Abstract
Background.
The heterogeneous biology of glioblastoma (GBM) emphasizes the need for imaging methods to assess tumor burden and assist in evaluating individual patients. The purpose of this study was to investigate early changes in metrics from 3D 1H magnetic resonance spectroscopic imaging (MRSI) data, compare them with anatomic lesion volumes, and determine whether they were associated with survival for patients with newly diagnosed GBM receiving a multimodality treatment regimen.
Methods.
Serial MRI and MRSI scans provided estimates of anatomic lesion volumes and levels of choline, creatine, N-acetylaspartate, lactate, and lipid. The association of metrics derived from these data with survival was assessed using Cox proportional hazards models with adjustments for age, Karnofsky performance score, and extent of resection. Temporal changes in parameters were evaluated using a Wilcoxon signed rank test.
Results.
Anatomic lesion volumes at the post-radiotherapy (RT) scan, metabolic lesion volume at mid-RT and post-RT scans, as well as metrics describing levels of choline, lactate, and lipid were associated with overall survival. There was a significant reduction in the enhancing lesion volume, increase in T2 lesion volume from mid-RT to post-RT, and decrease in parameters describing metabolite levels during these early time points.
Conclusion.
The MRSI data provided metrics that described the effects of treatment on the metabolic lesion burden and were associated with overall survival. This suggests that adding these parameters to standard assessments of changes in anatomic lesion volumes could contribute to making early decisions about the efficacy of such combination therapies.
Keywords: 1H MR spectroscopic imaging, metabolic parameters, newly diagnosed GBM, predicting outcome, survival, treatment.
Glioblastoma (GBM) is the most common primary malignant brain tumor in adults and typically has highly aggressive clinical behavior. Despite multimodality treatment, median survival is approximately 15 months with standard treatment, but there is wide variation among patients.1 The ability to make a connection between early changes in imaging parameters and clinical outcome would be valuable in determining how to adapt treatment strategies to individual patients.
Methods for monitoring response to therapy for brain tumors use a combination of clinical symptoms and morphological data from MRI. Assessment of changes in the cross-sectional diameter of the contrast enhancing T1-weighted lesion and the region of hyperintensity on T2-weighted images are key determinants for characterizing the effectiveness of therapy. The Macdonald criteria2 use the enhancing lesion as the metric of tumor growth, while the newer RANO (Response Assessment in Neuro-Oncology) criteria take into account cross-sectional diameter of both types of anatomic lesions.3
1H magnetic resonance spectroscopic imaging (MRSI) is a noninvasive tool that provides estimates of levels of metabolites, such as N-acetylaspartate (NAA), choline (Cho)-containing compounds, creatine, lactate, and lipid. This provides complementary data that can be directly compared with changes on morphologic images. Previous studies have indicated that presurgery and pretreatment MRSI data may be valuable for predicting survival outcomes from patients with GBM.4–7
The purpose of this study was to investigate whether early changes in MRSI data could provide metrics associated with subsequent treatment response and clinical outcome for a cohort of patients newly diagnosed with GBM and receiving a combination of radiation and temozolomide in conjunction with the investigational agent enzastaurin. The latter, a protein kinase C inhibitor, was shown in preclinical studies to have antiproliferative, pro-apoptotic, and anti-angiogenic properties.8,9 While the clinical results of the treatment have been compared with other combination therapies,10 the relationship of metabolic imaging parameters to outcome were not previously investigated for these patients.
Materials and Methods
In 43 patients, newly diagnosed GBM based upon World Health Organization criteria was evaluated in this study. Treatment included a 6-week cycle of fractionated external beam RT in conjunction with temozolomide and enzastaurin. Temozolomide was given at 75mg/m2 daily during RT and 200mg/m2 for 5 days every 28-day cycle after RT and enzastaurin was given at 250mg/day, both during and after RT. Patients who deviated from protocol due to side effects were excluded from the study. The study received approval from the institutional review board, and informed consent was obtained from all patients.
Assessment of Clinical Outcome
Overall survival (OS) was calculated as the time to death from the start of treatment and progression-free survival (PFS) as the time from the start of treatment to the examination at which the patient was determined to have progressed. To address cases of pseudoprogression,11 the clinical histories of all patients who progressed within 3 months of the completion of radiotherapy were re-reviewed by a neuroradiologist to determine whether the suspected progression was maintained. Patients who displayed stable exams that had been preceded by seemingly suspect exams were also re-reviewed to evaluate the longevity of the imaging findings.
MR Data Acquisition
Data were acquired using a 3 Tesla MR scanner (GE Healthcare) and a commercially available 8-channel phased array headcoil. Thirty-nine patients received a research imaging scan prior to RT (baseline), 29 received a mid-RT scan (1 mo), and 38 received a post-RT scan (2 mo). This resulted in a total of 106 scans being evaluated, with 25 of the subjects receiving research scans at all 3 time points. Acquired were an axial T2-weighted fluid attenuated inversion recovery (FLAIR) sequence with 3-mm slice thickness, as well as axial volumetric pre- and post-gadolinium T1-weighted inversion recovery spoiled gradient recalled images with 1.5-mm thickness.
A lactate-edited 3D point resolved spectroscopy (PRESS) MRSI sequence was applied for 99/106 scans. This used a flyback echo-planar readout gradient in the superior–inferior (SI) dimension and an over-PRESS factor of 1.5 with very selective saturation (VSS) bands to avoid chemical shift artifacts (repetition time/echo time = 1104/144ms, field of view = 16×16×16cm, voxel size = 1×1 × 1cm, total acquisition time = 9.5 minutes, 712 dwell points, and 988 Hz bandwidth).12 Anatomic images were used to define the PRESS region, which generally covered the lesion and 200–300 cc of normal tissue. Note that although there was some variability in the size of the PRESS region between examinations, the field of view and acquisition time were kept constant. Areas with sharply varying magnetic susceptibility and lipid contamination were avoided whenever possible. Additional VSS bands were utilized to suppress residual lipid signals.13
Image Processing
The data were transferred to an offline Linux workstation and processed to generate images in the same coordinate system. This involved coregistering anatomic images to the corresponding postcontrast T1-weighted scan.14 Volumetric regions of interest were defined by manual analysis to provide the contrast-enhancing (CE) lesion on T1-weighted postcontrast images and hyperintensity on FLAIR images minus the resection cavity (T2 lesion).
Reconstruction of MRSI Data
The methods used for reconstruction and postprocessing of the spectral data are fully automated and have been described in previous publications.15,16 In brief, the start point was the raw data file obtained from the lactate edited MRSI sequence. This comprised 8 channels of interleaved acquisitions (or cycles) obtained with radiofrequency pulses that modulated the phase of the lactate peak. The k-space time domain data for each cycle and channel were first filtered with a 4-Hz exponential function in the time domain, zero filled to 1024 points, and Fourier transformed to produce a k-space array of spectra. The next step was to apply the k-space Fourier transforms to produce 3D spatial arrays of spectral data, followed by combination using in-house developed software that weights the data by coil sensitivities estimated from low resolution proton density weighted images.16 Additional phase corrections were applied in the SI dimension to account for the flyback echo-planar readout gradient.17 The cycles were summed to produce an array of spectra containing choline, creatine, NAA, and lipid and subtracted to produce an array of spectra containing lactate. Spectra were baseline subtracted and phase and frequency corrected using parameters estimated from the summed array. Metabolite levels were obtained by estimating peak heights and integrated areas from the spectral arrays. Three-dimensional spectral arrays were viewed using the SIVIC software package.16 As shown in Fig. 1), color overlays of metabolite parameters such as the choline-NAA index (CNI) can also be viewed on the anatomic images. Datasets considered to be of limited quality due to poor coverage of the lesion or lipid contamination were excluded from analysis. This was the case for only 3 out of 99 total exams and resulted in metabolic data being included from n = 35 scans at pre-RT, n = 26 scans at mid-RT, and n = 35 scans at post-RT.
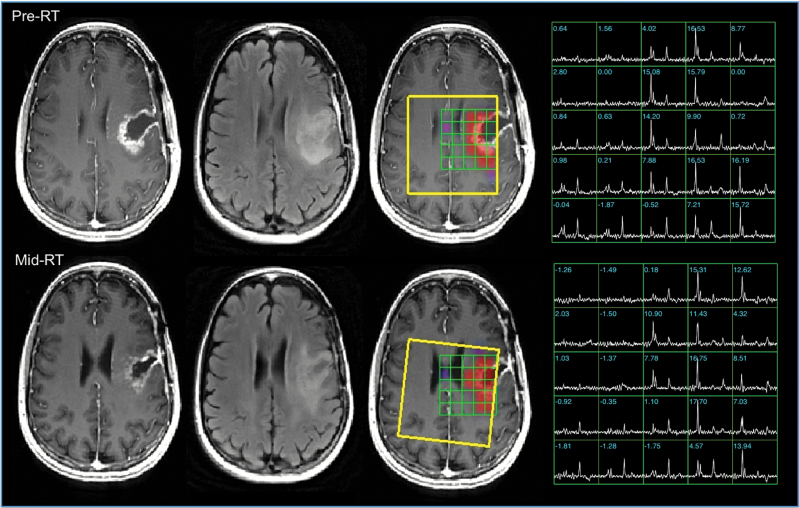
Fig. 1.
Anatomic images and metabolic data from patient A, a 41-year-old patient with KPS = 80, PFS = 67 days, and OS = 305 days. The pre-RT images indicate a subtotal resection with residual CE on the medial side of the cavity and a larger T2 lesion. The mid-RT images show that the cavity has contracted and there is reduced mass effect. The color overlays of the CNI parameters are thresholded to show only regions with CNI >2. The selected volume is highlighted in yellow. These and the spectral arrays highlight the region with abnormal metabolism, which has highly elevated choline, reduced NAA, with increased lipid in the cavity. Note that although the images and spectra were registered to improve visualization of the effects, the calculation of the quantitative parameters was done in the original coordinate frame.
Definition of the Metabolic Lesion
The CNI is a metric that was previously developed to describe the deviation of choline and NAA in regions of tumor relative to values in regions of normal brain tissue from the same individual.18,19 It is calculated from the choline and NAA levels in the spectral array with an iterative regression procedure to (i) eliminate outliers that correspond to tumor, (ii) estimate the ratio of Cho and NAA in “normal” brain from the slope of the line through the remaining voxels, (iii) determine the standard deviation (σ) of the distance of normal voxels from the regression line, and (iv) define the CNI for each voxel by its distance from the regression line divided by σ. Supplementary Fig. 1 shows this procedure schematically, and the corresponding supplementary text provides a more detailed discussion of the methodology. Once the CNI map had been calculated, the metabolic lesion (CNI2) was defined as the region having values greater than or equal to 2. The overlap between the CNI2 and the T2 lesion was also determined (CNI2t). The choline to creatine index (CCrI) was estimated in a similar manner, but with the definition of “normal” brain voxels being based upon those that had been identified as such during the CNI calculation.
Normalization of Metabolite Levels
To compare across serial exams, choline, creatine, and NAA intensities were normalized by their median value in voxels that had been identified during the CNI calculation as being from normal brain (nCho, nCr, and nNAA). Signals from lipid and lactate peaks were normalized with the median NAA from normal brain (nLac and nLip). To account for partial-voluming of tumor and normal tissue within the spectral voxels, the levels of excess choline relative to NAA (xsChoN = nCho-nNAA) and excess choline to creatine (xsChoCr = nCho-nCr) were calculated for each location.
Metrics Describing the Metabolic Lesion Burden
The metrics used to describe the size of the metabolic lesion were calculated for each examination separately and were therefore not dependent upon registration between scans. The parameters considered were the volumes of the CNI2 and CNI2t, as well as the maximum, median, and summation of values for the CNI, CCrI, xsChoN, xsChoCr, nLac, and nLip from voxels within the CNI2 regions at pre-, mid-, and post-RT. These times were chosen based upon previous analysis of 1H MRSI scans.6,20
Statistical Analysis
The primary analysis focused on the association of imaging parameters with PFS and OS using a Cox proportional hazards model that was adjusted for age, KPS, and extent of resection. Only tests with P values less than .05 were considered significant. The secondary analysis focused on exploring changes in anatomic and metabolic parameters among pre-, mid-, and post-RT scans. This was achieved by paired analysis within the groups of patients who had scans at all 3 time points using Wilcoxon signed rank tests. This included the evaluation of temporal changes in all subjects, as well as in subgroups with OS less than or greater than the median OS value.
Results
Population Parameters
The median age for the 43 patients considered was 57 years (range 27–80). One subject had a KPS of 60, another had a score of 70, and the rest were 80 or above. Median PFS was 234 days (no subjects censored) and median OS was 449 days (3 subjects censored). Of the patients followed on this study, 33/43 had known O6-DNA methylguanine-methyltransferase status, with 22 being unmethylated and 11 methylated. There was no significant difference in OS between these 2 subgroups (median of 480 days, 22 patients, 2 censored; and 529 days, 11 patients, none censored, respectively). The 2 subgroups considered in the secondary analysis had similar fractions of unmethylated versus methylated subjects to the overall population. Given the lack of significance for this parameter, it was not considered in the subsequent analysis of imaging parameters.
Examples of Data from Individual Patients
Figure 1 shows anatomic images, spectra, and metabolic imaging overlays from pre- and mid-RT for a 41-year-old patient who progressed at the post-RT time point (PFS = 67 days) with an OS of 305 days. Registration of images and spectral data between the 2 examinations was performed to allow for better visualization of the data. The anatomic images show a large initial resection cavity with contrast enhancement around the medial edge and some residual mass effect that decreased in the mid-RT scan. The surgical cavity reduced in size and the mass effect decreased at first, but the CE and T2 lesion volumes almost doubled in volume from pre- to post-RT (10.9 to 20.6 cc and 29.2 to 38.1 cc, respectively). The spectral arrays had regions with high choline peaks and low NAA around the cavity that extended beyond the CE lesion. Although the MRSI data were not available for the post-RT exam, the high maximum CNI values and increase from pre- and mid-RT (24.1 to 30.2) suggest that the tumor continued to grow during treatment. A second surgery confirmed the presence of residual/recurrent tumor.
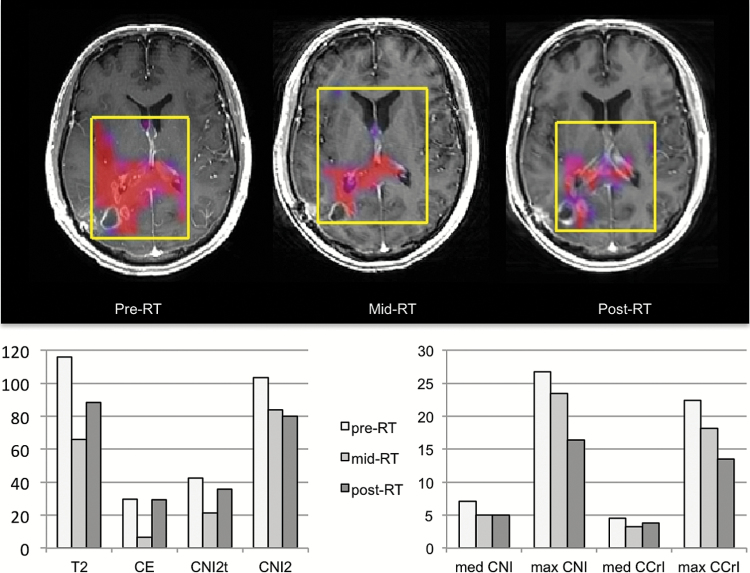
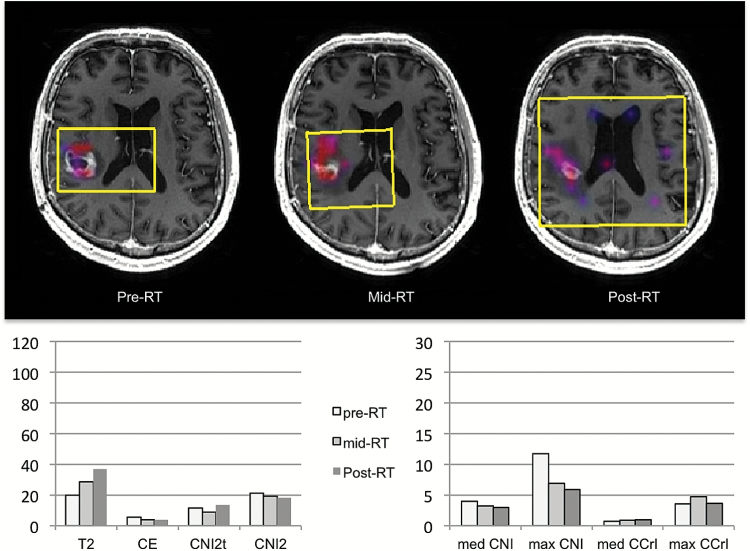
Figures 2 and 3 show examples of the coregistered anatomic images and color overlays of CNI, together with changes in imaging metrics for patients similar in age (57 or 58 y) and KPS (80 or 90) but with quite different PFS and OS (191 and 259 days for patient B versus 364 and 546 days for patient C). Of particular note is that the metabolic lesion was much larger than the CE lesion for patient B, suggesting that a large part of the tumor was nonenhancing (Fig. 2). The sum of nLac and the sum of nLip within the CNI2 lesion were also relatively high, with the former staying stable (27.1 to 27.0) and the latter increasing (22.6 to 40.3) from pre- to post-RT. Subsequent follow-up images showed the FLAIR lesion extending into the contralateral hemisphere. This is consistent with there being abnormal metabolism in this region at the early time points, which initially responded to therapy but then expanded again as the tumor progressed. For patient C (Fig. 3), the pre-RT anatomic and metabolic lesion volumes were much smaller, the CNI decreased from pre- to post-RT, and the initial maximum CNI and CCrI values were lower (11.7 and 3.5). The magnitudes of the sum of nLac and the sum of nLip within the CNI2 lesion were much lower (initial values of 5.4 and 4.9) and decreased further from pre- to post-RT.
Fig. 2.
Patient B was 58 years old with KPS = 80, PFS = 191 days, and OS = 269 days. Color overlays of CNI values on postcontrast T1-weighted images obtained at pre-, mid-, and post-RT scans and bar plots of changes in parameter values are shown. The overlays were thresholded to show only regions with CNI >2. The selected volumes are highlighted in yellow. The volumes of the T2 and CE lesions show an initial decrease and then an increase. The spatial extent of the metabolic lesion (CNI >2) and the maximum intensities of CNI and CCrI show a consistent decrease with time, suggesting an initial response to the treatment given.
Fig. 3.
Patient C was 50 years old with KPS = 90, PFS = 364 days, and OS = 546 days. Color overlays of CNI values on postcontrast T1-weighted images obtained at pre-, mid-, and post-RT scans are shown. The overlays were thresholded to show only regions with CNI >2. The selected volumes are highlighted in yellow. The spatial extent and magnitude of the regions with abnormal metabolism are much smaller than those for patient B, indicating much smaller metabolic lesions that further reduce with treatment. The patterns of changes in parameter values are seen in the bar graphs below.
Relationship of Imaging Parameters to Outcome
For anatomic imaging (see Table 1), only the volumes of the T2 and CE lesions at the post-RT scan were found to be significantly associated with PFS (P = .0076, hazard ratio [HR] 1.04 with 95% CI 1.01 and 1.07, and P = .016, HR 1.07 with 95% CI 1.01 and 1.14, 38 subjects, none censored) and OS (P = .0012, HR 1.05 with 95% CI 1.02 and 1.07, and P = .0027, HR 1.08 with 95% CI 1.02 and 1.14, 38 subjects, 3 censored). For metabolic imaging, the CNI2 lesion volume was associated with OS at both mid-RT (P = .015, 26 subjects, HR 1.05 with 95% CI 1.01 and 1.09, 3 censored) and post-RT (P < .0001, HR 1.08 with 95% CI 1.04 and 1.12, 35 subjects, 3 censored) but at only the post-RT scan for PFS (P = .039, HR 1.03 with 95% CI 1.00 and 1.06, 35 subjects, none censored). For the CNI2t volume, only the post-RT values showed an association with OS (P = .0021, HR 1.14 with 95% CI 1.06 and 1.23, 35 subjects with 3 censored), and there were no significant associations with PFS.
Table 1.
Association of imaging parameters with survival based upon Cox proportional hazards analysis, adjusting for age, KPS, and extent of resection. Metrics with P< .05 were considered to be significant and the P values marked with an asterisk are those that are less than .0024, which would be the cutoff for a Bonferroni correction for 21 independent statistical tests (corresponding to 7 types of parameters and 3 time points).
| Parameter | OS | PFS | ||||
|---|---|---|---|---|---|---|
| Pre-RT | Mid-RT | Post-RT | Pre-RT | Mid-RT | Post-RT | |
| Anatomic volumes | ||||||
| # Patients | 39 | 29 | 38 | 39 | 29 | 38 |
| T2 lesion | ns | ns | .0012* | ns | ns | .0076 |
| CE lesion | ns | ns | .0027 | ns | ns | .0160 |
| Metabolic volumes | ||||||
| # Patients | 35 | 26 | 35 | 35 | 26 | 35 |
| CNI >2 | ns | .0150 | <.0001* | ns | ns | .0390 |
| CNI >2 and T2 lesion | ns | ns | .0021* | ns | ns | ns |
| Choline relative to NAA in the same voxel | ||||||
| # Patients | 35 | 26 | 35 | 35 | 26 | 35 |
| median(CNI) | .0077 | .0099 | ns | ns | .021 | ns |
| maximum(CNI) | ns | .0008* | .0021* | ns | ns | ns |
| sum(CNI) | ns | .0028* | .0007* | ns | ns | ns |
| median(xsChoN) | .0010* | .0002* | .0006* | ns | .017 | .0340 |
| maximum xsChoN | .0180 | .0002* | .0004* | ns | ns | ns |
| sum(xsChoN) | ns | .0041 | .0007* | ns | ns | ns |
| Choline relative to creatine in the same voxel | ||||||
| # Patients | 35 | 26 | 35 | 35 | 26 | 35 |
| median(CCrI) | .0018* | .0046 | ns | ns | .017 | ns |
| maximum(CCrI) | .0230 | .0018* | .0056 | ns | ns | ns |
| sum(CCrI) | ns | .0031 | .0008* | ns | ns | ns |
| median(xsChoCr) | .0001* | .0110 | ns | ns | .028 | ns |
| maximum(xsChoCr) | .0250 | .0021* | .0120 | ns | ns | ns |
| sum(xsChoCr) | ns | .0068 | .0071 | ns | ns | ns |
| Lactate, lipid relative to nNAA in normal appearing brain | ||||||
| # Patients | 35 | 26 | 35 | 35 | 26 | 35 |
| median(nLac) | ns | ns | ns | ns | ns | ns |
| maximum(nLac) | ns | .0450 | ns | ns | ns | ns |
| sum(nLac) | ns | .0170 | .0007* | ns | ns | ns |
| median(nLip) | ns | ns | ns | ns | ns | ns |
| maximum(nLip) | ns | ns | ns | ns | ns | ns |
| sum(nLip) | ns | .0096 | .0191 | .0241 | ns | ns |
ns = P value not significant.
The magnitude and variations in the levels of choline relative to NAA and creatine were represented by the median, maximum, and sum of CNI, xsChoN, CCrI, and xsChoCr within the CNI2 lesion. All of these choline metrics showed a significant association with OS at the mid-RT time point (see Table 1), but only the maximum and sum values at the post-RT scan. The median values had a significant association with OS at the pre-RT scan. The sum of nLac and nLip at mid-RT and post-RT scans were associated with OS (see Table 1).
Serial Changes in Volumes of Anatomic Lesions
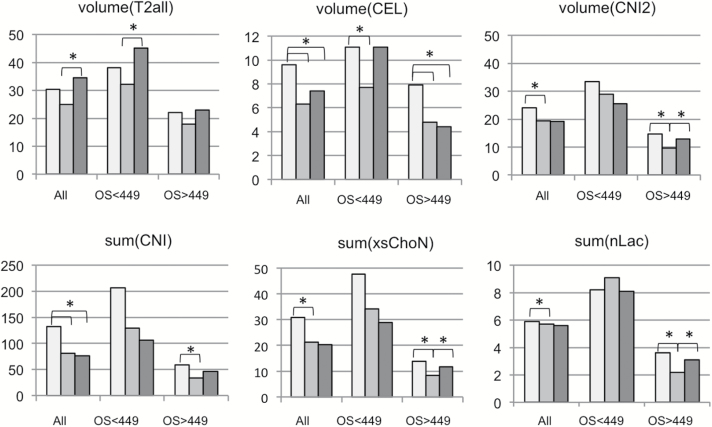
For the 25 patients with scans at all 3 time points, mean parameters at pre-RT and the percent changes in mean values from pre- to mid-RT and pre- to post-RT scans are seen in Table 2 and in the bar graphs in Fig. 4. The results include values for subgroups stratified by the median OS of 449 days. Overall, the T2 lesion volumes showed a downward trend from pre-RT to mid-RT, followed by a significant increase from mid- to post-RT (mean volumes of 30.4 cc, 24.9 cc, and 34.5 cc, respectively). Both the pre-RT mean T2 lesion volume and the percentage increase from pre-RT to post-RT were larger for the patients with OS <449 days (38.1 vs 22.0 cc, 18% vs 5%). The mean CE lesion volumes decreased significantly from pre- to mid-RT (P = .0001) and from pre- to post-RT (P = .0341), with the decrease being sustained from pre- to post-RT for the cohort with longer OS (P = .0034, 44% reduction). Note that the pre-RT mean volumes of the CE lesions for the patients with OS <449 days were larger compared with those with OS >449 days (11.1 vs 7.9 cc).
Table 2.
Temporal changes in anatomic lesion volumes and metabolic imaging parameters for patients with scans at all 3 time points, and those with OS either less than or greater than the median value of 449 days. These correspond to the graphs seen in Fig. 4. P values were obtained using a Wilcoxon signed rank test, P < .05 was considered significant. Because of the exploratory nature of the analysis, no corrections for multiple comparisons were applied, but it can be seen that many of the P values were less than .005.
| Parameter | Group | N | Pre-RTMean | Mid-RT Mean |
Post-RT Mean | Pre-RT to Mid-RT | Pre-RT to Post-RT | ||
|---|---|---|---|---|---|---|---|---|---|
| % change | P value | % change | P value | ||||||
| vol(T2 lesion) | All matched | 25 | 30.4 | 24.9 | 34.5 | –18% | ns | 13% | ns |
| OS <449 days | 13 | 38.1 | 32.1 | 45.1 | –16% | ns | 18% | ns | |
| OS >449 days | 12 | 22.0 | 17.9 | 23.0 | –19% | ns | 5% | ns | |
| vol(CE lesion) | All matched | 25 | 9.6 | 6.3 | 7.4 | –34% | .0001 | −23% | .0341 |
| OS <449 days | 13 | 11.1 | 7.7 | 11.1 | –31% | .0402 | 0% | ns | |
| OS >449 days | 12 | 7.9 | 4.8 | 7.9 | –39% | .0005 | –44% | .0034 | |
| vol(CNI >2) | All matched | 20 | 24.0 | 19.3 | 19.2 | –20% | .0117 | –20% | ns |
| OS <449 days | 10 | 33.4 | 28.9 | 25.5 | –13% | ns | –24% | ns | |
| OS >449 days | 10 | 14.6 | 9.7 | 12.8 | –34% | .0186 | –12% | ns | |
| sum(CNI) | All matched | 20 | 132.5 | 81.1 | 76.1 | –39% | .0028 | –43% | .0285 |
| OS <449 days | 10 | 206.4 | 128.9 | 106.5 | –38% | ns | –48% | ns | |
| OS >449 days | 10 | 58.7 | 33.2 | 46.2 | –43% | .0068 | –21% | ns | |
| sum(xsChoN) | All matched | 20 | 30.8 | 21.3 | 20.3 | –31% | .0035 | –34% | ns |
| OS <449 days | 10 | 47.4 | 34.1 | 28.9 | –29% | ns | –39% | ns | |
| OS >449 days | 10 | 13.8 | 8.4 | 11.7 | –29% | .0068 | –15% | ns | |
| sum(nLac) | All matched | 20 | 5.9 | 5.7 | 5.6 | –3% | .0371 | 5% | ns |
| OS <449 days | 10 | 8.2 | 9.1 | 8.1 | 11% | ns | –1% | ns | |
| OS >449 days | 10 | 3.6 | 2.2 | 3.1 | –39% | .0020 | –14% | ns | |
Fig. 4.
Bar plots of mean parameter values from pre-, mid-, and post-RT scans for all subjects with data from each time point and from subgroups with OS <449 days and OS >449 days. The parameters shown are T2 and CE lesion volume (n =25 subjects), the volume with CNI >2, the summed CNI, the summed xsChoN and the summed nLac within the CNI >2 region (n =20 subjects). Note that parameters and time points for which there is a large difference in values between the subgroups with short vs long survival are likely to be of particular interest for predicting outcome and indicating whether there has been a response to the treatment in a subset of patients. The light gray bars represent the pre-RT time point, the intermediate gray bars represent the mid-RT, and the dark gray bars represent the post-RT time point. The asterisk denotes a significant change between values at the indicated time points.
Serial Changes in Metabolite Imaging Metrics
Table 2 and the bar graphs in Fig. 4 also show changes in selected metabolic imaging parameters for the 20 subjects with MRSI data at all 3 time points. Note that many of the metabolic imaging metrics were more than 50% lower for the patients with OS >449 days compared with the patients having OS <449 days, indicating that the absolute values of these parameters were important in influencing outcome. The decreases in metabolic parameters from pre-RT to mid-RT were significant for the population overall, as well as for the subgroup with OS >449 days. This is consistent with there being an early reduction in tumor burden. Although the magnitude of the percentage change was similar from pre-RT to post-RT for the choline metrics, it was only significant for the summed CNI values (43%, P = .0285). Similar trends were seen for parameters describing changes in CCrI and xsChoCr. The summed lactate values showed a differential between subgroups of patients, with the mean summed nLac being 2.3-fold higher at pre-RT, 4.1-fold higher at mid-RT, and 2.6-fold higher at post-RT for the cohort with shorter OS.
Discussion
The analysis performed here shows that metrics derived from state-of-the-art MRSI provided early indicators of clinical outcome for patients undergoing treatment with radiation, temozolomide, and the protein kinase C inhibitor enzastaurin. From prior preclinical studies, this multimodality therapeutic approach was expected to act upon the tumor by directly damaging the DNA, impacting angiogenesis, reducing proliferation, and increasing apoptosis.8,9 The results of the quantitative analysis suggest that metabolic parameters at baseline, one month, and 2 months into treatment were able to capture alterations in tumor burden associated with these effects. The findings are consistent with prior studies using MRSI data to predict response to therapy,20–22 suggesting that it may be helpful for providing an early assessment of the utility of other combination therapies.
Previous studies have evaluated the relationship of anatomic imaging parameters to outcome for patients with newly diagnosed GBM receiving standard-of-care treatment with RT and temozolomide.23 These indicated that the T2 volume was associated with OS at pre- and post-RT and the CE lesion volume at only post-RT. While this is encouraging for the population as a whole, there have been numerous reports that RT and temozolomide can result in a temporary increase in the size of the CE lesion for a subset of patients (pseudoprogression). This poses a dilemma for the neuro-oncologist in terms of recommending whether to continue treatment or stop and seek alternative approaches.11 In the current study, the addition of enzastaurin further complicated the analysis, as it resulted in a substantial overall decrease in the volume of the CE lesion from pre- to mid-RT. This is presumably due to its anti-angiogenic effect.8 From mid-RT to post-RT, the CE lesion volume showed an increasing trend in the subgroup with shorter survival but was stable for the subgroup with longer survival. While the increase in the T2 lesion volume from mid-RT to post-RT in the subgroup with shorter survival may be caused by radiation effects or tumor progression, it appears that the combination treatment was less effective for these patients. Overall our results suggest that estimating changes in volumes of the CE and T2 lesion may be more sensitive than using visual assessments of changes in cross-sectional diameters, as is currently indicated by the RANO criteria.3
From the color overlays and the summary results shown in Table 2, it is clear that the metabolic lesion can extend beyond the region of contrast enhancement and can show a large decrease in magnitude and size with treatment. This is consistent with previous studies, which indicated that CNI values could identify nonenhancing tumor that was also different in shape and size than the T2 lesion.24 Using the MRSI data to define the spatial extent of tumor would change the target for RT,25–27 alter the definition of response to therapy, and assist in distinguishing residual and recurrent tumor from nonspecific treatment effects.28–30
Several different ways of representing the metabolic data were considered in this study. The CNI was developed and validated using image-guided tissue samples several years ago.18–19 As described in the Supplementary text, it has advantages over comparing changes in levels of the metabolites separately or considering the ratio of choline to NAA. This is because it provides values that represent the probability of differing from normal metabolite levels and can be used to automatically define regions that are likely to be abnormal. It is also robust in circumstances where the level of NAA is very low and could give a very high ratio value. The images shown in Figs. 1–3 provide examples of how using this metric as a color overlay can improve the visualization of the metabolic lesion. This is of particular interest for interpreting serial changes in tumor burden7,28,29 and avoiding complications associated with pseudoprogression and pseudoresponse.11
A similar algorithm was applied in our analysis to levels of choline and creatine in order to provide the CCrI. Experience in other studies has shown that the level of creatine can be quite variable in tumors, and so the CCrI may be less sensitive as a measure of the spatial extent of the lesion.23–28 The results shown in Table 1 indicate that if the CNI lesion is used as the reference region, metrics that describe both choline relative to NAA and choline relative to creatine are associated with OS. Table 2 highlights the early effects of treatment on metabolic parameters by showing that there were substantial reductions in choline metrics from pre- to mid-RT. These findings are consistent with the antiproliferative aspects of the combined therapies and explain the strong association of metabolic parameters from mid- and post-RT with OS.
The other metabolic parameters associated with OS were measures of excess normalized choline (xsChoN and xsChoCr) and ratios of lactate (nLac) and of lipid (nLip) relative to NAA in normal appearing brain. These findings are consistent with our previous analysis of metabolic parameters from patients being treated with RT and temozolomide.7 The 3D nature of the MRSI acquisition was important for including regions that were abnormal and providing voxels for estimating normal metabolite levels. The fact that so many of the choline metrics were associated with OS increases confidence in their value as predictors of outcome. The relatively small number of metabolic parameters associated with PFS may be due to the criteria for assessing progression having been based upon increases in the cross-sectional diameter of the CE lesion,2 which reflects differences in vessel permeability rather than changes in tumor metabolism.
The consideration of multiple time points and multiregion analysis was central to this study and helped to substantiate the interpretation of the data criteria. Of interest is whether the mid-RT scan, which is not done routinely in the clinic, adds to the information obtained. Although changes in anatomic and metabolic parameters could be detected at the post-RT scan, previous studies from our own and other groups have indicated that evaluating changes in diffusion or perfusion imaging parameters at this time point may be important for assessing early response to therapy.31,32 Combining these findings with metabolic data may provide even stronger predictors of outcome. Another factor of interest for future studies is whether voxel-by-voxel changes in metabolite levels may be more sensitive to detecting early changes in metabolite levels than observing summary values derived independently at each time point. This will be facilitated by recent developments in automatic prescription of the selected region, which can aid in defining similar regions for serial scans.33
Conclusion
This study demonstrated the use of MRSI data in assessing early treatment response in patients with newly diagnosed GBM undergoing treatment with a combination of radiation, temozolomide, and enzastaurin. The analysis highlights metrics describing the volume and levels of choline, lactate, and lipid at mid- and post-RT scans that were associated with OS. These results suggest that adding MRSI to routine follow-up scans and performing a quantitative analysis of the data obtained could aid clinicians in assessing the effects of other multimodality treatment regimens.
Supplementary Material
Supplementary material is available online at Neuro-Oncology (http://neuro-oncology.oxfordjournals.org/).
Funding
Funding was provided by NIH grants RO1 CA127612 and P01 CA118816.
Supplementary Material
Acknowledgments
The authors would like to thank Mary McPolin and Bert Jimenez for their assistance with data acquisition. Emma Essock-Burns, Adam Elkhaled, and Eugene Ozhinsky provided helpful input through numerous technical discussions.
Conflict of interest statement. The authors declare that they have no conflict of interest related to the material presented here.
References
- 1. Van Meir EG, Hadjipanayis CG, Norden AD, et al. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Macdonald DR, Cascino TL, Schold SC, Jr, et al. Response criteria for phase II studies of supratentorial malignant glioma. J. Clin. Oncol. 1990;8(7):1277–1280. [DOI] [PubMed] [Google Scholar]
- 3. Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol 2010;28:1963–72. [DOI] [PubMed] [Google Scholar]
- 4. Nelson SJ, Vigneron DB, Dillon WP. Serial evaluation of patients with brain tumors using volume MRI and 3D 1H MRSI. NMR Biomed. 1999;12(3):123–138. [DOI] [PubMed] [Google Scholar]
- 5. Crawford FW, Khayal IS, McGue C, et al. Relationship of pre-surgery metabolic and physiological MR imaging parameters to survival for patients with untreated GBM. J. Neurooncol. 2009;91(3):337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saraswathy S, Crawford FW, Lamborn KR, et al. Evaluation of MR markers that predict survival in patients with newly diagnosed GBM prior to adjuvant therapy. J. Neurooncol. 2009;91(1):69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Y, Lupo JM, Parvataneni R, et al. Survival analysis in patients with newly diagnosed glioblastoma using pre- and post-radiotherapy MR spectroscopic imaging. Neuro Oncol. 2013;15(5):607–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Teicher BA, Alvarez E, Menon K, et al. Antiangiogenic effects of a protein kinase Cbeta-selective small molecule. Cancer Chemother Pharmacol. 2002;49:69–77. [DOI] [PubMed] [Google Scholar]
- 9. Tabatabai G, Frank B, Wick A, et al. Synergistic antiglioma activity of radiotherapy and enzastaurin. Ann Neurol. 2007;61:153–161. [DOI] [PubMed] [Google Scholar]
- 10. Butowski N, Chang SM, Lamborn KR, et al. Phase II and pharmacogenomics study of enzastaurin plus temozolomide during and following radiation therapy in patients with newly diagnosed glioblastoma multiforme and gliosarcoma. Neuro Oncol. 2011;13(12):1331–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clarke JL, Chang S. Pseudoprogression and pseudoresponse: challenges in brain tumor imaging. Curr Neurol Neurosci Rep 2009;9:241–46. [DOI] [PubMed] [Google Scholar]
- 12. Park I, Chen AP, Zierhut ML, et al. Implementation of 3 T lactate-edited 3D 1H MR spectroscopic imaging with flyback echo-planar readout for gliomas patients. Ann Biomed Eng. 2011;39(1):193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ozhinsky E, Vigneron DB, Nelson SJ. Improved spatial coverage for brain 3D PRESS MRSI by automatic placement of outer-volume suppression saturation bands. J Magn Reson Imaging. 2011;33(4):792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. VTK: http://www.vtk.org
- 15. Li Y, Osorio JA, Ozturk-Isik E, et al. Considerations in applying 3D PRESS H-1 brain MRSI with an eight-channel phased-array coil at 3 T. Magnetic Resonance Imaging. 2006;24(10):1295–1302. [DOI] [PubMed] [Google Scholar]
- 16. Crane JC, Olson MP, Nelson SJ. SIVIC: Open-Source, Standards-Based Software for DICOM MR Spectroscopy Workflows. Int J Biomed Imaging. 2013;2013:169526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cunningham CH, Vigneron DB, Chen AP, et al. Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging. Magn Reson Med. 2005. Nov;54(5):1286–9. [DOI] [PubMed] [Google Scholar]
- 18. McKnight TR, Noworolski SM, Vigneron DB, et al. An automated technique for the quantitative assessment of 3D-MRSI data from patients with glioma. J Magn Reson Imaging. 2001;13(2):167–177. [DOI] [PubMed] [Google Scholar]
- 19. McKnight TR, von dem Bussche MH, Vigneron DB, et al. Histopathological validation of a three-dimensional magnetic resonance spectroscopy index as a predictor of tumor presence. J. Neurosurg. 2002;97(4):794–802. [DOI] [PubMed] [Google Scholar]
- 20. Muruganandham M, Clerkin PP, Smith BJ, et al. 3-Dimensional magnetic resonance spectroscopic imaging at 3 Tesla for early response assessment of glioblastoma patients during external beam radiation therapy. Int J Radiat Oncol Biol Phys. 2014;90(1):181–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laprie A, Catalaa I, Cassol E, et al. Proton magnetic resonance spectroscopic imaging in newly diagnosed glioblastoma: predictive value for the site of postradiotherapy relapse in a prospective longitudinal study. Int J Radiat Oncol Biol Phys. 2008;70(3):773–81. [DOI] [PubMed] [Google Scholar]
- 22. Lichy MP, Bachert P, Hamprecht F, et al. Application of (1)H MR spectroscopic imaging in radiation oncology: choline as a marker for determining the relative probability of tumor progression after radiation of glial brain tumors. Rofo. 2006;178(6):627–33. [DOI] [PubMed] [Google Scholar]
- 23. Li Y, Lupo JM, Polley MY, et al. Serial analysis of imaging parameters in patients with newly diagnosed glioblastoma multiforme. Neuro Oncol. 2011;13(5):546–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pirzkall A, Li X, Oh J, Chang S, Berger MS, Larson DA, Verhey LJ, Dillon WP, Nelson SJ. 3D MRSI for resected high-grade gliomas before RT: tumor extent according to metabolic activity in relation to MRI. Int J Radiat Oncol Biol Phys. 2004;59(1):126–37. [DOI] [PubMed] [Google Scholar]
- 25. Guo J, Yao C, Chen H, et al. The relationship between Cho/NAA and glioma metabolism: implementation for margin delineation of cerebral gliomas. Acta Neurochir (Wien). 2012;154(8):1361–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wright AJ, Fellows G, Byrnes TJ, et al. Pattern recognition of MRSI data shows regions of glioma growth that agree with DTI markers of brain tumor infiltration. Magn Reson Med. 2009;62(6):1646–51. [DOI] [PubMed] [Google Scholar]
- 27. Stadlbauer A, Buchfelder M, Doelken MT, et al. Magnetic resonance spectroscopic imaging for visualization of the infiltration zone of glioma. Cent Eur Neurosurg. 2011;72(2):63–9. [DOI] [PubMed] [Google Scholar]
- 28. Pirzkall A, McGue C, Saraswathy S, et al. Tumor regrowth between surgery and initiation of adjuvant therapy in patients with newly diagnosed glioblastoma. Neuro Oncol. 2009;11(6):842–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park I, Tamai G, Lee MC, et al. Patterns of recurrence analysis in newly diagnosed GBM following 3D conformal radiation therapy with respect to pre-RT MR spectroscopic findings. Int J Radiat Oncol Biol Phys. 2007;69(2):381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vöglein J, Tüttenberg J, Weimer M, et al. Treatment monitoring in gliomas: comparison of dynamic susceptibility-weighted contrast-enhanced and spectroscopic MRI techniques for identifying treatment failure. Invest Radiol. 2011;46(6):390–400. [DOI] [PubMed] [Google Scholar]
- 31. Mardor Y1, Pfeffer R, Spiegelmann R, et al. Early detection of response to radiation therapy in patients with brain malignancies using conventional and high b-value diffusion-weighted magnetic resonance imaging. J Clin Oncol. 2003;21(6):1094–100. [DOI] [PubMed] [Google Scholar]
- 32. Lemasson B, Chenevert TL, Lawrence TS, et al. Impact of perfusion map analysis on early survival prediction accuracy in glioma patients. Transl Oncol. 2013;6(6):766–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ozhinsky E, Vigneron DB, Chang SM, et al. Automated prescription of oblique brain 3D magnetic resonance spectroscopic imaging. Magn Reson Med. 2013;69(4):920–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.