDM6A) with low median allelic fraction of 0.11. Only one functional mutation in GBM genes (4%, NLRP5) belongs to this group with allelic fraction of 0.07.
Expression Profiling of mRNA
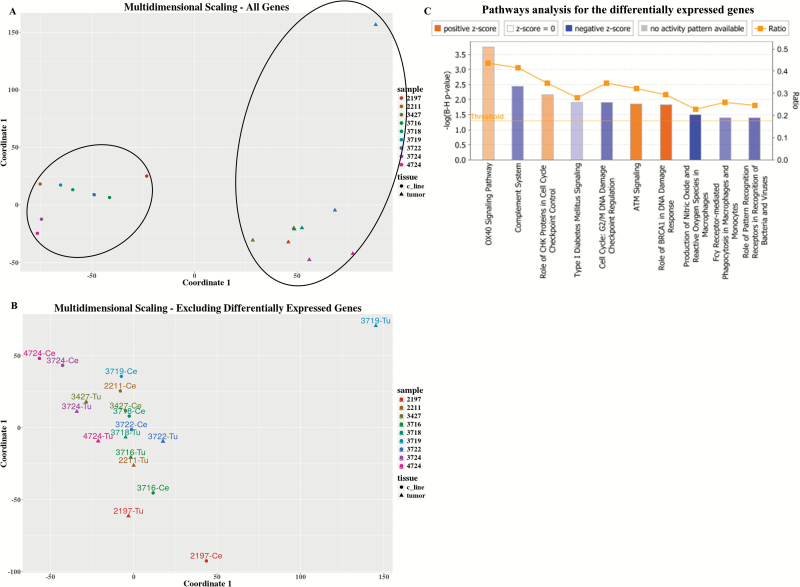
Transcription levels were measured for all tumor–PDCL pairs. One pair was excluded from the analysis due to low quality of tumor RNA. The GBM transcription subtype16 was maintained for 5/9 pairs (Supplementary Table 1). Multidimensional scaling (MDS) performed on all the measured genes resulted in distinct clusters: one for all the parental tumors and another for all the PDCLs (Fig. 3A).
Fig. 3.
Transcriptome (array data) landscape. (A). MDS for all measured genes. Each sample is denoted by a different color, tumors are marked as triangles and PDCLs as circles. (B) MDS for all genes excluding 2643 differentially expressed genes between the tumor and PDCL groups. (C) Ingenuity pathway analysis for the 2643 differentially expressed genes. Only biological pathways that are both significant and for which the activation/inhibition direction could be inferred are shown. Orange denotes pathway activation in PDCL compared with parental tumors and blue denoted pathway inhibition.
There were 2643 genes significantly differentially expressed between parental tumors and PDCLs. As shown in Fig. 3B, MDS performed after the exclusion of the differentially expressed genes resulted, as expected, in one unified cluster, but in addition the average distance between tumors and their paired PDCLs was shorter compared with the overall average pairwise distance (after exclusion of the outlier tumor 3719T, overall average Euclidean distance was 54 vs average tumor–PDCL pair distance of 34 on the MDS scale, P = .049, t-test).
In order to characterize the functional importance of this set of 2643 differentially expressed genes, pathway analysis was carried out. Fig. 3C and Supplementary Fig. 1 describe the pathways for which activation/inhibition state could be inferred. These pathways can be generally assigned as belonging to 3 groups: (i) immune pathways that were underexpressed in PDCL, such as “complement system”; (ii) cell cycle and DNA repair pathways that were activated, such as “ataxia telangiectasia mutated (ATM) signaling”; and (iii) a cell cycle and DNA repair pathway that was inhibited, such as “G2/M DNA damage checkpoint regulation.” Of note, the OX40 pathway is an immune pathway but surprisingly was measured as upregulated in PDCL. Detailed inspection of the genes’ group of this pathway shows that the classical immune genes (like human leukocyte antigen) were indeed strongly underexpressed, and several mitogen-activated protein kinase genes which are related to oncogenesis were overexpressed (Supplementary Table 2). Therefore, it seems that the immune part of the OX40 pathway is inhibited in GBM-PDCL in accordance with the other immune pathways.
The set of 2643 differentially expressed genes was analyzed for expression fingerprints identifying potential upstream regulators. Most of the top regulators were genes related to cancer, and their predicted activation seems pro-oncogenic in the PDCLs compared with the parental tumors (Supplementary Table 3). The most significant activation was for the fingerprint of the RABL6 gene (P = 3.7E-14), a Ras family–related protein (Supplementary Table 4). Only 2 upstream regulators were related to the immune system: (i) TGFB1 inhibition fingerprint (one of the major functions attributed to this gene in GBM is immunosuppression23) and (ii) the IL13 activation fingerprint, which showed a trend for activation. Of note were 2 fingerprints: (i) NURP1 (nuclear protein 1, a transcription factor activating the phosphatidylinositol-3 kinase/Akt pathway, an inhibition fingerprint in PDCL) and (ii) FBN1 (fibrillin-1, a structural glycoprotein related to extracellular matrix, an activation fingerprint in PDCL).
Pathway analysis was also carried out for the complete transcriptome (in contrast to the previously mentioned analysis for only the 2643 differentially expressed genes) using GSEA for the KEGG, Biocarta, and PID pathways sets (Supplementary Tables 5, 6). Many pathways involved in cell cycle regulation and DNA repair were shown to be upregulated in PDCL compared with parental tumors (eg, p53, BRCA, cell cycle, ATM). In addition, several metabolic pathways were overrepresented in PDCL compared with parental tumors (eg, pyruvate metabolism). Many immune-related pathways were underrepresented in PDCL compared with parental tumors (Fc gamma mediated phagocytosis and B-cell receptor signaling pathways). Thus, consistent with the pathway analysis noted above, several pathways involved in immunity were found enriched in parental tumors and several metabolic and cancer-related pathways were found enriched in PDCLs.
RNA Sequencing
RNA-seq analysis was performed for all parental tumor−PDCL pairs. Mean reads count was 197×106X±33. One sample was excluded from the analysis due to being an extreme outlier on principal component analysis. RNA-seq analysis aligned reads for 56638 Ensembl genes. Significantly differentially expressed were 15046 of the genes between tumors and PDCLs. Of the 2643 genes identified by the expression array analysis, 2005 (76%) were also identified here as differentially expressed. IPA analysis was also performed for the most significant differentially expressed genes (P < .001, FDR; n=6606 genes) (Supplementary Figs. 3–4). Comparable to the pathway analysis of expression array data: (i) several immune pathways were inhibited in PDCLs versus parental tumors, (ii) several pathways associated with cancer were activated in PDCLs versus parental tumors, and (iii) several cholesterol metabolic pathways were significantly differentially modulated in PDCLs versus parental tumors. Many pathways were similar between the 2 pathway analyses, specifically: (i) “dendritic cell maturation,” (ii) “complement maturation,” (iii) “role of BRCA1 in DNA damage response,” (iv) “ATM signaling,” and (v) several cholesterol metabolic pathways. Strikingly, upstream regulator analysis, conducted with RNA-seq data, revealed a list comparable to the one obtained with the expression profiling array data (Supplementary Table 7).
Fusion analysis was carried out for the RNA-seq data. No known GBM or cancer-related fusion12,24 was identified.
Clonal Analysis
Clonal analysis was carried out by Absolute algorithm21 for the 7 samples with available blood DNA whole exome sequencing data. A mutation that is estimated to be carried by >90% of cancer cells was defined as clonal, whereas the others were defined as subclonal. There were significantly more clonal mutations in tumors (65% clonal and 35% subclonal) and more subclonal mutations in PDCLs (56% clonal and 44% subclonal) for the complete mutation lists (P = 3.7E-5, chi-square test). Although not statistically significant, this trend was maintained for COSMIC and GBM driver gene mutation subgroups. Clonal status was not significantly different between protein changing mutations (eg, missense or nonsense mutations) and silent mutations.
The point mutation profiles of GBM driver genes (including intronic and silent mutations) are given in Table 2. Fifteen mutations were detected only in one tissue type (tumor or PDCL): (i) 6/15 were clonal, (ii) 8/15 were subclonal, and (iii) 1/15 was not classified by the algorithm. For the mutations that were detected in both tumors and PDCLs: (i) the predicted impact of the mutations is higher (P = .04, chi-square test), (ii) the cancer cell fraction was >90% for 11/12 mutations, and (iii) in the mutations for which clonal status could be formally tested by the algorithm, they were clonal in the tumor (5/5) but not necessarily in the PDCL.
Table 2.
Clonal profile for point mutations in GBM genes. All mutations in GBM driver genes (including intronic) are presented.
| Gene | Sample ID | Tissue | Formal Clone Status | Cell Fraction | Functional Impact |
|---|---|---|---|---|---|
| NLRP5 | 3724T | Tumor | Subclonal | 0.18 | Medium |
| DRD5 | 3427T | Tumor | Subclonal | 0.22 | None |
| DRD5 | 7015 | PDCL | Clonal | 0.83 | None |
| DRD5 | 7015 | PDCL | Subclonal | 0.73 | None |
| DRD5 | 7015 | PDCL | Subclonal | 0.27 | None |
| ABCC9 | 7142 | PDCL | Clonal | 1 | None |
| DRD5 | 7015 | PDCL | Subclonal | 0.73; 0.27 | None |
| LZTR1 | 7015 | PDCL | Clonal | 1 | None |
| PIK3R1 | N13-1520 | PDCL | Clonal | 1 | None |
| RB1 | 7015 | PDCL | Subclonal | 0.38 | Medium |
| SPTA1 | 7097 | PDCL | Clonal | 1 | Medium |
| TCHH | 7015 | PDCL | Subclonal | 0.21 | High |
| TCHH | 7015 | PDCL | Subclonal | 0.24 | Low |
| TP53 | 7015 | PDCL | Clonal | 1 | High |
| TP53 | N13-1520 | PDCL | – | 1 | High |
| ATRX | 3719T-7015 | Both | Clonal → clonal | 1 → 1 | High |
| EGFR | 3427T-6240 | Both | – | 1 → 1 | Medium |
| EGFR | 3722T-7060 | Both | Clonal → clonal | 1 → 1 | Low |
| GABRA6 | 4724T-N13-1520 | Both | – | 0.91 → 1 | Low |
| KEL | 3719T-7015 | Both | Clonal → subclonal | 1 → 0.64 | High |
| NF1 | 3719T-7014 | Both | – | 1 → 1 | High |
| NF1 | 4724T-N13-1520 | Both | – | 0.68 → 1 | Medium |
| PIK3CA | 3427T-6240 | Both | Clonal → subclonal | 1 → 0.82 | None |
| PIK3CA | 3719T-7014 | Both | – | 1 → 1 | Medium |
| PTEN | 3523T-7097 | Both | Clonal → clonal | 1 →1 | High |
| PTEN | 3724T-7142 | Both | – | 1 →1 | High |
| SPTA1 | 3427T-6240 | Both | – | 0.94 →0.84 | Medium |
Discussion
The use of PDCLs as preclinical models of GBM to investigate potential response to novel drugs is built on the assumption that major aspects of human tumor biology are recapitulated in the PDCLs. Hence, it seems important to examine to which extent indeed GBM-PDCL libraries represent the molecular landscape of human tumors.
This question was also addressed measuring multi-omic genomic profiles. In the original report of the Cancer Cell Line Encyclopedia (CCLE), the authors measured the genomic similarity of the reported CCL to published unpaired primary tumors of similar cancer types and concluded that with relatively few exceptions, the CCLE may provide representative molecular proxies for primary tumors in many cancer types.6 By contrast, Domcke et al compared the molecular landscapes of 47 CCLs of high-grade serous ovarian carcinoma from the CCLE and 316 tumor profiles from The Cancer Genome Atlas. They reported pronounced differences between commonly used ovarian CCLs and unpaired high-grade serous ovarian tumor samples. They identified several rarely used CCLs that more closely resemble cognate tumor profiles.25 Lee et al showed, based on expression profiles, that GBM cell lines were clustered remotely from their parental tumors compared with tumor stem cells.26 Nevertheless, these studies compared genomic landscapes of CCLs and unpaired human tumors. A major limitation of this approach is the inability to directly measure and quantitate the molecular change between primary tumors and CCLs due to the different biases of sample selection.
A recent study compared the molecular landscape of GBM-PDCLs and their paired parental tumors for CNV and expression landscape (see below).27 This study, however, did not quantify the focal CNV events agreement. Moreover, point mutations were not measured and transcriptome information was measured solely by expression array.
In the current study, paired GBM and PDCL samples' molecular characteristics were measured for CNV, point mutations, and transcriptome. There was generally good agreement between tumors and PDCLs for measures of gain/loss. CN-LOH agreements were low and could possibly represent real differences between tumors and PDCLs, or could be explained by algorithmic inaccuracies for CN-LOH. Importantly, GBM-specific chromosome gains and losses were comparable between parental tumors and PDCLs. For focal events: 10/15 characteristic GBM homozygous deletions and high-level amplifications were maintained. Of note, there were 2 CDKN2A homozygous deletions found in PDCL while not being detected in the paired parental tumor. Overall, CNV profiles are well maintained in PDCLs, but caution is advised for focal events which might be lost or gained in the transition from tumor to PDCL.
The agreement between tumors and their paired PDCLs for point mutations seems to depend on 3 major factors: (i) allelic fraction of the mutation: mutations that were maintained in tumors and their paired PDCLs were of higher allelic fraction (a plausible explanation for this is that low allelic fraction mutations have a higher probability to be lost in the passage from tumor to PDCL and are more difficult to detect due to sequencing and algorithmic reduced sensitivity for such mutations9); (ii) functional impact of a mutation: mutations that were maintained in tumors and their paired PDCLs were more frequently of functional impact; and (iii) relevance of genes in the context of GBM biology. Combining these 3 considerations, the median agreement between tumors and paired PDCLs was raised above the general median agreement of 41% and reached 67% and 79% for COSMIC genes and GBM driver genes, respectively. In the same manner, a minority of functional mutations were detected in tumors only for COSMIC and GBM genes and most have low allelic fraction. Of note, TP53 mutations of 2 PDCLs were not detected in their parental tumors. They might have existed in very low allelic fractions (Fig. 2D) in the tumors and positively selected in PDCLs. These PDCLs showed the lowest similarity with their parental tumors, and this possibly reflects that TP53 mutations allow acquisition of additional mutations in PDCLs.
The transcriptome analysis showed that GBM expression subtypes were maintained in 5/9 of the paired PDCLs. One putative reason for disagreement is that subtypes were defined for GBM tumors rather than in CCLs. The former may include transcription patterns typical of microenvironment cells and of interaction between tumor cells and the microenvironment.28 Indeed, Verhaak et al16 reported that the identification of corresponding CCL subtypes is not easily achievable. In addition, these differences can be attributed to intratumoral subtype heterogeneity.29,30 Tumors are distinct from PDCLs and usually cluster in 2 different groups. After exclusion of the differentially expressed genes, tumors clustered closely to their paired PDCLs. Accordingly, it seems that there is a strong expression pattern specific for tumors versus PDCLs; in addition, PDCLs maintain expression similarity of their parental tumors. The differences included the change of cell cycle regulators, underexpression of immune-related pathways, and change in lipid metabolism pathways in PDCLs. Upstream regulator analysis for the differentially expressed genes mostly revealed fingerprints associated with cancer-related genes. The cell cycle and DNA repair pathway changes seem to reflect enhanced proliferation of the PDCL. The underexpression of the immune pathways in PDCL seems to reflect the absence of immune cells in the PDCL microenvironment. Lipid metabolism changes possibly result from the radical change of metabolism of PDCLs in culture cell conditions. It seems, though, that the changes of cancer-related pathways in PDCLs compared with tumors may have significant influence on experiments performed on PDCLs. For example, response to cyclin dependent kinase inhibitors in PDCLs may not predict corresponding response in parental tumors, since cell cycle pathways are overexpressed in PDCLs. Further research is required in order to assess the importance of these changes. Analysis performed for RNA-seq data revealed comparable differences between PDCLs and tumors, and the confirmation of these results by an independent analysis method strengthens the above-mentioned conclusions.
Clonal analysis of the point mutations shows that there are more clonal mutations in tumors and more subclonal mutations in PDCLs. Clonal mutations in GBM driver genes were better maintained in tumors and PDCLs, and this supports the suitability of PDCLs as a model for GBM.
Our results are in agreement with the conclusion of Davis et al27 regarding the extent of recapitulation of major chromosomal CNV events and EGFR amplification. However, the general level of agreement of CNV between tumors and PDCLs seems higher in our dataset (median agreement of 85% versus correlation of 0.5). Moreover, our results show that there is good preservation of LOH but not of copy-neutral LOH. In terms of expression, Davis et al identified 63 differentially expressed genes that involve metabolic pathways. By contrast, our study revealed 2643 genes that are differentially expressed between tumors and PDCLs, inhibition of immune pathways, and change of several cell cycle and DNA repair pathways. In addition, changes in cancer-related master regulators expression fingerprints, specific for the PDCL population, were identified.
The results presented in the current report demonstrate relatively good agreement between tumors and their paired PDCLs and support their use as preclinical GBM models. However, there are some discrepancies between tumors and PDCLs which might represent real biological differences. We hypothesized several reasons for the discrepancies: (i) tumor heterogeneity which is well established for many tumors31 and specifically for GBM,29 (ii) different selective pressures during PDCL culturing in vitro compared with the tumor biological environment, and (iii) the microenvironment (ie, absence of immune cells) of PDCL culture conditions differ markedly from that of their paired parental tumors.21
Although the molecular analysis was comprehensive and included several omics modalities, the sample size is relatively small and hence a larger set of paired parental tumors–PDCLs would be necessary to confirm our findings and to provide additional insights.
To conclude, the current study supports the use of GBM-PDCL as a human GBM model. Indeed, the majority of functional tumor molecular alterations detected in the parental tumor are maintained in PDCL. Further analysis of molecular discrepancies between tumors and PDCLs may provide insights into GBM biology.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This work is part of the GlioTex (Glioblastoma and Experimental Therapeutics) project funded by the Foundation ARC pour la Recherche sur le Cancer; The Institut Universitaire de Cancérologie (IUC); OncoNeuroThèque; Hadassah France.
Conflict of interest statement. The authors disclose no potential conflicts of interest.
Supplementary Material
References
- 1. Prados MD, Byron SA, Tran NL, et al. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro Oncol. 2015;17(8):1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kwak EL, Bang Y-J, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358(11):1160–1174. [DOI] [PubMed] [Google Scholar]
- 4. Ciriello G, Miller ML, Aksoy BA, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45(10):1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lazar V, Rubin E, Depil S, et al. A simplified interventional mapping system (SIMS) for the selection of combinations of targeted treatments in non-small cell lung cancer. Oncotarget. 2015;6(16):14139–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li A, Liu Z, Lezon-Geyda K, et al. GPHMM: an integrated hidden Markov model for identification of copy number alteration and loss of heterozygosity in complex tumor samples using whole genome SNP arrays. Nucleic Acids Res. 2011;39(12):4928–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramos AH, Lichtenstein L, Gupta M, et al. Oncotator: cancer variant annotation tool. Hum. Mutat. 2015;36(4):E2423–E2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods. 2013;10(11):1081–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. My Cancer Genome 2013 http://www.mycancergenome.org Accessed May 21, 2015.
- 15. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reich M, Liefeld T, Gould J, et al. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. [DOI] [PubMed] [Google Scholar]
- 18. IPA, Qiagen Redwood City www.qiagen.com/ingenuity Accessed February 2, 2016.
- 19. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liberzon A. A description of the Molecular Signatures Database (MSigDB) Web site. Methods Mol. Biol. 2014;1150:153–160. [DOI] [PubMed] [Google Scholar]
- 21. Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olshen AB, Venkatraman ES, Lucito R, et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. [DOI] [PubMed] [Google Scholar]
- 23. Joseph JV, Balasubramaniyan V, Walenkamp A, et al. TGF-beta as a therapeutic target in high grade gliomas - promises and challenges. Biocheml Pharmacol. 2013;85(4):478–485. [DOI] [PubMed] [Google Scholar]
- 24. TCGA Fusion Gene Data Portal. 54.84.12.177/PanCanFusV2/ Accessed December 5, 2015.
- 25. Domcke S, Sinha R, Levine DA, et al. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 27. Davis B, Shen Y, Poon CC, et al. Comparative genomic and genetic analysis of glioblastoma-derived brain tumor-initiating cells and their parent tumors. Neuro Oncol. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Orr BA, Eberhart CG. Nature versus nurture in glioblastoma: microenvironment and genetics can both drive mesenchymal transcriptional signature. Am J Pathol. 2012;180(5):1768–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xie Y, Bergstrom T, Jiang Y, et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine. 2015;2(10):1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alizadeh AA, Aranda V, Bardelli A, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21(8):846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.