Abstract
Pneumocystis jirovecii is an unusual ascomycetous fungus that can be detected in the lungs of healthy individuals. Transmission from human to human is one of its main characteristics in comparison with other fungi responsible for invasive infections. P. jirovecii is transmitted through the air between healthy individuals, who are considered to be the natural reservoir, at least transiently. In immunocompromised patients, P. jirovecii multiplies, leading to subacute infections and acute life-threatening pneumonia, called Pneumocystis pneumonia [PCP]. PCP is caused by genotypically distinct mixtures of organisms in more than 90% of cases, reinforcing the hypothesis that there is constant inhalation of P. jirovecii from different contacts over time, although reactivation of latent organisms from previous exposures may be possible. Detection of P. jirovecii DNA without any symptoms or related radiological signs has been called “colonization”. This situation could be considered as the result of recent exposure to P. jirovecii that could evolve towards PCP, raising the issue of cotrimoxazole prophylaxis for at-risk quantitative polymerase chain reaction (qPCR)-positive immunocompromised patients. The more accurate way to diagnose PCP is the use of real-time quantitative PCR, which prevents amplicon contamination and allows determination of the fungal load that is mandatory to interpret the qPCR results and manage the patient appropriately. The detection of P. jirovecii in respiratory samples of immunocompromised patients should be considered for potential risk of developing PCP. Many challenges still need to be addressed, including a better description of transmission, characterization of organisms present at low level, and prevention of environmental exposure during immunodepression.
Keywords: Pneumocystis jirovecii, Pneumocystis pneumonia, PCP, fungal infections
An obligate organism of humans with a parasitic behavior
Pneumocystis species are fungal organisms that belong to the ascomycetous fungi based on phylogenetic studies on ribosomal 1, 2 and mitochondrial 3 DNA. These initial findings have been recently confirmed based on whole genome analysis 4, 5. Its closest phylogenetic organism is the plant pathogen Taphrina deformans 6. Pneumocystis spp. have been found in the lungs of most terrestrial mammals, and are considered to be specific to their host 7. Five species of Pneumocystis associated with specific hosts have been formally described so far: Pneumocystis murina in mice, Pneumocystis carinii and Pneumocystis wakefieldae in rats, Pneumocystis oryctolagi in rabbits, and Pneumocystis jirovecii in humans 7. It is important to note that most of the biological aspects discussed for P. jirovecii have actually been inferred from studies in other Pneumocystis species. Pneumocystis thrives at the surface of type I pneumocytes in the alveolar space 8 and is found as two main life cycle stages: the trophic form and the asci form (previously called “cysts”). Trophic forms represent the large majority of the whole Pneumocystis population and are thought to multiply through asexual binary fission 8, 9. Asci are presumably formed through the conjugation of two opposite mating type trophic forms via sexual reproduction, allowing the production of eight ascospore after maturation 10.
Three genomes of Pneumocystis spp. are available, with that of P. jirovecii recently released as 356 contigs 4. This makes comparative genomics studies possible, which are useful to understand how Pneumocystis evolved. One hallmark of Pneumocystis genomes is the lack of enzyme involved in chitin synthesis or degradation 5, although a cell wall is observed in asci, suggesting that it may not contain chitin, which seems rare in the fungal kingdom 11. In contrast, beta-glucan is present in the cell wall of asci as in other fungi 5. In addition, Pneumocystis seems to have lost thousands of genes involved in the biosynthesis of amino acids, lipids, thiamine, coenzyme A, myo-inositol, and biotin, assimilation of inorganic nitrogen and sulfur, and degradation of purines compared to Taphrina deformans 5, 12, 13. In contrast, proteases, RNA-processing proteins, and more specifically major surface glycoproteins (MSGs), unique to this genus, are expanded in the genome compared to other related fungal species 5. Interestingly, variation of the MSGs is known to induce changes in the surface antigens involved in the interface with host cells 14, which may facilitate evasion of the host immune response 15.
In total, these characteristics support the notion that P. jirovecii is a fungus that infects humans and exhibits parasitic behavior, meaning that P. jirovecii needs host nutrients to live and proliferate in the human respiratory tract. This hypothesis is reinforced by the current difficulties in cultivating Pneumocystis in vitro. Though low rates of multiplication of Pneumocystis have been reported using host cell and cell-free systems 16– 19, there is no in vitro culture that sustains the continuous growth of any Pneumocystis species.
Pathophysiology of Pneumocystis jirovecii pneumonia
P. jirovecii is responsible for a form of acute lung injury called PCP, which occurs mainly in immunocompromised patients, including those with HIV, and in non-HIV immunocompromised patients, such as those who have undergone solid organ transplantation, have hematological malignancies, or are receiving high-dose steroids 20, 21. The first description of PCP was described in cohorts of malnourished or premature infants by Vanek and Jirovec in the 1950’s 22, 23, initially establishing it mainly as a pediatric infection, which is not the case anymore. Overall, the pathology of Pneumocystis has been studied in animal models with animal-specific Pneumocystis species, with many findings secondarily translated into human pathology 24. In humans, genotyping of P. jirovecii is the main tool used to assess and study the epidemiology and the natural history of P. jirovecii infection with a specific focus on transmission 25.
Transmission
The hypothesis of an environmental reservoir of Pneumocystis has been investigated and is considered as very unlikely 24, 26. On the contrary, the transmission of Pneumocystis has long been understood to be between individuals via air from experiments performed in immunocompromised rats or nude mice 27, 28 with Pneumocystis DNA detected in the surrounding air of infected immunosuppressed or immunocompetent animals 29– 31. The same evidence has also been found in humans with the observation of P. jirovecii DNA detection in the surrounding air of patients or healthcare workers harboring P. jirovecii in their respiratory tract 32– 35, making airborne transmission the most likely route of acquisition in humans.
The airborne transmission of P. murina probably occurs through acquisition of the asci form of these fungi. Indeed, echinocandin-treated mice failed to transmit the disease because echinocandin treatment had decreased the number of cysts 36. A 12-hour contact was sufficient to allow transmission from rats infected with asci but not those infected only with trophic forms 37. Moreover, immunocompetent mice exposed to Pneumocystis-positive immunocompromised mice harbor and carry Pneumocystis after multiplication, although few respiratory symptoms (mild or asymptomatic infection) could be observed 38, 39. In addition, the Pneumocystis-exposed immunocompetent mice are able to transmit the disease to Pneumocystis-naïve immunocompetent mice 38, 39. Immunocompetent mice were able to resolve infection within 5 to 6 weeks after mild symptoms 40. This has also been observed in immunocompetent rabbits, with clearance of P. oryctolagi in about 90 days with an increase of anti- Pneumocystis IgG from day 21 to day 90 41. Together, these observations support the contention that immunocompetent hosts are likely to be a source of infection within 2 to 3 months after inoculation. Data obtained in humans corroborate this hypothesis with detection and modification of antibody titers over time of specific anti -P. jirovecii antibodies in healthy individuals 15, 42. Indeed, a serological survey of anti- P. jirovecii antibodies in 107 healthy infants showed a very rapid and early exposure to P. jirovecii over time with a maximum seroconversion rate (about 90%) at 18 months of life 43. Of note, P. jirovecii DNA had been detected in parallel in the respiratory tract of some of these infants as well 43. In addition, P. jirovecii has been found to be highly prevalent in immunocompetent individuals in a cohort of infants with sudden unexpected death 44 or adults who died from violent causes based on lung tissue sample analysis 45. Detection of P. jirovecii alone in immunocompetent infants with bronchiolitis supports the possibility that P. jirovecii could be associated with spontaneously resolved symptoms in infants 46. An increased incidence of P. jirovecii detection has been observed in pregnant women known to be physiologically immunosuppressed to some extent. This could reflect an increased exposure by older children within the family or a decreased immune control of P. jirovecii. Thus, pregnant women could be an important part of the reservoir of P. jirovecii 47.
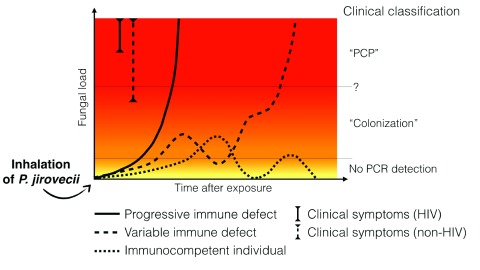
These facts support the hypothesis that immunocompromised infants or adults should be a reservoir of P. jirovecii and that immunocompetent individuals should also be, at least more transiently, a reservoir of this organism. Thus, immunocompetent individuals should serve as sources of constant circulation of the fungal organism in the population, with the risk of transmission to potentially immunocompromised hosts ( Figure 1).
Figure 1. The variable evolution of Pneumocystis fungal load after exposure depending on the host’s immune status.
Upon exposure, P. jirovecii multiplies at the surface of type I pneumocytes in the alveoli. At the very beginning of the infection, quantitative polymerase chain reaction (qPCR) may not be positive owing to very low or localized multiplication of P. jirovecii, even if a bronchoalveolar lavage (the most sensitive specimen) is performed. Depending on the host’s immune status, the evolution of the fungal load could differ: - Rapid and constant multiplication of P. jirovecii leading to Pneumocystis pneumonia (PCP) is typically observed in HIV patients or in patients under immunosuppressive regimens (regular line). The situation known as “PCP” is observed when a high fungal load is associated with symptoms. Note that symptoms appear for lower fungal loads in non-HIV patients compared to HIV patients. - Variable fungal load above or below the detection’s threshold is observed in patients with variable immune status with PCP occurring when immunosuppression increases. This situation is typically seen in hematology or cancer patients submitted to several courses of chemotherapy (dashed line). PCR detection in asymptomatic patients in the situation known as “colonization” could correspond to patients controlling the disease (decreasing fungal load) or to patients who will develop PCP in the near future. The threshold between PCP and “colonization” may be different in HIV-positive and HIV-negative patients. - Immunocompetent patients are potentially a reservoir of P. jirovecii with low multiplication rate due to rapid immune control and subsequent decrease of fungal load. Re-infection with a new genotype or reactivation could occur later on (dotted line).
Latent or recently acquired infection?
Regarding the hypothesis of constant airborne circulation of P. jirovecii and the early exposure in childhood, it is licit to hypothesize that PCP could be due to (i) the reactivation of organisms acquired in childhood, as demonstrated for other fungal diseases 48, 49, or (ii) the multiplication of recently acquired organisms. Reactivation of latent infection had been investigated in animal models and in humans. Animal studies in the lab suggested that reactivation occurred rarely after PCP (<25% of animals after 1 year) 50, 51. In humans, one study of recurrent pneumocystosis after treatment suggested new acquisition of P. jirovecii at the second episode 52. In this study, only five out of 10 patients harbored a different genetic pattern, suggesting that both newly acquired infection and reactivation may occur 52. However, this study used a genotyping method that is not sufficiently accurate to provide a definite answer to this question because it focused only on the diversity of one genetic marker (sequence of the mitochondrial large subunit ribosomal RNA gene) with variation of the sequence at only two positions.
Strong evidence for continuous acquisition of P. jirovecii comes from genotyping studies of PCP cases. Indeed, when PCP occurs, it has been shown that about 70% of the cases were composed of mixtures of organisms 53– 55. This proportion increased to more than 90% of cases if highly sensitive methods such as ultra-deep pyrosequencing were used 56. However, these mixtures can be the consequence of iterative inhalation of different genotypes over time or of a single exposure to different genotypes. Microevolution of P. jirovecii in a single host cannot be excluded.
The major evidence in favor of recent acquisition of P. jirovecii is the numerous description of outbreaks or clustered PCP cases in different settings including kidney transplant units 57, liver transplant units 58, pediatric oncology wards 59, hematology wards 60– 62, and wards of other medical specialties 55, 63. The detection of P. jirovecii DNA in the air surrounding individuals with or without active PCP and genotyping bridges the gap between the human source and the acquisition of a specific genotype from other individuals in a specific period and at a given location 32– 34. Transmission could also occur between healthy individuals such as healthcare workers and immunocompromised patients, as already suggested 32, 35, 64. Indeed, healthcare workers would be able to transmit P. jirovecii for quite a long time, since P. jirovecii was detected in specific individuals for up to 10 weeks 64. In immunocompromised patients, timing between exposure and disease (incubation time) has not been clearly defined. Therefore, it remains unanswered how long before PCP a possible transmission in the hospital setting may take place. Recently, transmission of a specific genotype in renal transplant recipients, hematology and cancer patients of a specific hospital have been studied over a period of 4 years. The median time between suspected exposure and PCP was 197 days (interquartile range: 57–342.5), suggesting that the incubation time of PCP is variable and can be as long as 3 months, even in hospitalized and immunocompromised individuals 55. However, in this study, patients harboring the outbreak genotype were more likely to be infected with a single genotype and not a mixture of genotypes 55. This suggests a recent acquisition and proliferation of this particular P. jirovecii genotype responsible for the outbreak in potentially P. jirovecii-naïve patients. In addition, genotypes found during PCP were more related to the place of PCP diagnosis rather than to the place of birth, reinforcing the hypothesis of temporarily related P. jirovecii acquisition in hospital settings 65– 67.
Therefore, it is conceivable that a P. jirovecii genotype acquired in the past could shape the host’s immune response. Indeed, immunity raised memory against the specific surface antigen of this specific genotype, resulting in equilibrium between host response and P. jirovecii multiplication. At this point, when other genotypes harboring other surface antigens are acquired, as already suggested 14, 15, 68, 69, this new organism could proliferate as long as the immune system has not raised an appropriate response to these new surface antigens.
Importantly, limitation of genotyping methods in samples harboring low fungal load, or the impossibility of following the natural history of PCP from P. jirovecii acquisition in childhood to PCP during immunosuppression, still prevents the acquisition of proof that reactivation of latent infection may occur in humans. However, it is likely that recent exposure to one or multiple P. jirovecii genotypes is a major route of infection in immunocompromised individuals. It cannot be excluded that both mechanisms could occur simultaneously: reactivation of P. jirovecii from old exposure and multiplication from recent P. jirovecii exposure. It is also still unknown if specific genotypes are more prone to give infection or more prone to proliferate compared to others or if some genotypes are more specific to a given host’s background.
The concept of “colonization”
The concept of “colonization” or “carriage” of P. jirovecii has been introduced as early as the point at which P. jirovecii DNA is detected in patients without any symptoms, although at risk for PCP 70. This concept has been extensively reviewed 71. HIV infection, malignancies, or solid organ transplantation or immunosuppression secondary to immunosuppressive drugs including steroids increase the prevalence of P. jirovecii detection in asymptomatic patients. However, the prevalence of this “colonization” is very variable from one study to another because of either the PCR method used or the patient cohorts studied having different levels of immunosuppression 71. P. jirovecii DNA has also been detected in non-immunosuppressed patients, such as those with chronic lung diseases, cigarette smokers, or pregnant women whose immunity could be considered as abnormal 71. Most of the authors assume that “colonization” is a specific nosological category, which has been brought into opposition with PCP 71. In contrast, one may consider that “colonized” patients who are either immunocompetent or immunocompromised could be considered as recently exposed to P. jirovecii and then serve as reservoirs of the organism, at least transiently, as discussed above. Consequently, we propose that “colonization” could be considered as a situation where people have recently encountered P. jirovecii. In those individuals, only the immune status will predict if infection with symptoms will occur. Thus, what is called “colonization” or “asymptomatic carriage” could be the starting point for PCP in immunocompromised patients and therefore should not be neglected, whatever the scenario retained (recent acquisition, or reactivation of previous contamination). For instance, Mori et al. have shown that patients receiving immunosuppressive therapy for rheumatoid arthritis with P. jirovecii DNA detection without respiratory symptoms have developed PCP in the next 2 to 4 weeks 72. Upon P. jirovecii exposure or reactivation in immunocompromised hosts, the fungal load initially not detectable will increase until a plateau at the stage of PCP 29. The rapidity of this increase should vary according to the immune status and the degree of immunosuppression ( Figure 1) 21, 73.
Therefore, the risk of PCP following P. jirovecii DNA detection underlies the need for prophylaxis in different clinical situations of immunosuppression 20, 74. The generalization of prophylaxis to asymptomatic carriers of P. jirovecii should decrease the fungal load or even eradicate the fungus in these patients, leading to fewer cases of PCP in populations at risk, but also reduce transmission to other immunocompromised patients who share the same specialized wards. The expected benefit would be to dramatically decrease the probability of nosocomial transmission in immunocompromised hosts. This latter point obviously needs clinical validation. The risk of wide use of cotrimoxazole prophylaxis might be an increase in cotrimoxazole resistance. However, resistance following full-dose cotrimoxazole appears to be extremely rare 75.
Practical considerations for the diagnosis of Pneumocystis pneumonia
Diagnostic tools
The most sensitive tools to detect P. jirovecii are PCR-based assays. Real-time qPCR is the only PCR format assuming both a low to null rate of false positivity and quantification of the fungal load 75. Microscopy with immunofluorescence is much less sensitive than qPCR, with the frequent observation of negative immunofluorescence in samples containing high quantities of DNA 75– 77. Indeed, real-time PCR has been recommended for PCP diagnosis by a European panel of experts at the recent European Conference on Infection in Leukemia (ECIL) 75.
The quantification of DNA is of the utmost importance to evaluate the risk of PCP. Indeed, the immunosuppression background influences the fungal load associated with PCP 21, 78, 79. HIV-positive patients harbor much higher fungal loads and have a much lower mortality rate (17–30%) 80. In contrast, HIV-negative patients harbor lower fungal loads associated with a higher mortality rate (28–58%) 80. This strongly suggests that the host’s immune status is one of the most important determinants that shape the ability to control PCP and the clinical presentation 21, 80. Consequently, a qPCR result must provide quantification, which is to be interpreted according to the underlying diseases and the presence of risk factors ( Figure 1).
Full-blown PCP with high fungal loads with or without microscopic evidence of cysts (high fungal load) does not raise any therapeutic hesitation. The detection of P. jirovecii DNA, whatever the fungal load, in symptomatic immunocompromised patients should be carefully considered to be responsible, at least partially, for the symptoms and be treated accordingly 75. The detection of low P. jirovecii loads in the respiratory tract of asymptomatic patients with ongoing or scheduled immunosuppressive therapy makes the patient at risk of developing PCP in the near future and at least prophylaxis should be considered. Abstention might be advised only in non-immunocompromised patients. However, even in non-immunocompromised patients, such as those with COPD, P. jirovecii can play a role in the development of COPD 81, 82 or at least in the severity of pulmonary symptoms 83, 84.
Some authors have proposed using a combination of qPCR results together with beta-glucan levels to help define PCP 85. In this report, it was demonstrated that a correlation existed between qPCR fungal load in respiratory samples and beta-glucan levels in serum. However, when intermediate fungal loads were detected, intermediate beta-glucan loads were observed, suggesting that beta glucan does not add more value for clinical decision making. On the other hand, pneumocystosis without the detection of glucan seems possible 86, although rare 75.
Specimens
The DNA quantification discussed above is highly dependent on the type and the quality of the respiratory sample. The use of bronchoalveolar lavage (BAL) has been recommended as the most sensitive specimen owing to the fact that BAL is the sample where the highest fungal load is expected compared to sputa or upper respiratory specimens (URS) 87– 89. Consequently, BAL is the only specimen able to exclude the diagnosis of PCP in case of negative qPCR results. In contrast, the absence of P. jirovecii detection in URS does not exclude the diagnosis if negative. If positive PCR in URS is detected, this supports the diagnosis of PCP knowing that the corresponding fungal load in the BAL should be higher than that observed in URS 75, 90.
Even if BAL is the specimen where the highest fungal load is expected, attention should be paid to how BAL is performed. In an autopsy study of healthy individuals, only 43% of lung tissue specimens were PCR positive, suggesting heterogeneity of the distribution of P. jirovecii in the lung 45. Indeed, P. jirovecii seems to be predominant in some areas of the lungs 91. Thus, P. jirovecii seems heterogeneously distributed and clustered in specific areas 92, as suggested from radiological analysis of PCP cases 93, 94. Therefore, some authors suggested sampling several lobes during the BAL procedure to increase the sensitivity of P. jirovecii detection 91. To assess the quality of the sample, it is possible to quantify, in parallel to P. jirovecii, human DNA in the sample 90.
Conclusion/perspectives
The concept of “colonization” as a specific state, which does not warrant treatment in opposition to PCP, seems outdated regarding the accumulated knowledge about the natural history of the disease. There is a constant back and forward evolution between the fungal load and the immune status of the patients and a constant risk of transmission between patients and healthcare staff. Therefore, the detection of P. jirovecii should not be discarded or considered as insignificant, even in asymptomatic patients. The risk of developing PCP for a patient should be interpreted as a function of the immune status of the patient not only at the time of diagnostic workup but also in the following days/weeks. For the best results, DNA detection should rely on qPCR assays performed on BAL, although URS can also be used for clinical decision-making if positive. Many questions remain to be addressed and should trigger future research including in the transmission and prevention of exposure in hospital settings. Isolation of patients in hospitals should be discussed, and one can imagine that, in wards taking care of immunocompromised patients, it will be recommended that patients and healthcare workers wear a mask to prevent transmission to other patients in cases of cough, which is already recommended for viral or bacterial pneumonia. One can also imagine that qPCR screening using non-invasive specimens (sputa, nasopharyngeal aspirates or swabs, oral washes) in patients and healthcare workers might help implement preventive measures such as isolation of patients and/or mask wearing 90. Generalized (to all patients at risk of PCP) or targeted (restricted to qPCR-positive patients upon screening) cotrimoxazole prophylaxis could also be discussed, although potential toxicity should be kept in mind. To go further in the description of transmission in humans, molecular tools able to characterize low to very low quantities of P. jirovecii DNA are needed. Ideally, these new tools would not be based on mitochondrial DNA, since mitochondrial heteroplasmy would prevent definite results 56. Loci from nuclear DNA should be more appropriate, although present as a single copy, limiting the sensitivity of their detection. Eventually, new diagnostic tools based on RNA detection and quantification allowing a more precise description of the active or resting state of P. jirovecii in a given patient would help clinicians in deciphering whether the patient should be considered to have PCP if the multiplication is active or whether other etiological agents should be incriminated when facing resting microorganisms. These tools are under investigation and could be available in the near future 95.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Sergio L Vargas, University of Chile, Santiago, Chile
Melanie T. Cushion, University of Cincinnati College of Medicine, Cininnati, OH, USA
Francis Gigliotti, University of Rochester Medical Center, Rochester, NY, USA
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; referees: 3 approved]
References
- 1. Edman JC, Kovacs JA, Masur H, et al. : Ribosomal RNA sequence shows Pneumocystis carinii to be a member of the fungi. Nature. 1988;334(6182):519–22. 10.1038/334519a0 [DOI] [PubMed] [Google Scholar]
- 2. Stringer SL, Stringer JR, Blase MA, et al. : Pneumocystis carinii: sequence from ribosomal RNA implies a close relationship with fungi. Exp Parasitol. 1989;68(4):450–61. 10.1016/0014-4894(89)90130-6 [DOI] [PubMed] [Google Scholar]
- 3. Pixley FJ, Wakefield AE, Banerji S, et al. : Mitochondrial gene sequences show fungal homology for Pneumocystis carinii. Mol Microbiol. 1991;5(6):1347–51. 10.1111/j.1365-2958.1991.tb00781.x [DOI] [PubMed] [Google Scholar]
- 4. Cissé OH, Pagni M, Hauser PM: De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio. 2012;4(1):e00428–12. 10.1128/mBio.00428-12 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 5. Ma L, Chen Z, Huang da W, et al. : Genome analysis of three Pneumocystis species reveals adaptation mechanisms to life exclusively in mammalian hosts. Nat Commun. 2016;7: 10740. 10.1038/ncomms10740 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Cissé OH, Almeida JM, Fonseca A, et al. : Genome sequencing of the plant pathogen Taphrina deformans, the causal agent of peach leaf curl. mBio. 2013;4(3):e00055–13. 10.1128/mBio.00055-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Redhead SA, Cushion MT, Frenkel JK, et al. : Pneumocystis and Trypanosoma cruzi: nomenclature and typifications. J Eukaryot Microbiol. 2006;53(1):2–11. 10.1111/j.1550-7408.2005.00072.x [DOI] [PubMed] [Google Scholar]
- 8. Aliouat-Denis CM, Martinez A, Aliouat el M, et al. : The Pneumocystis life cycle. Mem Inst Oswaldo Cruz. 2009;104(3):419–26. 10.1590/S0074-02762009000300004 [DOI] [PubMed] [Google Scholar]
- 9. Tamburrini E, Mencarini P, Visconti E, et al. : Imbalance between Pneumocystis carinii cysts and trophozoites in bronchoalveolar lavage fluid from patients with pneumocystosis receiving prophylaxis. J Med Microbiol. 1996;45(2):146–8. 10.1099/00222615-45-2-146 [DOI] [PubMed] [Google Scholar]
- 10. Thomas CF, Jr, Limper AH: Current insights into the biology and pathogenesis of Pneumocystis pneumonia. Nat Rev Microbiol. 2007;5(4):298–308. 10.1038/nrmicro1621 [DOI] [PubMed] [Google Scholar]
- 11. Lenardon MD, Munro CA, Gow NA: Chitin synthesis and fungal pathogenesis. Curr Opin Microbiol. 2010;13(4):416–23. 10.1016/j.mib.2010.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cissé OH, Pagni M, Hauser PM: Comparative genomics suggests that the human pathogenic fungus Pneumocystis jirovecii acquired obligate biotrophy through gene loss. Genome Biol Evol. 2014;6(8):1938–48. 10.1093/gbe/evu155 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Porollo A, Sesterhenn TM, Collins MS, et al. : Comparative genomics of Pneumocystis species suggests the absence of genes for myo-inositol synthesis and reliance on inositol transport and metabolism. mBio. 2014;5(6):e01834. 10.1128/mBio.01834-14 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Kutty G, Maldarelli F, Achaz G, et al. : Variation in the major surface glycoprotein genes in Pneumocystis jirovecii. J Infect Dis. 2008;198(5):741–9. 10.1086/590433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daly KR, Fichtenbaum CJ, Tanaka R, et al. : Serologic responses to epitopes of the major surface glycoprotein of Pneumocystis jiroveci differ in human immunodeficiency virus-infected and uninfected persons. J Infect Dis. 2002;186(5):644–51. 10.1086/341565 [DOI] [PubMed] [Google Scholar]
- 16. Aliouat el-M, Dujardin L, Martinez A, et al. : Pneumocystis carinii growth kinetics in culture systems and in hosts: involvement of each life cycle parasite stage. J Eukaryot Microbiol. 1999;46(5):116S–7. [PubMed] [Google Scholar]
- 17. Schildgen V, Mai S, Khalfaoui S, et al. : Pneumocystis jirovecii can be productively cultured in differentiated CuFi-8 airway cells. mBio. 2014;5(3):e01186–14. 10.1128/mBio.01186-14 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Cushion MT, Walzer PD: Preclinical drug discovery for new anti- pneumocystis compounds. Curr Med Chem. 2009;16(20):2514–30. 10.2174/092986709788682038 [DOI] [PubMed] [Google Scholar]
- 19. Cushion MT, Collins MS: Susceptibility of Pneumocystis to echinocandins in suspension and biofilm cultures. Antimicrob Agents Chemother. 2011;55(10):4513–8. 10.1128/AAC.00017-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cordonnier C, Cesaro S, Maschmeyer G, et al. : Pneumocystis jirovecii pneumonia: still a concern in patients with haematological malignancies and stem cell transplant recipients. J Antimicrob Chemother. 2016;71(9):2379–85. 10.1093/jac/dkw155 [DOI] [PubMed] [Google Scholar]
- 21. Roux A, Canet E, Valade S, et al. : Pneumocystis jirovecii pneumonia in patients with or without AIDS, France. Emerg Infect Dis. 2014;20(9):1490–7. 10.3201/eid2009.131668 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Gajdusek DC: Pneumocystis carinii; etiologic agent of interstitial plasma cell pneumonia of premature and young infants. Pediatrics. 1957;19(4 pt 1):543–65. [PubMed] [Google Scholar]
- 23. Vanek J, Jirovec O: Parasitare Pneumonie. Interstitielle Plasmazellenpneumonie der Fruhgeborenen, verursacht durch Pneumodystis Carinii. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg. 1952;158:120–7. [PubMed] [Google Scholar]
- 24. Chabé M, Hugot JP, Dei-Cas E: Pneumocystis molecular phylogeny: a way to understand both pneumocystosis natural history and host taxonomy. New Frontiers of Molecular Epidemiology of Infectious Diseases.Springer;2012;149–78. 10.1007/978-94-007-2114-2_8 [DOI] [Google Scholar]
- 25. Hauser PM: The development of a typing method for an uncultivable microorganism: the example of Pneumocystis jirovecii. Infect Genet Evol. 2004;4(3):199–203. 10.1016/j.meegid.2004.01.011 [DOI] [PubMed] [Google Scholar]
- 26. Peterson JC, Cushion MT: Pneumocystis: not just pneumonia. Curr Opin Microbiol. 2005;8(4):393–8. 10.1016/j.mib.2005.06.010 [DOI] [PubMed] [Google Scholar]
- 27. Hughes WT: Natural mode of acquisition for de novo infection with Pneumocystis carinii. J Infect Dis. 1982;145(6):842–8. 10.1093/infdis/145.6.842 [DOI] [PubMed] [Google Scholar]
- 28. Walzer PD, Schnelle V, Armstrong D, et al. : Nude mouse: a new experimental model for Pneumocystis carinii infection. Science. 1977;197(4299):177–9. 10.1126/science.301657 [DOI] [PubMed] [Google Scholar]
- 29. Choukri F, Aliouat el M, Menotti J, et al. : Dynamics of Pneumocystis carinii air shedding during experimental pneumocystosis. J Infect Dis. 2011;203(9):1333–6. 10.1093/infdis/jir018 [DOI] [PubMed] [Google Scholar]
- 30. Menotti J, Emmanuel A, Bouchekouk C, et al. : Evidence of airborne excretion of Pneumocystis carinii during infection in immunocompetent rats. Lung involvement and antibody response. PLoS One. 2013;8(4):e62155. 10.1371/journal.pone.0062155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Latouche S, Olsson M, Polack B, et al. : Detection of Pneumocystis carinii f. sp. in air samples collected in animal rooms. J Eukaryot Microbiol. 1997;44(6):46S–7. 10.1111/j.1550-7408.1997.tb05768.x [DOI] [PubMed] [Google Scholar]
- 32. Valade S, Azoulay E, Damiani C, et al. : Pneumocystis jirovecii airborne transmission between critically ill patients and health care workers. Intensive Care Med. 2015;41(9):1716–8. 10.1007/s00134-015-3835-9 [DOI] [PubMed] [Google Scholar]
- 33. Choukri F, Menotti J, Sarfati C, et al. : Quantification and spread of Pneumocystis jirovecii in the surrounding air of patients with Pneumocystis pneumonia. Clin Infect Dis. 2010;51(3):259–65. 10.1086/653933 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Le Gal S, Damiani C, Rouillé A, et al. : A cluster of Pneumocystis infections among renal transplant recipients: molecular evidence of colonized patients as potential infectious sources of Pneumocystis jirovecii. Clin Infect Dis. 2012;54(7):e62–71. 10.1093/cid/cir996 [DOI] [PubMed] [Google Scholar]
- 35. Miller RF, Ambrose HE, Wakefield AE: Pneumocystis carinii f. sp. hominis DNA in immunocompetent health care workers in contact with patients with P. carinii pneumonia. J Clin Microbiol. 2001;39(11):3877–82. 10.1128/JCM.39.11.3877-3882.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cushion MT, Linke MJ, Ashbaugh A, et al. : Echinocandin treatment of pneumocystis pneumonia in rodent models depletes cysts leaving trophic burdens that cannot transmit the infection. PLoS One. 2010;5(1):e8524. 10.1371/journal.pone.0008524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martinez A, Halliez MC, Aliouat el M, et al. : Growth and airborne transmission of cell-sorted life cycle stages of Pneumocystis carinii. PLoS One. 2013;8(11):e79958. 10.1371/journal.pone.0079958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gigliotti F, Harmsen AG, Wright TW: Characterization of transmission of Pneumocystis carinii f. sp. muris through immunocompetent BALB/c mice. Infect Immun. 2003;71(7):3852–6. 10.1128/IAI.71.7.3852-3856.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Chabé M, Dei-Cas E, Creusy C, et al. : Immunocompetent hosts as a reservoir of pneumocystis organisms: histological and rt-PCR data demonstrate active replication. Eur J Clin Microbiol Infect Dis. 2004;23(2):89–97. 10.1007/s10096-003-1092-2 [DOI] [PubMed] [Google Scholar]
- 40. An CL, Gigliotti F, Harmsen AG: Exposure of immunocompetent adult mice to Pneumocystis carinii f. sp. muris by cohousing: growth of P. carinii f. sp. muris and host immune response. Infect Immun. 2003;71(4):2065–70. 10.1128/IAI.71.4.2065-2070.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Tamburrini E, Ortona E, Visconti E, et al. : Pneumocystis carinii infection in young non-immunosuppressed rabbits. Kinetics of infection and of the primary specific immune response. Med Microbiol Immunol. 1999;188(1):1–7. 10.1007/s004300050098 [DOI] [PubMed] [Google Scholar]
- 42. Bishop LR, Kovacs JA: Quantitation of anti- Pneumocystis jiroveci antibodies in healthy persons and immunocompromised patients. J Infect Dis. 2003;187(12):1844–8. 10.1086/375354 [DOI] [PubMed] [Google Scholar]
- 43. Vargas SL, Hughes WT, Santolaya ME, et al. : Search for primary infection by Pneumocystis carinii in a cohort of normal, healthy infants. Clin Infect Dis. 2001;32(6):855–61. 10.1086/319340 [DOI] [PubMed] [Google Scholar]
- 44. Vargas SL, Ponce CA, Gallo M, et al. : Near-universal prevalence of Pneumocystis and associated increase in mucus in the lungs of infants with sudden unexpected death. Clin Infect Dis. 2013;56(2):171–9. 10.1093/cid/cis870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ponce CA, Gallo M, Bustamante R, et al. : Pneumocystis colonization is highly prevalent in the autopsied lungs of the general population. Clin Infect Dis. 2010;50(3):347–53. 10.1086/649868 [DOI] [PubMed] [Google Scholar]
- 46. Nevez G, Totet A, Pautard JC, et al. : Pneumocystis carinii detection using nested-PCR in nasopharyngeal aspirates of immunocompetent infants with bronchiolitis. J Eukaryot Microbiol. 2001;48(Suppl):122S–3. 10.1111/j.1550-7408.2001.tb00479.x [DOI] [PubMed] [Google Scholar]
- 47. Vargas SL, Ponce CA, Sanchez CA, et al. : Pregnancy and asymptomatic carriage of Pneumocystis jiroveci. Emerg Infect Dis. 2003;9(5):605–6. 10.3201/eid0905.020660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Garcia-Hermoso D, Janbon G, Dromer F: Epidemiological evidence for dormant Cryptococcus neoformans infection. J Clin Microbiol. 1999;37(10):3204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Alanio A, Vernel-Pauillac F, Sturny-Leclère A, et al. : Cryptococcus neoformans host adaptation: toward biological evidence of dormancy. mBio. 2015;6(2): pii: e02580-14. 10.1128/mBio.02580-14 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Vargas SL, Hughes WT, Wakefield AE, et al. : Limited persistence in and subsequent elimination of Pneumocystis carinii from the lungs after P. carinii pneumonia. J Infect Dis. 1995;172(2):506–10. 10.1093/infdis/172.2.506 [DOI] [PubMed] [Google Scholar]
- 51. Chen W, Gigliotti F, Harmsen AG: Latency is not an inevitable outcome of infection with Pneumocystis carinii. Infect Immun. 1993;61(12):5406–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Keely SP, Stringer JR, Baughman RP, et al. : Genetic variation among Pneumocystis carinii hominis isolates in recurrent pneumocystosis. J Infect Dis. 1995;172(2):595–8. 10.1093/infdis/172.2.595 [DOI] [PubMed] [Google Scholar]
- 53. Hauser PM, Blanc DS, Sudre P, et al. : Genetic diversity of Pneumocystis carinii in HIV-positive and -negative patients as revealed by PCR-SSCP typing. AIDS. 2001;15(4):461–6. [DOI] [PubMed] [Google Scholar]
- 54. Parobek CM, Jiang LY, Patel JC, et al. : Multilocus microsatellite genotyping array for investigation of genetic epidemiology of Pneumocystis jirovecii. J Clin Microbiol. 2014;52(5):1391–9. 10.1128/JCM.02531-13 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Gits-Muselli M, Peraldi M, de Castro N, et al. : New Short Tandem Repeat-Based Molecular Typing Method for Pneumocystis jirovecii Reveals Intrahospital Transmission between Patients from Different Wards. PLoS One. 2015;10(5):e0125763. 10.1371/journal.pone.0125763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Alanio A, Gits-Muselli M, Mercier-Delarue S, et al. : Diversity of Pneumocystis jirovecii during Infection Revealed by Ultra-Deep Pyrosequencing. Front Microbiol. 2016;7:733. 10.3389/fmicb.2016.00733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. de Boer MG, de Fijter JW, Kroon FP: Outbreaks and clustering of Pneumocystis pneumonia in kidney transplant recipients: a systematic review. Med Mycol. 2011;49(7):673–80. 10.3109/13693786.2011.571294 [DOI] [PubMed] [Google Scholar]
- 58. Desoubeaux G, Dominique M, Morio F, et al. : Epidemiological Outbreaks of Pneumocystis jirovecii Pneumonia Are Not Limited to Kidney Transplant Recipients: Genotyping Confirms Common Source of Transmission in a Liver Transplantation Unit. J Clin Microbiol. 2016;54(5):1314–20. 10.1128/JCM.00133-16 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Yiannakis EP, Boswell TC: Systematic review of outbreaks of Pneumocystis jirovecii pneumonia: evidence that P. jirovecii is a transmissible organism and the implications for healthcare infection control. J Hosp Infect. 2016;93(1):1–8. 10.1016/j.jhin.2016.01.018 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Robin C, Alanio A, Gits-Muselli M, et al. : Molecular Demonstration of a Pneumocystis Outbreak in Stem Cell Transplant Patients: Evidence for Transmission in the Daycare Center. Front Microbiol. 2017;8:700. 10.3389/fmicb.2017.00700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Helweg-Larsen J, Tsolaki AG, Miller RF, et al. : Clusters of Pneumocystis carinii pneumonia: analysis of person-to-person transmission by genotyping. QJM. 1998;91(12):813–20. 10.1093/qjmed/91.12.813 [DOI] [PubMed] [Google Scholar]
- 62. Ong YL, Jones FG: A cluster of suspected Pneumocystis carinii Pneumonia following intensive chemotherapy in a Belfast haematology unit. Ulster Med J. 1998;67(2):104–9. [PMC free article] [PubMed] [Google Scholar]
- 63. Depypere M, Saegeman V, Lagrou K: Typing of Pneumocystis jirovecii by multilocus sequencing: evidence of outbreak? Eur J Clin Microbiol Infect Dis. 2016;35(6):911–6. 10.1007/s10096-016-2615-y [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Durand-Joly I, Soula F, Chabé M, et al. : Long-term colonization with Pneumocystis jirovecii in hospital staffs: a challenge to prevent nosocomial pneumocystosis. J Eukaryot Microbiol. 2003;50(Suppl):614–5. 10.1111/j.1550-7408.2003.tb00650.x [DOI] [PubMed] [Google Scholar]
- 65. Beard CB, Carter JL, Keely SP, et al. : Genetic variation in Pneumocystis carinii isolates from different geographic regions: implications for transmission. Emerg Infect Dis. 2000;6(3):265–72. 10.3201/eid0603.000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Miller RF, Lindley AR, Copas A, et al. : Genotypic variation in Pneumocystis jirovecii isolates in Britain. Thorax. 2005;60(8):679–82. 10.1136/thx.2004.039818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dohn MN, White ML, Vigdorth EM, et al. : Geographic clustering of Pneumocystis carinii pneumonia in patients with HIV infection. Am J Respir Crit Care Med. 2000;162(5):1617–21. 10.1164/ajrccm.162.5.9707101 [DOI] [PubMed] [Google Scholar]
- 68. Gigliotti F: Antigenic variation of a major surface glycoprotein of Pneumocystis carinii. J Protozool. 1991;38(6):4S–5S. [PubMed] [Google Scholar]
- 69. Stringer JR: Antigenic variation in pneumocystis. J Eukaryot Microbiol. 2007;54(1):8–13. 10.1111/j.1550-7408.2006.00225.x [DOI] [PubMed] [Google Scholar]
- 70. Leigh TR, Wakefield AE, Peters SE, et al. : Comparison of DNA amplification and immunofluorescence for detecting Pneumocystis carinii in patients receiving immunosuppressive therapy. Transplantation. 1992;54(3):468–70. 10.1097/00007890-199209000-00016 [DOI] [PubMed] [Google Scholar]
- 71. Morris A, Norris KA: Colonization by Pneumocystis jirovecii and its role in disease. Clin Microbiol Rev. 2012;25(2):297–317. 10.1128/CMR.00013-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mori S, Cho I, Sugimoto M: A followup study of asymptomatic carriers of Pneumocystis jiroveci during immunosuppressive therapy for rheumatoid arthritis. J Rheumatol. 2009;36(8):1600–5. 10.3899/jrheum.081270 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 73. Ahn IE, Jerussi T, Farooqui M, et al. : Atypical Pneumocystis jirovecii pneumonia in previously untreated patients with CLL on single-agent ibrutinib. Blood. 2016;128(15):1940–3. 10.1182/blood-2016-06-722991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stern A, Green H, Paul M, et al. : Prophylaxis for Pneumocystis pneumonia (PCP) in non-HIV immunocompromised patients. Cochrane Database Syst Rev. 2014; (10):CD005590. 10.1002/14651858.CD005590.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Alanio A, Hauser PM, Lagrou K, et al. : ECIL guidelines for the diagnosis of Pneumocystis jirovecii pneumonia in patients with haematological malignancies and stem cell transplant recipients. J Antimicrob Chemother. 2016;71(9):2386–96. 10.1093/jac/dkw156 [DOI] [PubMed] [Google Scholar]
- 76. Mühlethaler K, Bögli-Stuber K, Wasmer S, et al. : Quantitative PCR to diagnose Pneumocystis pneumonia in immunocompromised non-HIV patients. Eur Respir J. 2012;39(4):971–8. 10.1183/09031936.00095811 [DOI] [PubMed] [Google Scholar]
- 77. Valero C, Buitrago MJ, Gits-Muselli M, et al. : Copy Number Variation of Mitochondrial DNA Genes in Pneumocystis jirovecii According to the Fungal Load in BAL Specimens. Front Microbiol. 2016;7:1413. 10.3389/fmicb.2016.01413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thomas CF, Jr, Limper AH: Pneumocystis pneumonia: clinical presentation and diagnosis in patients with and without acquired immune deficiency syndrome. Semin Respir Infect. 1998;13(4):289–95. [PubMed] [Google Scholar]
- 79. Limper AH, Offord KP, Smith TF, et al. : Pneumocystis carinii pneumonia. Differences in lung parasite number and inflammation in patients with and without AIDS. Am Rev Respir Dis. 1989;140(5):1204–9. 10.1164/ajrccm/140.5.1204 [DOI] [PubMed] [Google Scholar]
- 80. Cordonnier C, Alanio A, Cesaro S, et al. : Pneumocystis jirovecii pneumonia: still a concern in patients with haematological malignancies and stem cell transplant recipients-authors' response. J Antimicrob Chemother. 2017;72(4):1266–8. 10.1093/jac/dkw580 [DOI] [PubMed] [Google Scholar]
- 81. Shipley TW, Kling HM, Morris A, et al. : Persistent pneumocystis colonization leads to the development of chronic obstructive pulmonary disease in a nonhuman primate model of AIDS. J Infect Dis. 2010;202(2):302–12. 10.1086/653485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Christensen PJ, Preston AM, Ling T, et al. : Pneumocystis murina infection and cigarette smoke exposure interact to cause increased organism burden, development of airspace enlargement, and pulmonary inflammation in mice. Infect Immun. 2008;76(8):3481–90. 10.1128/IAI.00165-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Morris A, Alexander T, Radhi S, et al. : Airway obstruction is increased in pneumocystis-colonized human immunodeficiency virus-infected outpatients. J Clin Microbiol. 2009;47(11):3773–6. 10.1128/JCM.01712-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Morris A, Sciurba FC, Lebedeva IP, et al. : Association of chronic obstructive pulmonary disease severity and Pneumocystis colonization. Am J Respir Crit Care Med. 2004;170(4):408–13. 10.1164/rccm.200401-094OC [DOI] [PubMed] [Google Scholar]
- 85. Damiani C, Le Gal S, Da Costa C, et al. : Combined quantification of pulmonary Pneumocystis jirovecii DNA and serum (1->3)-β-D-glucan for differential diagnosis of pneumocystis pneumonia and Pneumocystis colonization. J Clin Microbiol. 2013;51(10):3380–8. 10.1128/JCM.01554-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kamada T, Furuta K, Tomioka H: Pneumocystis pneumonia associated with human immunodeficiency virus infection without elevated (1 → 3)-β-D glucan: A case report. Respir Med Case Rep. 2016;18:73–5. 10.1016/j.rmcr.2016.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 87. Elvin KM, Björkman A, Linder E, et al. : Pneumocystis carinii pneumonia: detection of parasites in sputum and bronchoalveolar lavage fluid by monoclonal antibodies. BMJ. 1988;297(6645):381–4. 10.1136/bmj.297.6645.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hazen KC: Essential Procedures for Clinical Microbiology. Isenberg HD, editor. Washington, DC: ASM press;1998. [Google Scholar]
- 89. Jorgensen JH, Pfaller MA, Versalovic J, et al. : Manual of Clinical Microbiology. 11th Edition. Washington, D.C.: ASM Press;2015. 10.1128/9781555817381 [DOI] [Google Scholar]
- 90. Guigue N, Alanio A, Menotti J, et al. : Utility of adding Pneumocystis jirovecii DNA detection in nasopharyngeal aspirates in immunocompromised adult patients with febrile pneumonia. Med Mycol. 2015;53(3):241–7. 10.1093/mmy/myu087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Levine SJ, Kennedy D, Shelhamer JH, et al. : Diagnosis of Pneumocystis carinii pneumonia by multiple lobe, site-directed bronchoalveolar lavage with immunofluorescent monoclonal antibody staining in human immunodeficiency virus-infected patients receiving aerosolized pentamidine chemoprophylaxis. Am Rev Respir Dis. 1992;146(4):838–43. 10.1164/ajrccm/146.4.838 [DOI] [PubMed] [Google Scholar]
- 92. Helweg-Larsen J, Lundgren B, Lundgren JD: Heterogeneity and compartmentalization of Pneumocystis carinii f. sp. hominis genotypes in autopsy lungs. J Clin Microbiol. 2001;39(10):3789–92. 10.1128/JCM.39.10.3789-3792.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Crans CA, Jr, Boiselle PM: Imaging features of Pneumocystis carinii pneumonia. Crit Rev Diagn Imaging. 1999;40(4):251–84. 10.3109/10408379991249194 [DOI] [PubMed] [Google Scholar]
- 94. Kanne JP, Yandow DR, Meyer CA: Pneumocystis jiroveci pneumonia: high-resolution CT findings in patients with and without HIV infection. AJR Am J Roentgenol. 2012;198(6):W555–61. 10.2214/AJR.11.7329 [DOI] [PubMed] [Google Scholar]
- 95. Alanio A, Bergeron A, Sturny-Leclère A, et al. : Diagnosis of Pneumocystis pneumonia: switching from DNA quantification to RNA expression analysis. 7th Trends in Medical Mycology Lisboa.2015. [Google Scholar]