Cerebrospinal fluid (CSF) is a potential source of tumor-derived DNA in patients with primary or secondary CNS tumors. Selected mutations in CSF circulating tumoral cell-free DNA (cfDNA) have been identified by PCR detection techniques, and more recently panels of genes have been assessed by targeted next-generation sequencing.1,2
As a matter of fact, different clinically validated platforms for mutational analysis with distinct analytical sensitivities are currently available in routine diagnostic practice. In our study we tracked the mutational repertoire in 2 cases of leptomeningeal metastatic disease in synchronous samples of circulating cfDNA derived from plasma and CSF using the mass spectrometry technology Sequenom MassARRAY and fast COLD-PCR followed by Sanger sequencing.
In Case 1, a 58-year-old male presenting with progressive hearing loss was diagnosed with a multifocal moderately differentiated lung adenocarcinoma with a right temporal brain metastatic lesion. Following metastasis resection and whole-brain radiation therapy, pulmonary lobectomy with lymphadenectomy was performed. Before further treatments the patient developed neoplastic meningitis with nodular lumbar enhancement on MRI without recurrence at the previous supratentorial metastatic site. CSF and plasma samples were obtained from the patient, and intrathecal chemotherapy with liposomal cytarabine was started. In the following weeks the patient’s performance status quickly deteriorated and death occurred 7 months after the initial diagnosis.
The patient in Case 2 was a 64-year-old female who presented with headache, vertigo, and drowsiness. CT and MRI demonstrated a right hemispheric cerebellar contrast-enhancing lesion diagnosed after resection as a lung adenocarcinoma metastasis. Staging assessments identified a lung neoplasm in the superior left lobe associated with mediastinal lymphadenopathies. Following neoadjuvant chemotherapy, pulmonary lobectomy with lymphadenectomy was performed, thus confirming a poorly differentiated lung adenocarcinoma. After 6 months, multiple cerebellar metastases developed and were treated with radiotherapy, obtaining a partial response. Three months later, symptoms suggestive for neoplastic meningitis were confirmed by MRI (infratentorial leptomeningeal nodules). Intrathecal chemotherapy with liposomal cytarabine was started; however, due to poor patient performance this treatment was stopped in favor of palliative care.
In both cases, DNA was extracted from the tumor resection specimen and from synchronous samples of plasma obtained from peripheral blood and of CSF obtained from a lumbar puncture performed before starting intrathecal chemotherapy.
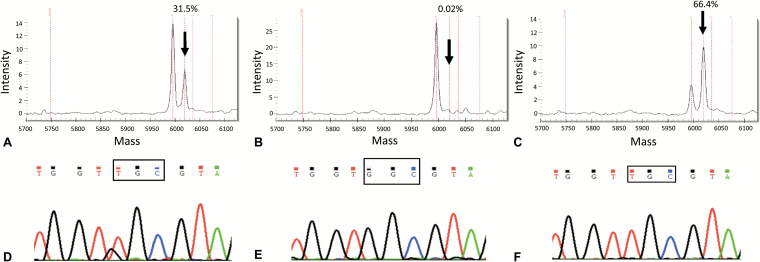
First, we carried out Sequenom using the Lung Status kit, which identifies the main nucleotide substitutions affecting EGFR and KRAS in lung adenocarcinoma and explores mutations at multiple codons of other relevant genes. The primary tumors of Case 1 and Case 2 harbored a p.G13C (c.37G>T, Fig. 1A) and a p.G12F (c.34_35delGGinsTT, Supplementary Fig. 1A) KRAS mutation, respectively. These KRAS mutations were absent in plasma but detectable in CSF cfDNA (0.02% vs 66.4% for Case 1, Fig. 1B–C; 0.02% vs 45% for Case 2, Supplementary Fig. 1B–C).
Fig. 1.
Detection of DNA alterations in the tumor tissue and in liquid biopsies of Case 1. The KRAS mutation c.37G>T, pG13C was diagnosed by Sequenom (Myriapod Lung Cancer kit, Diatech Pharmacogenetics) in the tumor resection sample (A); the estimated mutant allele frequency was 30% (arrow), suggesting a homozygous KRAS mutation, since the tumor cell content in the selected tumor area accounted for about 30%. The same driver mutation was not detected in plasma cfDNA samples (B), but was identified in CSF (C). The mutant allele frequency of >50% found in the CSF sample (arrow) suggested a likely homozygous pattern, in a way akin to the primary tumor. The highly sensitive fast COLD-PCR assay detected the same mutation in the tissue sample (D), confirming also the negativity of plasma (E) and positivity of CSF (F), using DNA input comprised in the range of absolute assay reproducibility.3
To explore the G>A or G>T KRAS mutations at codons 12/13 with higher analytical sensitivity than Sequenom, we then assessed all samples by fast COLD-PCR followed by Sanger sequencing. The DNA quantities were all comprised in a range of absolute reproducibility to detect a heterozygous mutation.3 The p.G13C as well as the p.G12F KRAS mutations identified by Sequenom were detected by fast COLD-PCR in tissue and in CSF-derived DNA (Fig. 1D, F; Supplementary Fig. 1D, 1F), while no KRAS mutations were identified in plasma (Fig. 1E and Supplementary Fig. 1E).
Finally, we measured the extent of DNA fragmentation by calculating a ratio between the concentrations of 247/115 bp real-time PCR-amplified products, suggestive of apoptosis or necrosis for ratios tending to 1 or 0, respectively.4 The ratios observed in plasma and CSF samples (51% vs 35% for Case 1 and 100% for both in Case 2) indicate a heterogeneous degree of apoptosis in the samples, confirming there is no consensus on the origin of the cfDNA (necrosis vs apoptosis).5,6
One may argue that we could not detect the KRAS mutation in plasma cfDNA due to the analytical sensitivity of our sequencing platform. For KRAS mutations the limit of detection estimated for Sequenom is near 5%, provided that a DNA input not below 1 ng is available. Notably, only the DNA inputs from CSF were below the cutoff (0.8 ng for Case 1 and 0.022 ng for Case 2), nevertheless we detected the mutation in these samples with a mutant allele frequency much higher than the sensitivity cutoff of 5% (ie, 66% for Case 1 and 45% for Case 2). Most importantly, no KRAS mutations were identified in plasma cfDNA even when analyzed with a 50-fold more sensitive detection method (fast COLD-PCR).
Our results provide another line of evidence that cfDNA derived from metastatic deposits in the brain with clinical features of meningeal carcinomatosis is more abundant in CSF compared with plasma. This is a phenomenon likely mediated by the intimate contact of CSF with tumor cells and by the compartmentalization of CSF from plasma due to the blood–brain barrier. Although accrued only in 2 patients, our data are the first to suggest that the relative enrichment of CSF in tumoral cfDNA compared with plasma can reliably allow for comprehensive sequencing of a panel of genes by Sequenom MassARRAY in a way akin to targeted panels of massively parallel sequencing. In addition, a second-level analysis of specific KRAS mutations by fast COLD-PCR followed by Sanger sequencing can be employed to guarantee a higher enrichment of specific mutant alleles with G>T or G>A substitutions.
Taken together, these results corroborate the notion that CSF likely represents a preferable source of representative liquid biopsy in brain metastatic lesions featuring meningeal carcinomatosis, at least when no extra-CNS localizations are evident. Liquoral liquid biopsies can help monitor changes in metastatic deposits in the CNS and may complement the diagnosis of meningeal carcinomatosis.2 Although a lumbar puncture is a more invasive procedure than a blood draw, the possible lack of representative tumoral cfDNA in the plasma may delay molecular diagnosis or lead to nonconclusive results in this delicate subset of patients. Otherwise, liquoral liquid biopsy can allow the identification of either actionable genetic alterations or a mutation correlated to resistance to targeted therapies leading to crucial changes in the treatment decision making.
In both of the presented cases the detected mutations were not strictly propaedeutic to tailor patient treatment (ie, not actionable); however, in lung cancer patients KRAS mutations have been (i) repeatedly implicated as markers of poor prognosis7,8 and (ii) shown to significantly correlate with brain metastatic disease.9 Finally, since preclinical evidence suggests the existence of at least 2 subgroups of mutant KRAS lung tumors with distinct genetic/metabolic signatures and unique therapeutic susceptibilities assessable on the basis of their relative mutant allelic content,10 combined quantitative and qualitative KRAS locus assessment may hold both prognostic and therapeutic usefulness.
Supplementary Material
Supplementary material is available online at Neuro-Oncology (http://neuro-oncology.oxfordjournals.org/).
Funding
This work was supported by the Italian Association of Cancer Research (AIRC, MFAG13310 to C.M.), the Ministry of University (ex 60% and 2015HAJH8E to C.M. and ex-60% P.C.) and Rete Oncologica Piemonte e Valle d’Aosta (to P.C.).
Conflict of interest statement: None to declare.
Supplementary Material
References
- 1. Pentsova EI, Shah RH, Tang J, et al. Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncol. 2016;34(20):2404–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. De Mattos-Arruda L, Mayor R, Ng CK, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mariani S, Di Bello C, Bonello L, et al. Flexible lab-tailored cut-offs for suitability of formalin-fixed tumor samples for diagnostic mutational analyses. PLoS One. 2015;10(4):e0121815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Umetani N, Giuliano AE, Hiramatsu SH, et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24(26):4270–4276. [DOI] [PubMed] [Google Scholar]
- 5. Giacona MB, Ruben GC, Iczkowski KA, et al. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17(1):89–97. [DOI] [PubMed] [Google Scholar]
- 6. Yörüker EE, Özgür E, Keskin M, et al. Assessment of circulating serum DNA integrity in colorectal cancer patients. Anticancer Res. 2015;35(4):2435–2440. [PubMed] [Google Scholar]
- 7. Lococo F, Gandolfi G, Rossi G, et al. Deep sequencing analysis reveals that KRAS mutation is a marker of poor prognosis in patients with pulmonary sarcomatoid carcinoma. J Thorac Oncol. 2016;11(8):1282–1292. [DOI] [PubMed] [Google Scholar]
- 8. Pan W, Yang Y, Zhu H, et al. KRAS mutation is a weak, but valid predictor for poor prognosis and treatment outcomes in NSCLC: a meta-analysis of 41 studies. Oncotarget. 2016;7(7):8373–8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao N, Wilkerson MD, Shah U, et al. Alterations of LKB1 and KRAS and risk of brain metastasis: comprehensive characterization by mutation analysis, copy number, and gene expression in non-small-cell lung carcinoma. Lung Cancer. 2014;86(2):255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kerr EM, Gaude E, Turrell FK, et al. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531(7592):110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.