Abstract
Background
The 15q11.2 BP1-BP2 microdeletion (Burnside-Butler susceptibility locus) is an emerging condition with over 200 individuals reported in the literature. TUBGCP5, CFYIP1, NIPA1 and NIPA2 genes are located in this chromosome 15 region and when disturbed individually are known to cause neurological, cognitive or behavioural problems as well as playing a role in both Prader-Willi and Angelman syndromes. These syndromes were the first examples in humans of genomic imprinting and typically caused by a deletion but involving the distal chromosome 15q11-q13 breakpoint BP3 and proximally placed breakpoints BP1 or BP2 of different parental origin. The typical 15q11-q13 deletion involves BP1 and BP3 and the typical type II deletion at BP2 and BP3. Several studies have shown that individuals with the larger type I deletion found in both Prader-Willi and Angelman syndromes are reported with more severe neurodevelopmental symptoms compared to those individuals with the smaller type II deletion.
Methods
The literature was reviewed and clinical and cytogenetic findings summarised in 200 individuals with this microdeletion along with the role of deleted genes in diagnosis, medical care and counseling of those affected and their family members.
Results
Reported findings in this condition include developmental delays (73% of cases) and language impairment (67%) followed by motor delay (42%), ADD/ADHD (35%) and autism spectrum disorder (27%). The de novo frequency has been estimated at 5 to 22% with low penetrance possibly related to subclinical manifestation or incomplete clinical information on family members. A prevalence of 0.6 to 1.3% has been identified in one study for patients with neurological or behavioural problems presenting for genetic services and chromosomal microarray analysis.
Conclusions
The summarised results indicate that chromosome 15q11.2 BP1-BP2 microdeletion is emerging as one of the most common cytogenetic abnormalities seen in individuals with intellectual impairment, autism spectrum disorder and other related behavioural or clinical findings but more research is needed.
INTRODUCTION
The 15q11.2 BP1–BP2 Microdeletion: Clinical Description
Individuals with a microdeletion of the 15q11.2 BP1-BP2 region or Burnside-Butler susceptibility locus can present with a wide range of clinical findings including intellectual disabilities and language delays found in greater than two-thirds of the individuals with this deletion along with neurodevelopmental behavioural disturbances (autism, ataxia, poor coordination, ADD, seizures), psychiatric problems (schizophrenia) and/or mild dysmorphic features (Murthy et al. 2007; Doornbos et al. 2009; Burnside et al. 2011; Abdelmoity et al. 2012; Jerkovich & Butler 2014; Cafferkey et al. 2013; Cox & Butler 2015). Not all individuals with this microdeletion will present with known clinical manifestations, an active area of study to further explain the non-penetrance nature of this emerging microdeletion disorder. The 15q11.2 BP1-BP2 microdeletion encompasses a 500 kb region located between proximal long arm breakpoints BP1 and BP2 in the centromeric side of the chromosome 15q11-q13 region involving four genes (i.e., TUBGCP5, CYFIP1, NIPA1, NIPA2) (Christian et al. 1995; Pujana et al. 2002; Chai et al. 2003; Bittel & Butler 2005; Cox & Butler 2015). This emerging cytogenetic disorder has an estimated prevalence ranging from 0.6%–1.3% of patients presenting for genetic services and chromosomal microarray analysis usually due to unexplained behaviour, cognitive and/or psychiatric problems (Burnside et al. 2011; de Wolf et al. 2013; Rosenfeld et al. 2013; Chaste et al 2014; Cox & Butler 2015; Hashemi et al. 2015). The prevalence rate is estimated at two to four fold increase compared with controls without a known or recognised phenotype (Itsara et al. 2009; de Kovel et al. 2010; Rosenfeld et al. 2013; Cox & Butler 2015).
A literature review and summary of clinical features from 200 individuals with an identified 15q11.2 BP1-BP2 microdeletion were reported by Cox and Butler (2015) and grouped into five categories: 1) developmental (73% of cases) and speech (67%) delays; 2) dysmorphic ears (46%) and palatal anomalies (46%); 3) writing (60%) and reading (57%) difficulties, memory problems (60%) and verbal IQ scores ≤75 (50%); 4) general behavioural problems, unspecified (55%) and 5) abnormal brain imaging (43%). Other clinical features were not considered as common including seizures/epilepsy (26%), autism spectrum disorder (27%), attention deficit disorder (ADD)/attention deficit hyperactivity disorder (ADHD) (35%), schizophrenia/paranoid psychosis (20%) and motor delay (42%) (see Table 1). Not all individuals with the deletion were clinically affected, yet the collection of findings appeared to share common biological pathways and therefore presumed genetic mechanisms. Neuropsychiatric and behaviour disturbances with mild dysmorphic features were associated with genomic imbalances of the 15q11.2 BP1-BP2 region, including the microdeletion and showed both incomplete penetrance or variable expressivity in those harboring this chromosome deletion requiring more research (Burnside et al. 2011; Cox & Butler 2015).
Table 1.
Summary of Growth and Development, Dysmorphic, Cognitive, Behavioural and Psychiatric Features with Other Related Medical Concerns Reported in 15q11.2 BP1-BP2 Microdeletion Disorder
| CATEGORY* | Total (%) |
|---|---|
| Growth and Development | |
| Microcephaly | 9/38 (24) |
| Developmental delay (general) | 126/172 (73) |
| Motor delay | 66/158 (42) |
| Speech delay | 99/148 (67) |
| Dysmorphic Features | |
| Dysmorphism, unspecified | 55/141 (39) |
| Broad forehead | 8/39 (21) |
| Dysmorphic ears | 9/39 (46) |
| Palatal abnormalities | 9/39 (46) |
| Intelligence and Academic Achievement | |
| ID with FSIQ ≤75/special education | 43/116 (37) |
| Verbal IQ ≤75 | 2/4 (50) |
| Performance IQ ≤75 | 1/3 (33) |
| Reading difficulties | 4/7 (57) |
| Writing difficulties | 3/5 (60) |
| Memory problems | 3/5 (60) |
| Behavioural and Psychiatric Problems | |
| General behaviour problems, unspecified | 75/136 (55) |
| Autism spectrum disorder | 43/161 (27) |
| Schizophrenia/paranoid psychosis | 4/20 (20) |
| OCD | 18/68 (26) |
| ODD | 16/68 (24) |
| ADD/ADHD | 28/80 (35) |
| Self-injurious behaviours | 18/68 (26) |
| Other Related Medical Concerns | |
| Seizures/epilepsy | 57/216 (26) |
| Ataxia/balance issues | 22/78 (28) |
| Abnormal brain imaging | 32/75 (43) |
Found in at least 20% of reported individuals and modified from Cox and Butler, 2015
The 15q11.2 BP1-BP2 Microdeletion: Clinical Cytogenetic Findings
The human chromosome 15 is classified as an acrocentric and contains five common breakpoint sites located in the proximal long arm of the chromosome (15q11-q13) and referred to as BP1–BP5 (Christian et al. 1995; Pujana et al. 2002; Chai et al. 2003). Breakpoints BP1 and BP2 are located at the proximal end of the 15q11-q13 region while breakpoints BP3, BP4 and BP5 are located more distally in the region. A cluster of low copy DNA repeats are found within this region and facilitate mis-pairing of homologous chromosome 15s during meiosis leading to an abnormal or non-allelic chromosome alignment or recombination and chromosome anomalies (Pujana et al. 2002; Locke et al. 2004). These low copy repeat sequences are called duplicons and contain pseudogenes (Eichler 1998; Pujana et al. 2002). These duplicons found within breakpoints BP1, BP2 and BP3 are characterised by the presence of the HERC2 gene at BP3 and HERC2 pseudogenes at breakpoints BP1 and BP2 (Pujana et al. 2002; Visser et al. 2012).
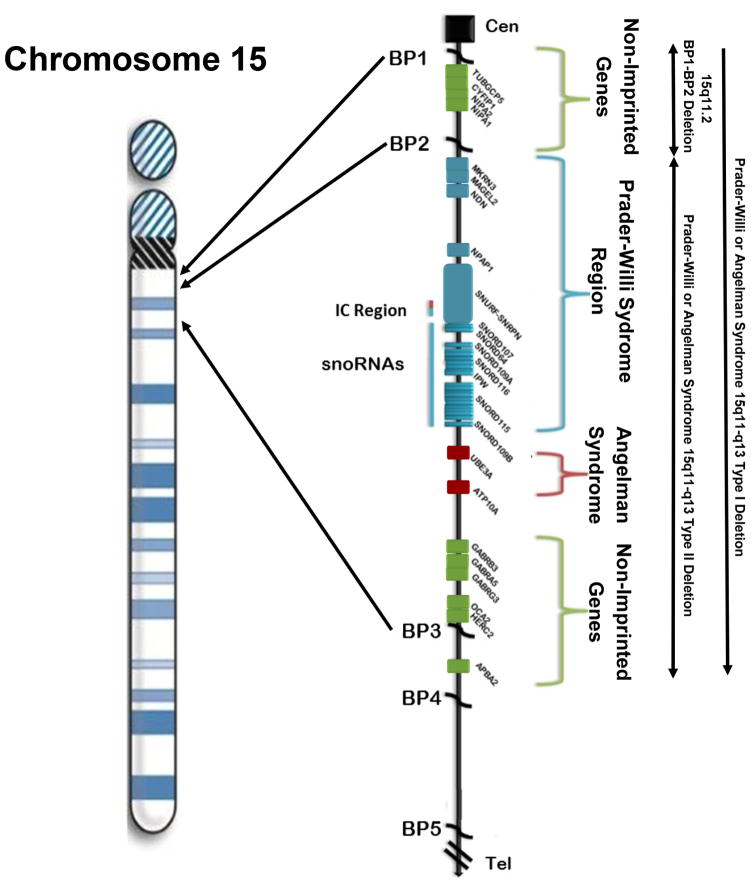
The first examples of errors in genomic imprinting in humans are Prader-Willi syndrome (PWS) and Angelman syndrome (AS) (Nicholls et al. 1989; Butler 1990; Butler et al. 2006; Williams et al. 2010; Cassidy et al. 2012; Angulo et al. 2015; Butler 2016). The two syndromes are entirely different clinically and most frequently caused by a deletion of either the paternal chromosome 15q11-q13 region in PWS or a maternal deletion of the same chromosome 15 region in AS. The different parental origins of the 15q11-q13 deletion in both syndromes involve the distal breakpoint BP3 and either the proximal BP1 or proximal BP2 breakpoints. Typical deletions of the 15q11-q13 in region are now classified as type I involving breakpoints BP1 and BP3 or type II involving BP2 and BP3 (Bittel & Butler 2005; Butler et al. 2006; Williams et al. 2010; Cassidy et al. 2012; Angulo et al. 2015; Butler 2016) (see Figure 1). The larger type I deletion is approximately 6.6 Mb in size and the smaller type II deletion is 5.3 Mb in size (Butler et al. 2008). Individuals with the larger type I deletion seen in both Prader-Willi and Angelman syndromes often have more learning and behavioural problems compared to those individuals with the smaller typical type II deletion (Butler et al 2004; Hartley et al. 2005; Milner et al. 2005; Varela et al. 2005; Bittel et al. 2006; Sahoo et al. 2007; Zarcone et al. 2007; Valente et al. 2013). Particularly more compulsions, self-injurious and other maladaptive behaviours and lower cognitive, reading and math scores are seen in those with type II deletion and PWS and more impaired speech and seizure activity in those individuals with AS.
Figure 1.
Chromosome 15 ideogram illustrates the gene pattern with symbols in linear order from the centromeric (Cen) end in the 15q11-q13 region. Genes in non-imprinted regions are shown in green, the Prader-Willi syndrome region in blue and the Angelman syndrome region in red. Chromosome 15q breakpoints (BP1, BP2, BP3, BP4, BP5) are shown with locations of the three recognized chromosome 15q deletions including 15q11.2 BP1-BP2.
The four genes found in the 15q11.2 BP1-BP2 region are NIPA1, NIPA2, CYFIP1 and TUBGCP5 which are found between breakpoints BP1 and BP2 which span approximately 500 kb and considered key genes causing behavioural and academic differences seen between Type I and Type II deletion genetic subtypes in individuals with these two rare genetic disorders (PWS and AS) (Butler et al 2004; Bittel & Butler 2005; Hartley et al. 2005; Milner et al. 2005; Varela et al. 2005; Bittel et al. 2006; Sahoo et al. 2007; Zarcone et al. 2007; Valente et al. 2013). The four genes are highly conserved and biallelically expressed with the NIPA1 (non-imprinted in Prader-Willi/Angelman syndrome 1) gene being studied most often and causes autosomal dominant hereditary spastic paraplegia and postural disturbance when mutated (Rainier et al. 2003; Chen et al. 2005; Goytain et al. 2007; Rainier et al. 2012). This gene is also known to mediate Mg2+ transport and is highly expressed in the brain (Goytain et al. 2007). The second gene is NIPA2 (non-imprinted in Prader-Willi/Angelman syndrome 2) and its encoded protein plays a role in renal Mg2+ transport (Goytain et al. 2007) and when mutated, causes childhood absence epilepsy (Jiang et al. 2012). The TUBGCP5 (tubulin gamma complex associated protein 5) gene is associated with attention deficit hyperactivity disorder (ADHD) and obsessive compulsive disorder (OCD) (de Wolf et al. 2013). The CYFIP1 (cytoplasmic fragile X mental retardation 1 FMR1 interacting protein 1) is a gene whose product interacts with FMRP, the protein coded by the FMR1 gene and associated with fragile X syndrome (FXS), the most common cause of intellectual disabilities in families (Hagerman & Hagerman 2002). FXS primarily affects males. The protein products of these two interrelated genes located on different chromosomes (i.e., X and 15) play important roles in the regulation of mRNAs found in the brain (Bozdagi et al. 2012; de Wolf et al. 2013). In addition, recent studies using stem cell research from patients with the 15q11.2 BP1-BP2 microdeletion showed abnormalities of dendritic spine formation indicating a role in neurodevelopment (Das et al. 2015).
A pattern of disturbed gene expression and associated behavioural findings in subjects with either PWS or Angelman syndrome having different genetic subtypes implicated genes within the 15q11.2 BP1 and BP2 genomic region (Butler et al. 2004; Bittel et al. 2006; Valente et al. 2013). Hence, Burnside et al. (2011) summarised findings from a large cohort of patients presenting for genetic services at a large commercial genetic testing laboratory for high resolution chromosomal microarrays and found that 0.86% of the approximate 17,000 individuals studied had an abnormality, either a deletion or duplication of the 15q11.2 BP1-BP2 region. They reported 69 subjects with the 15q11.2 microdeletion and 77 subjects with a microduplication which further supported that this genomic area is a susceptibility site or locus for neurological dysfunction and impaired development. More specifically, the 15q11.2 BP1-BP2 microdeletion correlated with language or motor delays, autism, behavioural problems, seizures and on occasion dysmorphism or congenital anomalies (Wong et al. 2013; Jerkovich & Butler 2014; Usrey et al. 2014). Findings from this large cohort of patients with the microdeletion identified by microarray analysis supported the original observations by Butler et al. (2004) of differences in behavioural patterns found in those individuals with PWS having the larger type I deletion compared with those PWS subjects having the smaller type II deletion. More recently, Ho et al. (2016) summarised ultra-high resolution chromosomal microarray results on 10,351 consecutive patients presenting for genetic services over a four year time period and reported that this emerging microdeletion syndrome was the leading finding of those with ASD only (N=5,694 patients) and those with ASD having other problems such as intellectual disability and/or multiple congenital anomalies (N=2,844 patients).
A report of genetic disturbances in the 15q11.2 BP1-BP2 region without PWS was first made by Murthy et al. (2007) in two individuals and later reported by Doornbos et al. (2009) in nine individuals. Most of those individuals in the two studies had behavioural related or neurological problems. Later, a cohort of 1,654 pediatric patients presenting with neurological problems reported by Abdelmoity et al. (2012) found that 21 (1.27%) of the subjects had a 15q11.2 BP1-BP2 defect or microdeletion. In their report, 87.5% of the 21 patients with the defect or microdeletion presenting for genetic services had the deletion and developmental delay or learning problems. Following this study a report by Cafferkey et al. (2012) showed that in 14,605 patients referred for microarray analysis, 83 (0.57%) had the 15q11.2 BP1-BP2 microdeletion; the vast majority presented with behavioural or developmental problems such as motor delays with deletions of NIPA1, NIPA2, TUBGCP5, and CYFIP1 genes - all found in the deleted region prone to both microdeletions or microduplications.
The 15q11.2 BP1-BP2 Microdeletion: Implications for Clinical Cytogenetics
Although both microdeletion or microduplications are recognised in the 15q11.2 BP1-BP2 region, not all individuals share an apparent clinical phenotype or clinically affected and contain genes showing incomplete or low penetrance for pathogenicity and with variable expressivity reminiscent of dominantly inherited genes. Data summarised in the literature from 66,462 individuals have shown that 0.25% of individuals with the 15q11.2 BP1-BP2 microdeletion (Itsara et al. 2009; de Kovel et al. 2010; Cooper et al. 2011; Cafferkey et al. 2013; Rosenfeld et al. 2013) have penetrance estimates at 10.4% representing an estimated two fold increase compared with the general population which could be explained by paucity or incomplete inheritance data (Rosenfeld et al. 2013). The estimated penetrance is low compared to other microdeletion syndromes such as the 16p11.2 deletion with a penetrance of 62% (Rosenfeld et al., 2013). A higher penetrance is often seen in copy number variants that have more de novo frequencies while low penetrance estimates may reflect subclinical presentation or manifestation of features that are recognized as components of disorders including neuropsychiatric disturbances in parents of affected individuals or in control cohorts.
This cytogenetic region when disturbed is associated with autism and other behaviour, learning or neurodevelopmental problems and one of the most common cytogenetic regions harboring genes for these clinical problems supported by a recent study of 10,351 consecutive patients (7,422 M; 2,929 F or 2.5:1 sex ratio) presenting for genetic services and microarray analysis (Ho et al. 2016). They reported that 5,694 patients had ‘any ASD’ designation as the sole testing indicator or in combination with any other testing indication. Those patients were further grouped as ‘ASD only’ (2,850 patients or 27.5%) or as ‘ASD+’ (2,844 or 27.4%). The ‘ASD+’ category represents those with ASD and another testing indication such as seizures, developmental delay and/or intellectual disability or multiple congenital anomalies and ‘ASD only’ refers to those with ASD as the only testing indication. The 15q11.2 BP1-BP2 microdeletion syndrome was found in 9% of the top 85 cytogenetic findings in their study. More males were found in their total study cohort of patients but more females showed the microdeletion compared with males. No apparent clinical differences were described based on gender and no parent of origin effect was identified or discussed. More detailed neuropsychiatric and behaviour assessments are needed from other family members (and parents) of those with the 15q11.2 BP1-BP2 deletion as well as more sophisticated genetic testing to better characterise the relationship between the cytogenetic defect and clinical presentation (variability and severity). This testing may include next generation (exome) sequencing or targeted sequencing of the four genes in the 15q11.2 BP1-BP2 region (or associated interactive genes in common pathways combined with detailed behaviour/psychiatric/cognitive measures of “affected” and “non-affected” family members with and without the microdeletion. This would allow determination of the coding genetic status of the “normal” or non-deleted alleles in families. In addition, clinical description and phenotypic findings may be incomplete poorly characterised or unavailable for parents and other family members having the microdeletion without concerted efforts required to undertake such testing and detailed assessments needed to thoroughly address these issues and for genotype-phenotype relationships.
The 15q11.2 BP1-BP2 deletion is reported to be de novo and ranges from 5% to 22% in individuals while the inheritance data to date for this microdeletion was studied by Cafferkey et al. (2013) in 43 subjects with this microdeletion and 22 individuals (51%) inherited the chromosome finding from a parent without known health or learning/behaviour problems. Ten of 29 (35%) of individuals inherited the microdeletion from an abnormal parent. Hence, modifying genes outside of the chromosome region may play a role and will require further investigations by examining interacting genes or pathways impacting on the phenotype and clinical presentation.
RESULTS AND DISCUSSION
Clinical Description
Patients with the 15q11.2 BP1-BP2 microdeletion do present regularly for genetic services with a range of clinical findings, most commonly intellectual disability and language delay seen in more than two-thirds of affected individuals with this deletion. Other clinical features or reported associated findings include neurobehavioural disturbances (poor coordination, ataxia, attention deficit disorder, seizures), autism, psychotic problems (schizophrenia) and dysmorphic features (reviewed by Cox & Butler 2015). Due to incomplete penetrance and variable expressivity, not all individuals with this microdeletion will present with these typical clinical manifestations and more research is needed on causation and understanding pathophysiology.
Genetic Laboratory Testing
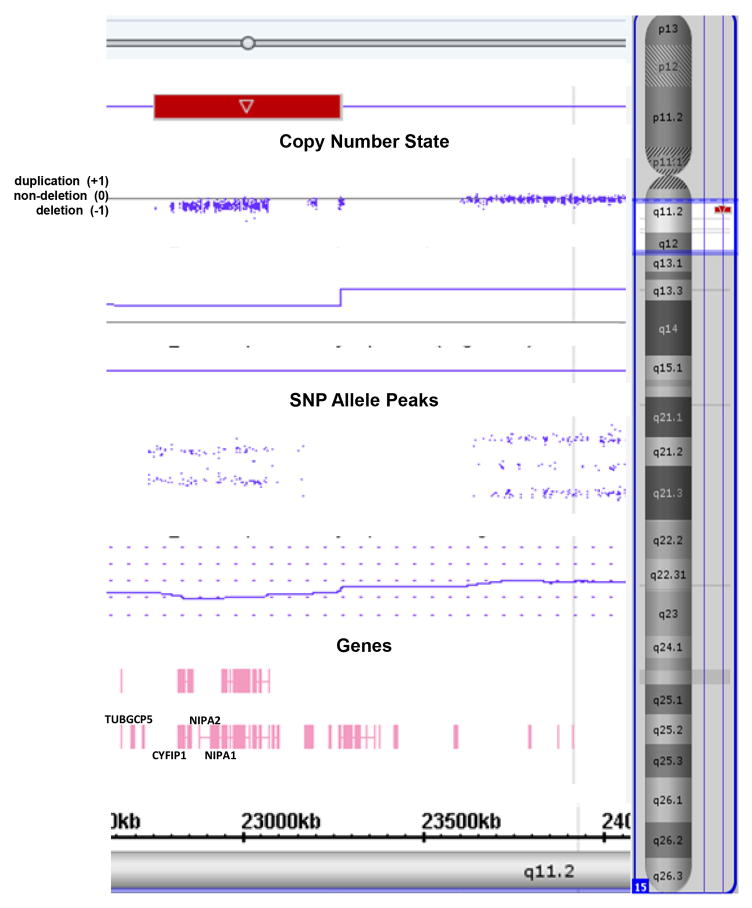
The 15q11.2 BP1-BP2 microdeletion encompasses a 500kb region located between the centromeric 15q11-q13 region breakpoints, BP1 and BP2 in the proximal long arm of chromosome 15 (Christian et al. 1995; Pujana et al. 2002; Chai et al. 2003). Four non-imprinted genes are found in this genomic region (i.e., TUBGCP5, CYFIP1 NIPA1, NIPA2) and are known to play a role in neurodevelopment and function when disturbed individually and contribute to seizures, spasticity and ataxia with cognitive and behavioural problems. This emerging microdeletion disorder containing neurodevelopmental susceptibility loci has a prevalence ranging from 0.6 to 1.3% in patients presenting primarily with behavioural, cognitive and/or psychiatric problems necessitating genetic testing. The estimated prevalence range is two to four-fold increased compared with controls without these findings (Burnside et al. 2011; Cafferkey et al. 2013; Rosenfeld et al. 2013; Cox & Butler 2015). There are several laboratory methods available to identify this cytogenetic anomaly including a chromosomal microarray (CMA) using oligonucleotide probes, high resolution SNP microarrays containing over 2 million genome-wide probes, genotyping informative chromosome 15q11.2 markers and specific FISH probes from this region for cytogenetic or diagnostic purposes. However, genotyping and FISH techniques are not as powerful or informative as newly developed and validated high resolution SNP microarrays and use of bioinformatics with computer software programs produced to analyse the generated large genetic datasets (see Figure 2).
Figure 2.
Chromosomal microarray analysis showing the typical 15q11.2 BP1-BP2 microdeletion (red rectangle) involving chromosome 15 breakpoints BP1 and BP2 with loss of four genes (TUBGCP5, CYFIP1, NIPA1, NIPA2) courtesy of Lineagen, Inc. (Salt Lake City, Utah). The copy number state determined by CNV probes and allele peaks by SNP probes confirm the deletion status of the 15q11.2 BP1-BP2 region.
Establishing the Diagnosis
Individuals reported with the 15q11.2 BP1-BP2 microdeletion may present with a wide range of clinical features and presentation. No formal diagnostic criteria has been established in this newly emerging microdeletion disorder. Individuals with the deletion have no obvious or distinctive clinical findings that are unique or specifically seen in the disorder, but they can present with neurodevelopmental behaviour problems or physical anomalies. More detailed and comprehensive studies are warranted to further characterise the molecular mechanism accounting for the low or incomplete penetrance and variable expressivity, common in this disorder and range of clinical presentation.
Genomic coordinates represent the minimum deletion size associated with 15q11.2 BP1-BP2 microdeletion or Burnside-Butler susceptibility locus and border the four genes (CYFIP1, TUBGCP5, NIPA1, NIPA2) in the genomic region. Coordinates for deletions may vary based on arrays used, designed and validated by the testing laboratory and level of genetic technology. The genomic coordinates can also change with updates from one human genome build to another (e.g. hg18 vs hg19). The size of the microdeletion calculated from the genomic positions given may also vary slightly from the expected microdeletion size due to the presence or absence of segmental duplications near the proximal breakpoints BP1 or BP2 of the 15q11-q13 region.
The diagnosis of the 15q11.2 BP1-BP2 microdeletion is established by having the deletion of the 500 kb heterozygous deletion at the proximal long arm of chromosome 15 between breakpoints BP1 and BP2 detected using several types of genetic testing methods (see Table 2). The four genes in the 15q11.2 BP1 and BP2 region are highly conserved and biallelically expressed with prone to formation of either microdeletions or macroduplications in the offspring due to common DNA low copy repeats or duplicons at these proximal breakpoint sites leading to misalignment of the homologous chromosome 15s in meiosis resulting in deletions or duplications of the region (Bittel & Butler 2005; Butler et al. 2006; Burnside et al. 2011; Cassidy et al. 2012; Cox & Butler 2015; Butler 2016). These genes then become candidates individually or collectively for causing the behavioural problems and clinical changes seen in this emerging cytogenetic disorder seen using high resolution microarrays and not visible with routine or high resolution chromosome studies without the use of new genetic technology and bioinformatics approach assisted with computer based programs for analysis of several hundred thousands to millions of genome-wide DNA probes, both copy number and single nucleotide polymorphisms (SNPs).
Table 2.
Genetic Testing, Characteristics and Identification of the 15q11.2 BP1-BP2 Microdeletion
| Test Method | Deletion, Location and Size | Sensitivity and Detection | |
|---|---|---|---|
| Proband | At-risk Family Members | ||
| FISH or MLPA using specific 15q11.2 probes | 15q11.2 BP1-BP2; 500 kb | 100% | 100% with appropriate probes (identify deletion/duplication) |
| High resolution chromosomal SNP microarray | Genome wide or 15q11.2 BP1-BP2 region | 100% | 100% identified copy number variants (deletion/duplication) and loss of heterozygosity |
| MLPA alone | 15q11.2 BP1-BP2 only | 100% | 100% identified copy number variants (deletion/duplication) |
| Chromosomal (oligonucleotide) microarray | Genome wide or 15q11.2 BP1-BP2 region | 100% | 100% identified cytogenetic deletion |
| Genotyping of polymorphic DNA probes from 15q11.2 BP1-BP2 region using patient and parental DNA | 15q11.2 BP1-BP2 region only | 100% | 100% identified with appropriate informative DNA probes and parental DNA sources |
Management and Treatment
Treatment is directed to address the specific features or clinical problems seen in each affected individual based on clinical genetics evaluation, neuropsychological, cognitive and developmental-behaviour assessments by trained clinical geneticists, psychologists, psychometrists and developmental experts.
Features seen in this microdeletion syndrome when present may require attention by healthcare providers and specialists to treat on a case by case basis. The reported features include developmental and speech delays found in the majority of subjects with reading, writing and memory difficulties and low verbal and performance IQs and may require educational intervention. Approximately one-half of reported subjects show general behavioural problems and abnormal brain imaging. These individuals should be evaluated by speech, behavioural and educational experts with skills in those with learning impairment. Additional findings include seizures, motor delay and postural disturbances due to brain development or abnormalities and will require neurological services and possibly include physical, occupational and rehabilitation therapies.
Congenital abnormalities have been reported (ear and palatal abnormalities) and surgical, ear, nose and throat specialists and dental care services may be needed. Behavioural and psychiatric disturbances are commonly reported including ASD, schizophrenia/paranoid psychosis, OCD and ADHD and these findings will require attention by members of the multidisciplinary health care team of providers. Hence, medical interventions and psychiatric/behavioural therapies will be implicated in affected individuals depending on the neurodevelopmental, cognitive and behavioural problems identified, their age of onset and family history. As indicated, care of the other clinical features such as dysmorphism and congenital anomalies (e.g., cleft palate), speech and language delay, balance problems, hyperactivity, ADD, autism and cognitive impairment, seizures and psychiatric (schizophrenia) will be required and dependent on age at onset, severity and duration. Clinical genetics and genetic counseling of family members and genetic testing of those at-risk are warranted. Currently, the chromosome 15q11.2 BP1-BP2 microdeletion can be detected prenatally by chromosomal microarray analysis and once identified, then genetic counseling to supply information regarding this disorder and examination for associated features such as congenital anomalies should be undertaken and questions entertained.
Surveillance and Consultation
Developmental, speech and behaviour assessments are indicated at regular intervals depending on the age at diagnosis and severity of clinical problems and onset. Referrals to health related experts and specialists are made for early intervention and enrollment in specialised developmental programs and interventions to address these concerns, if required and to treat the problems. The services of other specialists are needed such as neurology, if seizures and balance problems are present, child psychology and psychiatry for therapies and medication needs if behavioural problems, autism and schizophrenia exist and surgical consultation and intervention for congenital anomalies (e.g., cleft palate) along with developmental problems including physical limitations or impairments.
Genetic Services and Genetic Counseling
With advances in genetic technology and readily available testing, the 15q11.2 BP1-BP2 microdeletion is emerging as a relatively common and recognised condition with a susceptibility locus related to a collection of clinical findings and presentations with a low penetrance estimated at about 10%. Approximately one half of the defects are inherited from an apparently healthy parent while 35% of parents will have an abnormal presentation. The reported de novo deletion frequency is less than 25%. The children of a parent with this deletion will have a 50% chance of inheriting the deletion (Burnside et al. 2011; Cafferkey et al. 2013; Cox & Butler 2015). Prenatal testing can be accomplished by detecting the microdeletion, but cannot predict the phenotype based on the penetrance information and variable expressivity status known to date regarding this condition More research is needed to assist in gaining a better understanding of the genetics and causation, gene interaction and pathway description and analysis which should lead to improved recognition and potential treatment along with more accurate genetic counseling for families with affected family members.
The 15q11.2 BP1-BP2 microdeletion should be under consideration in individuals with microcephaly and motor/language delay. In addition, dysmorphic features including craniofacial and, palatal defects along with contractures/arthrogryposis are seen in about 40% of affected individuals (Wong et al. 2013; Jerkovich & Butler 2014; Usrey et al. 2014; Cox & Butler 2015; Ho et al. 2016). General behaviour problems are reported in greater than 55% of individuals in a recent review of the literature, specifically autism spectrum disorder; schizophrenia/paranoid psychosis; OCD, oppositional defiant disorder, ADD, ADHD; self-injury and anxiety (Cafferkey et al. 2013; Cox & Butler 2015). Affected individuals may present with only a subset of the clinical findings including intellectual disabilities or aberrant behaviour and without obvious dysmorphic or growth anomalies.
As discussed earlier, multiple laboratory methods can identify the microdeletion including FISH (Butler et al. 2006; Butler 2016), MS-MLPA (Bittel et al. 2007; Henkhaus et al. 2012) and microarray analyses (Roberts et al. 2014; Butler et al. 2014; Ho et al. 2016). Genotyping of appropriate informative DNA markers within the genomic region can also be used to identify the 15q11.2 BP1-BP2 microdeletion in at-risk family members. Access to parental samples is important for genetic testing to determine the recurrence risk estimates by utilizing the above genetic testing methods but no parent of origin effects are known for the four non-imprinted genes found in this cytogenetic region.
Similarly-related Genetic Disorders - The 15q11.2 BP1-BP2 Microduplication
The 15q11.2 BP1-BP2 chromosome region is reported in individuals with a separate similarly-related chromosome anomaly, the reciprocal microduplication that occurs at the same proximal breakpoint sites due to misalignment of homologous chromosome 15s in meiosis from the same mechanism that leads to the microdeletion. This phenomenon is also reported for other chromosome microdeletion/microduplication syndromes such as the 7q11.2 region involved with the 7q11.2 microduplication syndrome (Abbas et al. 2016) and the 7q11.2 microdeletion causing Williams syndrome (Pober 2010). The individuals with the 15q11.2 BP1-BP2 microduplication have a similar list of neuropsychiatric and behavioural findings as seen in those with the microdeletion involving the same four genes but more investigators are needed to further characterize the similarities (and differences) in the two cytogenetic conditions. These findings include intellectual disabilities, motor and speech delay, autism spectrum disorder; ADHD, coordination problems (ataxia), seizures, schizophrenia and occasionally growth anomalies with dysmorphic features (Burnside et al. 2011; Cox & Butler 2015). Specifically, phenotypic features in common include developmental delay/intellectual disability (at 40%), ADHD (at 38%), behavioural/neurological problems (at 62%), autism spectrum disorders (at 43%), hypotonia (at 13%), speech delay (at 49%), motor delay (at 27%), ataxia/balance problems (at 23%), seizures (at 12%), dysmorphic features (at 42%) as summarised by a review of 52 subjects in the report by Burnside et al. (2011). The duplication can either be de novo or inherited from an apparently unaffected parent, as seen in the families with affected members having the microdeletion of the same chromosome region.
Advanced Genomic Testing Methods
The microdeletion of the 15q11.2 BP1-BP2 region is not detected by routine G-banded chromosome analysis or other routine cytogenetic banding techniques. Copy number variation in this region is more readily detected by chromosomal microarray analysis, targeted deletion by fluorescence in situ hybridization (FISH), methylation specific-multiplex ligation dependent problem amplification (MS-MLPA) or high resolution chromosomal SNP arrays. Genotyping using DNA probes from this region with parental DNA can identify the deletion as well as its parental origin but not at the same level of detail as with high resolution microarrays.
CONCLUSION
Chromosomal microarrays using oligonucleotide probes or more recently validated arrays using SNP probes are now the standard of care approach to detect small submicroscopic deletions or duplications in the genome in individuals presenting for genetic services with developmental/language delay, behavioural problems/disorders and mild dysmorphic or growth anomalies, specifically the 15q11.2 BP1-BP2 microdeletion. Prior to 2011, the identification of the microdeletion was not interpreted as having clinical significance but now with over 200 individuals analysed in over 20 reports in the literature with many more identified in recent studies of individuals presenting for genetic services, this microdeletion disorder is emerging at the molecular and clinical level and appears to be the most common or one of the most common cytogenetic causes of autism and/or neurodevelopmental deficits in humans. More research is needed to further characterise and understand the genetic causation and gene interaction and for development of treatment plans for affected individuals and accurate genetic counseling.
Acknowledgments
The National Institute of Child Health and Human Development (NICHD) and grant number HD02528 is acknowledged.
References
- Abbas E, Cox DM, Smith T, Butler MG. The 7q11.23 microduplication syndrome: a clinical report with selective mutism and review of literature. Journal of Pediatric Genetics. 2016;5(3):129–40. doi: 10.1055/s-0036-1584361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmoity AT, LePichon JB, Nyp SS, Soden SE, Daniel CA, Yu S. 15q11.2 proximal imbalances associated with a diverse array of neuropsychiatric disorders and mild dysmorphic features. Journal of Developmental & Behavioral Pediatrics. 2012;33:570–576. doi: 10.1097/DBP.0b013e31826052ae. [DOI] [PubMed] [Google Scholar]
- Angulo MA, Butler MG, Cataletto ME. Prader-Willi syndrome: a review of clinical, genetic, and endocrine findings. Journal of Endocrinological Investestigation. 2015;38(12):1249–1263. doi: 10.1007/s40618-015-0312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel DC, Butler MG. Prader-Willi syndrome: clinical genetics, cytogenetics and molecular biology. Expert Review of Molecular Medicine. 2005;7(14):1–20. doi: 10.1017/S1462399405009531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel DC, Kibiryeva N, Butler MG. Expression of 4 genes between chromosome 15 breakpoints 1 and 2 and behavioral outcomes in Prader-Willi syndrome. Pediatrics. 2006;118:e1276–e1283. doi: 10.1542/peds.2006-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel DC, Kibiryeva N, Butler MG. Methylation-specific multiplex ligation-dependent probe amplification analysis of subjects with chromosome 15 abnormalities. Genetic Testing. 2007;11(4):467–75. doi: 10.1089/gte.2007.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Dorr N, Pilorge M, Takahashi N, Buxbaum JD. Haploinsufficiency of Cyfip1 produces fragile X-like phenotypes in mice. PLoS One. 2012;7(8):e42422. doi: 10.1371/journal.pone.0042422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside RD, Pasion R, Mikhail FM, Carroll AJ, Robin NH, Youngs EL, et al. Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: A susceptibility region for neurological dysfunction including developmental and language delay. Human Genetics. 2011;130:517–528. doi: 10.1007/s00439-011-0970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG. Prader-Willi syndrome: current understanding of cause and diagnosis. American Journal of Medical Genetics. 1990;35(3):319–332. doi: 10.1002/ajmg.1320350306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG. Single gene and syndromic causes of obesity: Illustrative examples. Progressive Molecular Biology and Translational Science. 2016;140:1–45. doi: 10.1016/bs.pmbts.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Bittel DC, Kibiryeva N, Talebizadeh Z, Thompson T. Behavioral differences among subjects with Prader-Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics. 2004;113:565–573. doi: 10.1542/peds.113.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Fischer W, Kibiryeva N, Bittel DC. Array comparative genomic hybridization (aCGH) analysis in Prader-Willi syndrome. American Journal of Medical Genetics A. 2008;146A:854–860. doi: 10.1002/ajmg.a.32249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Lee PDK, Whitman BY. Management of Prader-Willi Syndrome. New York: Springer-Verlag; 2006. [Google Scholar]
- Butler MG, Usrey K, Roberts JL, Schroeder SR, Manzardo AM. Clinical presentation and microarray analysis of Peruvian children with atypical development and/or aberrant behavior. Genetics Research International. 2014;2014:408516. doi: 10.1155/2014/408516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafferkey M, Ahn JW, Flinter F, Ogilvie C. Phenotypic features in patients with 15q11.2 (BP1-BP2) deletion: Further delineation of an emerging syndrome. American Journal of Medical Genetics A. 2013;164A:1916–1922. doi: 10.1002/ajmg.a.36554. [DOI] [PubMed] [Google Scholar]
- Cassidy SB, Schwartz S, Miller JL, Driscoll DJ. Prader-Willi syndrome. Genetics in Medicine. 2012;14(1):10–26. doi: 10.1038/gim.0b013e31822bead0. [DOI] [PubMed] [Google Scholar]
- Chai JH, Locke DP, Greally JM, Knoll JH, Ohta T, Dunai J, et al. Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader-Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. American Journal of Human Genetics. 2003;73:898–925. doi: 10.1086/378816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaste P, Sanders SJ, Mohan KN, Klei L, Song Y, Murtha MT, et al. Modest impact on risk for autism spectrum disorder of rare copy number variants at 15q11.2, specifically breakpoints 1 to 2. Autism Research. 2014;7:355–362. doi: 10.1002/aur.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Song C, Guo H, Xu P, Huang W, Zhou Y, et al. Distinct novel mutations affecting the same base in the NIPA1 gene cause autosomal dominant hereditary spastic paraplegia in two Chinese families. Human Mutation. 2005;2(5):135–141. doi: 10.1002/humu.20126. [DOI] [PubMed] [Google Scholar]
- Christian SL, Robinson WP, Huang B, Mutirangura A, Line MR, Nakao M, et al. Molecular characterization of two proximal deletion breakpoint regions in both Prader-Willi and Angelman syndrome patients. American Journal of Human Genetics. 1995;57(1):40–48. [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nature Genetics. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DM, Butler MG. The 15q11.2 BP1-BP2 microdeletion syndrome: a review. International Journal of Molecular Sciences. 2015;16(2):4068–4082. doi: 10.3390/ijms16024068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das DK, Tapias V, D’Aiuto L, Chowdari KV, Francis L, Zhi Y, et al. Genetic and morphological features of human iPSC-derived neurons with chromosome 15q11.2 (BP1-BP2) deletions. Molecular Neuropsychiatry. 2015;1(2):116–123. doi: 10.1159/000430916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wolf V, Brison N, Devriendt K, Peeters H. Genetic counseling for susceptibility loci and neurodevelopmental disorders: The del15q11.2 as an example. American Journal of Medical Genetics A. 2013;161A:2846–2854. doi: 10.1002/ajmg.a.36209. [DOI] [PubMed] [Google Scholar]
- Doornbos M, Sikkema-Raddatz B, Ruijvenkamp CA, Dijkhuizen T, Bijlsma EK, Gijsbers AC, et al. Nine patients with a microdeletion 15q11.2 between breakpoints 1 and 2 of the Prader-Willi critical region, possibly associated with behavioral disturbances. European Journal of Medical Genetics. 2009;52:108–115. doi: 10.1016/j.ejmg.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Eichler EE. Masquerading repeats: Paralogous pitfalls of the human genome. Genome Research. 1998;8:758–762. doi: 10.1101/gr.8.8.758. [DOI] [PubMed] [Google Scholar]
- Goytain A, Hines RM, El-Hussein A, Quamme GA. NIPA1 (SPG6), the basis for autosomal dominant form of hereditary spastic paraplegia, encodes a functional Mg2+ transporter. Journal of Biological Chemistry. 2007;282:8060–8068. doi: 10.1074/jbc.M610314200. [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Hagerman PJ. Fragile X syndrome: Diagnosis, treatment and research. 3. The John Hopkins University Press; Baltimore, MD, USA: 2002. [Google Scholar]
- Hartley SL, Maclean WE, Jr, Butler MG, Zarcone J, Thompson T. Maladaptive behaviors and risk factors among the genetic subtypes of Prader-Willi syndrome. American Journal of Medical Genetics A. 2005;136:140–145. doi: 10.1002/ajmg.a.30771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi B, Bassett A, Chitayat D, Chong K, Feldman M, Flanagan J, et al. Deletion of 15q11.2 (BP1-BP2) region: further evidence for lack of phenotypic specificity in a pediatric population. American Journal of Medical Genetics A. 2015;167A(9):2098–2102. doi: 10.1002/ajmg.a.37134. [DOI] [PubMed] [Google Scholar]
- Henkhaus RS, Kim SJ, Kimonis VE, Gold JA, Dykens EM, Driscoll DJ, et al. Methylation-specific multiplex ligation-dependent probe amplification and identification of deletion genetic subtypes in Prader-Willi syndrome. Genetic Testing and Molecular Biomarkers. 2012;16(3):178–86. doi: 10.1089/gtmb.2011.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KS, Wassman ER, Baxter AL, Hensel CH, Martin MM, Prasad A, et al. Chromosomal Microarray Analysis of Consecutive Individuals with Autism Spectrum Disorders Using an Ultra-High Resolution Chromosomal Microarray Optimized for Neurodevelopmental Disorders. Int J Mol Sci. 2016;17(12) doi: 10.3390/ijms17122070. pii: E2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, et al. Population analysis of large copy number variants and hotspots of human genetic disease. American Journal of Human Genetics. 2009;84(2):148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerkovich AM, Butler MG. Further phenotypic expansion of 15q11.2 BP1-BP2 microdeletion (Burnside-Butler) syndrome. Journal of Pediatric Genetics. 2014;3:41–44. doi: 10.3233/PGE-14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Zhang Y, Zhang P, Sang T, Zhang F, Ji T, et al. NIPA2 located in 15q11.2 is mutated in patients with childhood absence epilepsy. Human Genetics. 2012;131:1217–1224. doi: 10.1007/s00439-012-1149-3. [DOI] [PubMed] [Google Scholar]
- Locke DP, Segraves R, Nicholls RD, Schwartz S, Pinkel D, Albertson DG, et al. BAC microarray analysis of 15q-q13 rearrangements and the impact of segmental duplications. Journal of Medical Genetics. 2004;41:175–182. doi: 10.1136/jmg.2003.013813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner KM, Craig EE, Thompson RJ. Prader-Willi syndrome: Intellectual abilities and behavioural features by genetic subtype. Journal of Child Psychology and Psychiatry. 2005;46:1089–1096. doi: 10.1111/j.1469-7610.2005.01520.x. [DOI] [PubMed] [Google Scholar]
- Murthy SK, Nygren AO, El Shakankiry HM, Schouten JP, Al Khayat Al, Ridha A, et al. Detection of a novel familial deletion of four genes between BP1 and BP2 of the Prader-Willi/Angelman syndrome critical region by oligo-array CGH in a child with neurological disorder and speech impairment. Cytogenetics and Genome Research. 2007;116(1–2):135–140. doi: 10.1159/000097433. [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Knoll JH, Butler MG, Karam S, Lalande M. Genetic imprinting suggested by maternal heterodisomy in nondeletion Prader-Willi syndrome. Nature. 1989;342(6247):281–285. doi: 10.1038/342281a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober BR. Williams-Beuren syndrome. New England Journal of Medicine. 2010;362(3):239–352. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- Pujana MA, Nadal M, Guitart M, Armengol L, Gratacos M, Estivill X. Human chromosome 15q11-q14 regions of rearrangements contain clusters of LCR15 duplicons. European Journal of Human Genetics. 2002;10:26–35. doi: 10.1038/sj.ejhg.5200760. [DOI] [PubMed] [Google Scholar]
- Rainier S, Chai JH, Tokarz D, Nicholls RD, Fink JK. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) American Journal of Human Genetics. 2003;73:967–971. doi: 10.1086/378817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainier S, Chai JH, Tokarz D, Nicholls RD, Fink JK. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) American Journal of Human Genetics. 2012;73:967–971. doi: 10.1086/378817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JL, Hovanes K, Dasouki M, Manzardo AM, Butler MG. Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders or learning disability presenting for genetic services. Gene. 2014;535(1):70–78. doi: 10.1016/j.gene.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Coe BP, Eichler EE, Cuckle H, Shaffer LG. Estimates of penetrance for recurrent pathogenic copy-number variants. Genetics in Medicine. 2013;15:478–481. doi: 10.1038/gim.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T, Bacino CA, German JR, Shaw CA, Bird LM, Kimonis V, et al. Identification of novel deletions of 15q11q13 in Angelman syndrome by array-CGH: Molecular characterization and genotype-phenotype correlations. European Journal of Human Genetics. 2007;15:943–949. doi: 10.1038/sj.ejhg.5201859. [DOI] [PubMed] [Google Scholar]
- Usrey KM, Williams CA, Dasouki M, Fairbrothe LC, Butler MG. Congenital arthrogryposis: An extension of the 15q11.2 BP1-BP2 microdeletion syndrome? Case Report Genetics. 2014;2014:127258. doi: 10.1155/2014/127258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente KD, Varela MC, Koiffmann CP, Andrade JQ, Grossmann R, Kok F, et al. Angelman syndrome caused by deletion: A genotype-phenotype correlation determined by breakpoint. Epilepsy Research. 2013;105:234–239. doi: 10.1016/j.eplepsyres.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Varela MC, Kok F, Setian N, Kim CA, Koiffmann CP. Impact of molecular mechanisms, including deletion size, on Prader-Willi syndrome phenotype: Study of 75 patients. Clinical Genetics. 2005;67:47–52. doi: 10.1111/j.1399-0004.2005.00377.x. [DOI] [PubMed] [Google Scholar]
- Visser M, Kayser M, Palstra RJ. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Research. 2012;22(3):446–455. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CA, Driscoll DJ, Dagli AI. Clinical and genetic aspects of Angelman syndrome. Genetics in Medicine. 2010;12(7):385–395. doi: 10.1097/GIM.0b013e3181def138. [DOI] [PubMed] [Google Scholar]
- Wong D, Johnson SM, Young D, Iwamoto L, Sood S, Slavin TP. Expanding the BP1-BP2 15q11.2 microdeletion phenotype: Tracheoesophageal fistula and congenital cataracts. Case Report Genetics. 2013;2013:801094. doi: 10.1155/2013/801094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarcone J, Napolitano D, Peterson C, Breidbord J, Ferraioli S, Caruso-Anderson M, et al. The relationship between compulsive behavior and academic achievement across the three genetic subtypes of Prader-Willi syndrome. Journal of Intellectual Disability Research. 2007;51(Pt.6):478–487. doi: 10.1111/j.1365-2788.2006.00916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]