Abstract
The three-dimensional structure of bovine lens leucine aminopeptidase (EC 3.4.11.1) complexed with bestatin, a slow-binding inhibitor, has been solved to 3.0-A resolution by the multiple isomorphous replacement method with phase combination and density modification. In addition, the structure of the isomorphous native enzyme has been refined at 2.7-A resolution, and the current crystallographic R factor is 0.169 for a model that includes the two zinc ions and all 487 amino acid residues comprising the asymmetric unit. The enzyme is physiologically active as a hexamer, which has 32 symmetry and is triangular in shape with a triangle edge length of 115 A and maximal thickness of 90 A. The monomers are crystallographically equivalent and each is folded into two unequal alpha/beta domains connected by an alpha-helix to give a comma-like shape with approximate maximal dimensions of 90 x 55 x 55 A3. The secondary structural composition is 40% alpha-helix and 19% beta-strand. The N-terminal domain (160 amino acids) mediates trimer-trimer interactions and does not appear to participate directly in catalysis. The C-terminal domain (327 amino acids) is responsible for catalysis and binds the two zinc ions, which are 2.88 A apart. The pair of metal ions is located near the edge of an eight-stranded, saddle-shaped beta-sheet. One zinc ion is coordinated by carboxylate oxygen atoms of Asp-255, Asp-332, and Glu-334 and the carbonyl oxygen of Asp-332. The other zinc ion is coordinated by the carboxylate oxygen atoms of Asp-255, Asp-273, and Glu-334. The active site also contains two positively charged residues, Lys-250 and Arg-336. The six active sites are themselves located in the interior of the hexamer, where they line a disk-shaped cavity of radius 15 A and thickness 10 A. Access to this cavity is provided by solvent channels that run along the twofold symmetry axes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen M. P., Yamada A. H., Carpenter F. H. Kinetic parameters of metal-substituted leucine aminopeptidase from bovine lens. Biochemistry. 1983 Aug 2;22(16):3778–3783. doi: 10.1021/bi00285a010. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Carpenter F. H., Harrington K. T. Intermolecular cross-linking of monomeric proteins and cross-linking of oligomeric proteins as a probe of quaternary structure. Application to leucine aminopeptidase (bovine lens). J Biol Chem. 1972 Sep 10;247(17):5580–5586. [PubMed] [Google Scholar]
- Carpenter F. H., Vahl J. M. Leucine aminopeptidase (Bovine lens). Mechanism of activation by Mg 2+ and Mn 2+ of the zinc metalloenzyme, amino acid composition, and sulfhydryl content. J Biol Chem. 1973 Jan 10;248(1):294–304. [PubMed] [Google Scholar]
- Cuypers H. T., van Loon-Klaassen L. A., Egberts W. T., de Jong W. W., Bloemendal H. The primary structure of leucine aminopeptidase from bovine eye lens. J Biol Chem. 1982 Jun 25;257(12):7077–7085. [PubMed] [Google Scholar]
- Durbin R. M., Burns R., Moulai J., Metcalf P., Freymann D., Blum M., Anderson J. E., Harrison S. C., Wiley D. C. Protein, DNA, and virus crystallography with a focused imaging proportional counter. Science. 1986 May 30;232(4754):1127–1132. doi: 10.1126/science.3704639. [DOI] [PubMed] [Google Scholar]
- Harrison S. C., Winkler F. K., Schutt C. E., Durbin R. M. Diffraction methods for biological macromolecules. Oscillation method with large unit cells. Methods Enzymol. 1985;114:211–237. doi: 10.1016/0076-6879(85)14021-8. [DOI] [PubMed] [Google Scholar]
- Henson H., Frohne M. Crystalline leucine aminopeptidase from lens (alpha-aminoacyl-peptide hydrolase; EC 3.4.11.1). Methods Enzymol. 1976;45:504–520. doi: 10.1016/s0076-6879(76)45045-0. [DOI] [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurnak F., Rich A., van Loon-Klaassen A., Bloemendal H., Taylor A., Carpenter F. H. Preliminary X-ray study of leucine aminopeptidase (bpvine lens), and oligomeric metalloenzyme. J Mol Biol. 1977 May 5;112(1):149–153. doi: 10.1016/s0022-2836(77)80162-9. [DOI] [PubMed] [Google Scholar]
- Kantrowitz E. R., Lipscomb W. N. Escherichia coli aspartate transcarbamylase: the relation between structure and function. Science. 1988 Aug 5;241(4866):669–674. doi: 10.1126/science.3041592. [DOI] [PubMed] [Google Scholar]
- Kiselev N. A., Stel'mashchuk V. Y., Tsuprun V. L., Ludewig M., Hanson H. Electron microscopy of leucine aminopeptidase. J Mol Biol. 1977 Sep;115(1):33–43. doi: 10.1016/0022-2836(77)90244-3. [DOI] [PubMed] [Google Scholar]
- Phillips W. C., Rayment I. A systematic method for aligning double-focusing mirrors. Methods Enzymol. 1985;114:316–329. doi: 10.1016/0076-6879(85)14025-5. [DOI] [PubMed] [Google Scholar]
- Ramachandran G. N., Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Remington S., Wiegand G., Huber R. Crystallographic refinement and atomic models of two different forms of citrate synthase at 2.7 and 1.7 A resolution. J Mol Biol. 1982 Jun 15;158(1):111–152. doi: 10.1016/0022-2836(82)90452-1. [DOI] [PubMed] [Google Scholar]
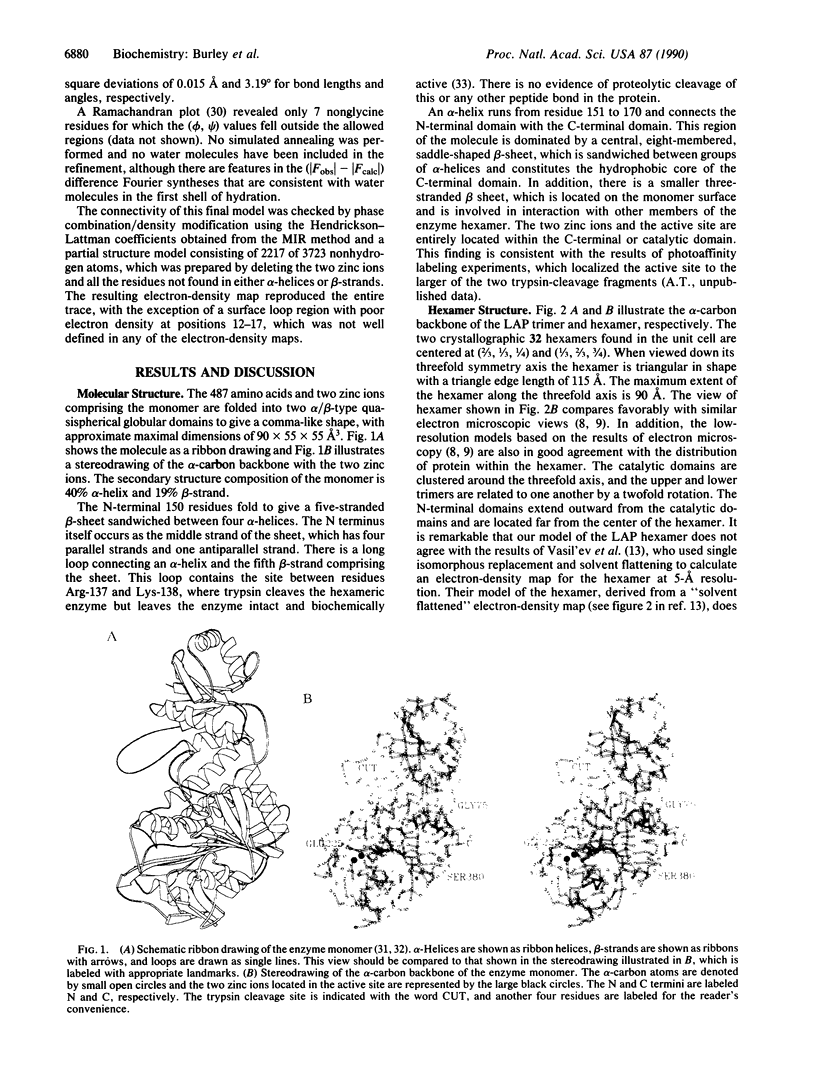
- Richardson J. S. Schematic drawings of protein structures. Methods Enzymol. 1985;115:359–380. doi: 10.1016/0076-6879(85)15026-3. [DOI] [PubMed] [Google Scholar]
- Robbins A. H., Stout C. D. The structure of aconitase. Proteins. 1989;5(4):289–312. doi: 10.1002/prot.340050406. [DOI] [PubMed] [Google Scholar]
- Roderick S. L., Matthews B. W. Crystallization of methionine aminopeptidase from Escherichia coli. J Biol Chem. 1988 Nov 15;263(32):16531–16531. [PubMed] [Google Scholar]
- Sheriff S., Hendrickson W. A., Smith J. L. Structure of myohemerythrin in the azidomet state at 1.7/1.3 A resolution. J Mol Biol. 1987 Sep 20;197(2):273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]
- Taylor A., Carpenter F. H., Wlodawer A. Leucine aminopeptidase (bovine lens): an electron microscopic study. J Ultrastruct Res. 1979 Jul;68(1):92–100. doi: 10.1016/s0022-5320(79)90145-x. [DOI] [PubMed] [Google Scholar]
- Taylor A., Volz K. W., Lipscomb W. N., Takemoto L. J. Leucine aminopeptidase from bovine lens and hog kidney. Comparison using immunological techniques, electron microscopy, and X-ray diffraction. J Biol Chem. 1984 Dec 10;259(23):14757–14761. [PubMed] [Google Scholar]
- Umezawa H., Aoyagi T., Suda H., Hamada M., Takeuchi T. Bestatin, an inhibitor of aminopeptidase B, produced by actinomycetes. J Antibiot (Tokyo) 1976 Jan;29(1):97–99. doi: 10.7164/antibiotics.29.97. [DOI] [PubMed] [Google Scholar]
- Van Loon-Klaassen L., Cuypers H. T., Bloemendal H. Limited tryptic digestion of bovine eye lens leucine aminopeptidase. FEBS Lett. 1979 Nov 15;107(2):366–370. doi: 10.1016/0014-5793(79)80409-3. [DOI] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]