Abstract
Background.
Patients with meningiomas have widely divergent clinical courses. Some entirely recover following surgery alone, while others have relentless tumor recurrences. This clinical conundrum is exemplified by rhabdoid meningiomas, which are designated in the World Health Organization Classification of Tumours as high grade, despite only a subset following an aggressive clinical course. Patient management decisions are further exacerbated by high rates of interobserver variability, biased against missing possibly aggressive tumors. Objective molecular determinants are needed to guide classification and clinical decision making.
Methods.
To define genomic aberrations of rhabdoid meningiomas, we performed sequencing of cancer-related genes in 27 meningiomas from 18 patients with rhabdoid features and evaluated breast cancer [BRCA]1–associated protein 1 (BAP1) expression by immunohistochemistry in 336 meningiomas. We assessed outcomes, germline status, and family history in patients with BAP1-negative rhabdoid meningiomas.
Results.
The tumor suppressor gene BAP1, a ubiquitin carboxy-terminal hydrolase, is inactivated in a subset of high-grade rhabdoid meningiomas. Patients with BAP1-negative rhabdoid meningiomas had reduced time to recurrence compared with patients with BAP1-retained rhabdoid meningiomas (Kaplan–Meier analysis, 26 mo vs 116 mo, P < .001; hazard ratio 12.89). A subset of patients with BAP1-deficient rhabdoid meningiomas harbored germline BAP1 mutations, indicating that rhabdoid meningiomas can be a harbinger of the BAP1 cancer predisposition syndrome.
Conclusion.
We define a subset of aggressive rhabdoid meningiomas that can be recognized using routine laboratory tests. We implicate ubiquitin deregulation in the pathogenesis of these high-grade malignancies. In addition, we show that familial and sporadic BAP1-mutated rhabdoid meningiomas are clinically aggressive, requiring intensive clinical management.
Keywords: BAP1, exome sequencing, rhabdoid meningiomas
Importance of the Study
Meningiomas with rhabdoid features represent a class of potentially aggressive tumors with high rate of recurrence, though not all meningiomas with these histologic findings display the same natural history. To understand the molecular signature that discriminates this spectrum of clinical course, we performed a genomic characterization of rhabdoid meningiomas. We show that the tumor suppressor gene BAP1 is inactivated in 6 high-grade, rhabdoid meningiomas. In addition to patients with somatic BAP1 loss, 2 patients carried germline mutations, indicating that such meningiomas can arise as part of the BAP1 cancer predisposition syndrome. Furthermore, we demonstrate that BAP1 loss can be detected by immunohistochemistry, and the addition of this routine test can help risk-stratify which patients require intensive clinical management with close surveillance and consideration for adjuvant therapies.
Meningiomas are the most common primary tumor of the CNS and comprise over a dozen subtypes.1 The genetic aberrations that drive tumorigenesis have been identified for many of the common benign subtypes, but not for some of the rare World Health Organization (WHO) grade II and III subtypes for which achieving surgical cure is less likely.2–7 While many meningiomas are sporadic, some arise as part of tumor predisposition syndromes such as neurofibromatosis type 2 due to mutations in NF28 and familial multiple meningioma syndrome due to mutations in SMARCE1.6,9 Identifying patients with inherited forms of meningiomas can illuminate the pathogenesis of these tumors as well as guide genetic counseling.
Recent studies have demonstrated that the patterns of mutations and chromosomal aberrations in many sporadic meningiomas are strongly associated with distinct histologic subtypes as well as the location of the tumors within the CNS.3,4,7 Anterior skull base meningiomas often harbor mutations in SMO or in the ubiquitin ligase TRAF7. Mutations in AKT1, PIK3CA, or KLF4 often co-occur with TRAF7 mutations.4,7 Posterior skull base meningiomas and meningioma of the cerebral convexities often harbor sporadic mutations in NF2, and such meningiomas often display fibroblastic histology. Angiomatous meningiomas generally lack NF2 mutations but have multiple chromosomal polysomies.2
Rhabdoid meningioma is a meningioma subtype10,11 codified in the WHO Classification of Tumours as a highly aggressive grade III malignancy with high rates of recurrence and mortality. However, clinical experience suggests that meningiomas with rhabdoid features are biologically heterogeneous; some tumors have anaplastic high-grade histologic features, while others lack overt features of malignancy.12 Those lacking anaplastic features appear to follow a benign clinical course, even if rhabdoid features are well developed and extensive throughout the tumor.12 Defining the rhabdoid meningioma subtype is also confounded by their rarity and heterogeneity, leading to significant variations in diagnosis,13 with attendant implications for determining which patients require adjuvant therapy. Therefore, understanding the genetic drivers of high-grade rhabdoid meningiomas would more precisely facilitate diagnosis, prognosis, and management for patients with meningiomas.
Methods
Pathologic Examination and Clinical History
Histopathologic diagnosis and tumor purity were confirmed by review of the hematoxylin and eosin stained sections by 2 neuropathologists (S.S., M.A.) and a subset of cases were reviewed by R.A.V, A.P, and C.G. Information of the clinical history was collected from the patients’ electronic medical records. This study was approved by the human subject institutional review board of the Dana-Farber/Brigham and Women’s Cancer Center, Massachusetts General Hospital, the Mayo Clinic, and the University of California–San Francisco.
Immunohistochemistry
Immunohistochemistry (IHC) was performed using a commercially available mouse monoclonal antibody to BAP1 (C-4; Santa Cruz #sc-28383) on paraffin-embedded formalin-fixed tissue. Staining details can be found in the Supplementary materials. Cases were scored as negative for BAP1 when staining was lost in essentially all or nearly all (>95%) cells that were overtly tumor cells. Staining was deemed technically successful in cases with BAP1 immunoreactive stromal cells such as fibroblasts, lymphocytes, and endothelial cells, which serve as a robust and reliable positive control for staining.
DNA Sample Preparation
DNA was prepared using standard techniques as described in the Supplementary materials.
Targeted Exome Sequencing
Samples RM-6, RM-15, RM-16, and RM-5924 were sequenced using one of 2 different assays as previously described2,14 (see Supplementary materials). Sequencing was performed to a mean depth of 80X and analysis was performed as previously described.2,14 Raw sequencing data were processed using the Picard tools pipeline and the Genome Analysis Toolkit (GATK).15,16 Mutation analysis for single nucleotide variants (SNVs) was performed using MuTect v1.1.417; indel calling was performed using the GATK SomaticIndelDetector tool; SNVs and indels were annotated using Oncotator. To analyze somatic copy number alterations from whole exome data, we used the ReCapSeg algorithm, which assesses homologue-specific copy ratios (HSCRs) from segmental estimates of multipoint allelic copy ratios at heterozygous loci incorporating the statistical phasing software (Beagle) and population haplotype panels (HapMap3).18–20
Whole Exome Sequencing and Phylogenetic Analysis
Whole exome sequencing for RM-23 and RM-31 and matched normal DNA from the corresponding patients was performed using Broad Institute platforms as previously described.3 Libraries were constructed and sequenced on Illumina HiSeq 2000 using 76 bp paired-end reads as previously described.21,22 Reads were aligned using the Burrows-Wheeler Aligner, de-multiplexed with Picard tools, and sorted using Samtools. Quality control and germline single nucleotide polymorphism fingerprinting were conducted using the previously described firehose pipeline.21 Somatic mutations were called using MuTect22 and Strelka.23 Somatic copy number alterations for individual alleles were inferred from sequencing read depth of whole exome sequencing data, and cancer-cell fraction values for each mutation were inferred from allelic fractions and corresponding copy number alterations using Absolute v1.2 as previously described.20 Phylogenetic trees were generated based on somatic mutations only using a branched-sibling evolutionary model under the assumption that related cancer tissues are descended from a common ancestral clone.24 Data, including sequence data and analyses, will be available for download from the database of Genotypes and Phenotypes (dbGaP).
Array-Based Comparative Genomic Hybridization Analysis
To confirm the single copy loss of a large portion of 3p that was inferred from sequencing data, we performed array-based comparative genomic hybridization on samples from RM-23 using a stock 1 × 1M Agilent SurePrint G3 Human CGH Microarray chip in a lab certified by the Clinical Laboratory Improvement Amendments as previously described.2,25
Loss of Heterozygosity Analysis
Loss of heterozygosity (LOH) analysis was performed using standard methodologies as described in the Supplementary materials.
Statistical Analysis
We performed statistical analyses using standard methodologies as described in the Supplementary materials.
Results
We sequenced 560 cancer-associated genes in 14 meningiomas that had been diagnosed as rhabdoid meningioma or meningioma with some degree of rhabdoid features and performed LOH analysis (Supplementary Table 1, Supplementary Fig. 1). This discovery cohort reflected the nosologic uncertainty prevalent in this tumor subtype. In some cases, rhabdoid features were suggestive but not definitive; in others, focal clusters of rhabdoid cells were noted; and in others rhabdoid cells with well-developed features were widespread and associated with grade I, II, or III features. We determined whether the rhabdoid features were present in <20%, 20%–50%, or >50% of the tumor cells.
We detected a total of 749 nonsynonymous variants and 47 insertions/deletions (Supplementary Tables 2, 3). In 8 samples without grade III anaplastic features but with rhabdoid or “rhabdoid-like” cells, we identified chromosome 22 LOH (Supplementary Fig. 1). We detected nonsynonymous variants in the NF2 gene in 5 of these 8 specimens and the AKT1 E17K mutation in 4/14 mutually exclusive specimens (Supplementary Table 2; Supplementary Fig. 1). None of these specimens was noted to have mutations in the TERT promoter. Thus, the principal genetic aberrations in this diverse collection of meningiomas with rhabdoid features overlapped with those already reported in more common subtypes.3,4
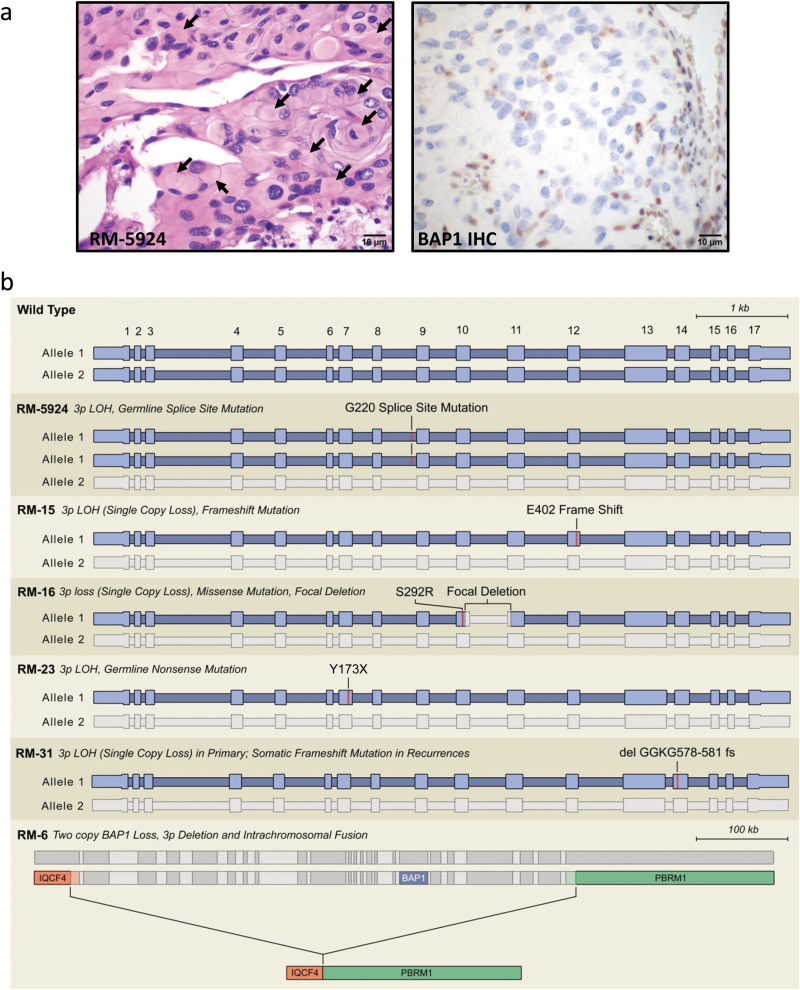
However, in one sample (RM-5924) with distinct anaplastic histology including well-developed rhabdoid cells and poorly differentiated cells (Supplementary Fig. 2), we detected a splice-site mutation in the BAP1 gene (p.G220_splice; Fig. 1; Supplementary Tables 2, 3), a tumor suppressor gene on chromosome 3p21.26–28 This tumor had copy neutral chromosome 3 LOH, maintaining 2 copies of mutant BAP1 consistent with endoreduplication (Supplementary Fig. 3).
Fig. 1.
BAP1 loss in a high-grade rhabdoid meningioma. (A) Hematoxylin and eosin staining and BAP1 immunohistochemistry on sample RM-5924. Arrows highlight several of the globular paranuclear inclusions. (B) Schematic of BAP1 genetic aberrations resulting in BAP1 inactivation in syndromic and sporadic high-grade rhabdoid meningiomas. A list of the mutation calls made for these tumors is presented in Supplementary Tables 2, 6–8).
Furthermore, we analyzed BAP1 protein expression using IHC in this cohort of 14 meningiomas with rhabdoid features. Samples lacking genetic aberrations in BAP1 had intact BAP1 expression. However, in sample RM-5924, BAP1 was lost in the tumor cells in both the rhabdoid and poorly differentiated tumor nuclei (Fig. 1a; Supplementary Fig. 2), with retained expression in nonneoplastic nuclei providing a positive internal control.
Germline mutations in BAP1 result in a tumor predisposition syndrome, which confers a high risk for developing a variety of tumors, including uveal and cutaneous melanoma, lung adenocarcinoma and mesothelioma, renal cell carcinoma, and papillary thyroid carcinoma.26,29–32 When we reviewed the medical record of the patient affected by rhabdoid meningioma RM-5924, we found that her father had a mesothelioma. Consistent with this family history, we detected the p.G220 splice-site BAP1 mutation in the patient’s constitutional DNA (Supplementary Table 1).
To assess the frequency of BAP1 loss by IHC in rhabdoid meningiomas, we collected a set of 57 samples from 47 patients who had been diagnosed with rhabdoid meningioma or meningioma with some degree of rhabdoid features determined by the reviewing surgical pathologist (Supplementary Table 4). Similar to our discovery set, the tumors had considerably heterogeneous histology. We performed BAP1 IHC on these tumors and on 265 additional meningiomas of diverse subtypes and grades (Supplementary Table 5), including 26 anaplastic grade III samples lacking any rhabdoid cells. BAP1 was expressed in all 265 non-rhabdoid meningiomas. However, among the 47 patients, we identified 5 with BAP1-negative rhabdoid cells (Supplementary Figs. 4–6). Each of these five patients had tumors with distinctly well-developed rhabdoid cytomorphology and all tumors had high-grade histology.
Across these 57 samples there was a correlation between loss of BAP1 and the extent of rhabdoid features (BAP1 was lost in 3 of the 37 samples composed of ≤50% rhabdoid cells; BAP1 was lost in 9 of 20 samples composed of >50% rhabdoid cells; chi-square statistic = 10.6; P = .0011), between loss of BAP1 and mitotic rate (BAP1 was not lost in any of the 40 samples with <5 mitoses per 10 high powered fields; BAP1 was lost in 12 of 17 samples with ≥5 mitoses per 10 high powered fields; chi-square = 35.8; P < .0001) and between loss of BAP1 and WHO grade (chi-square = 12.3; P < .0021).
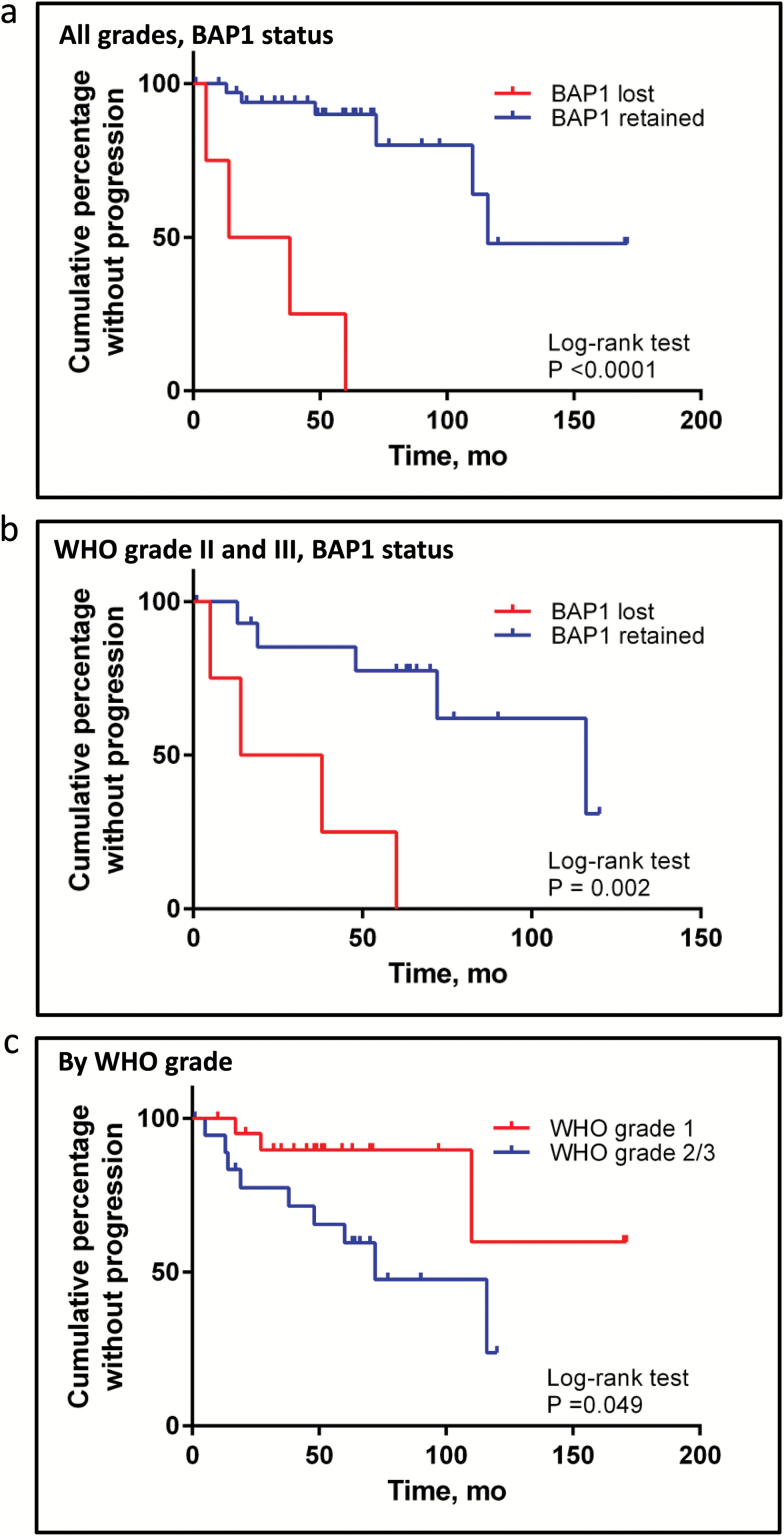
Clinical follow-up was available for 4 of these 5 patients and showed that BAP1 loss correlated closely with poor outcomes. Two patients died of disease (RM-6, RM-31), one had 3 recurrences and remains alive with significant morbidity (RM-23), and the other had 2 recurrences, including systemic metastases (RM-15). Thus, BAP1-deficient meningiomas were clinically aggressive and more likely to recur compared with BAP1-retained grades II and III meningiomas (hazard ratio [HR] = 12.9 in all grades; HR = 6.0 in grade II/III; Kaplan–Meier analysis log-rank test P < .001 for all grades, P = .002 for higher grades (Fig. 2; Supplementary Table 4). Whereas WHO grades II and III meningiomas as a group had a median time to progression of 72 months, the median time to progression for patients with BAP1 intact grades II and III meningiomas was 116 months but only 26 months for patients with BAP1-deficient grades II and III meningiomas. The association of BAP1 and recurrence was independent of grade in a multivariate model (P = .015). Further multivariate model building was limited by small numbers.
Fig. 2.
Kaplan–Meier survival analysis. Plot of the cumulative percentage without recurrence among the validation cohort of “meningioma with rhabdoid features” (BAP1 lost vs BAP1 retained) for patients with clinical follow-up harboring meningiomas of all grades (A) and only those with higher-grade (WHO II and III) meningiomas (B). Time to first progression used in analyses except if primary had documented intact BAP1 (eg, RM-31-a) in which time to progression after BAP1 loss is used (eg, time to RM-31-c from RM-31-b). Curves were compared using the log-rank (Mantel–Cox) test. P-value is displayed. Hazard ratio (HR) for BAP1-deficient tumors is listed in the text (log-rank): 12.9 when assessing all grades and 6.0 when assessing only higher grades (WHO grade II and III). (C) Plot of the cumulative percentage without recurrence among the validation cohort of WHO grade I meningioma and WHO grades II and grade III meningioma with rhabdoid features.
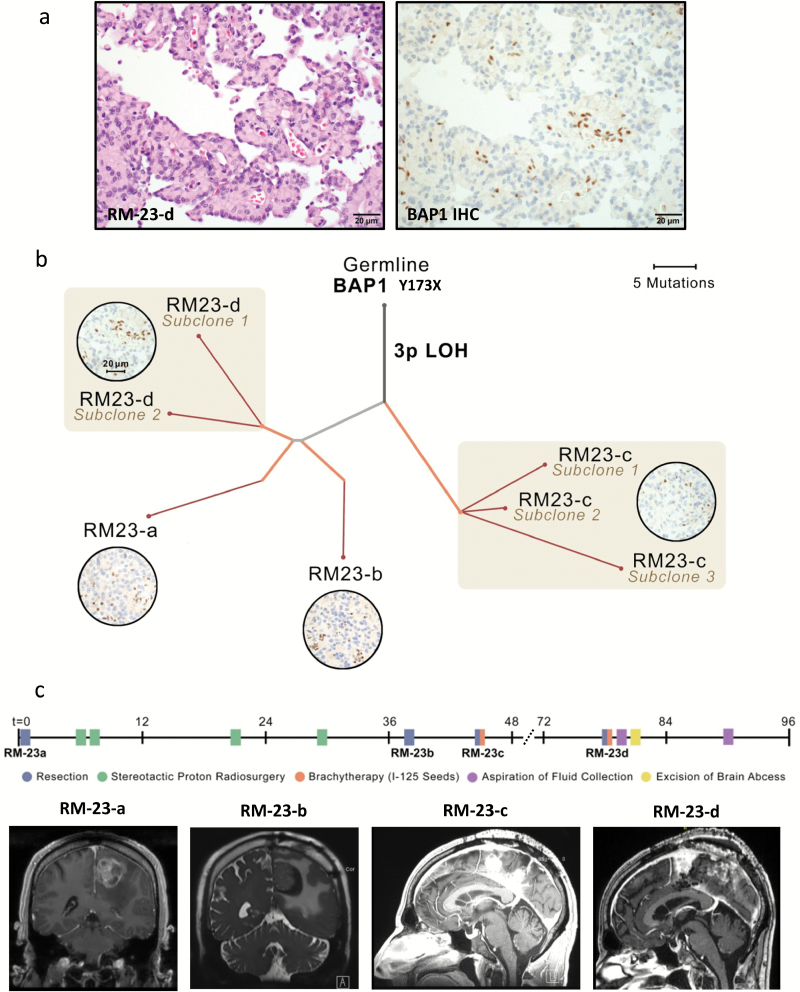
To identify the genomic aberrations in BAP1 that underlie the BAP1-negative rhabdoid meningiomas, we performed whole exome sequencing or focused sequencing of all exons from 300 cancer-associated genes from 12 available samples from these 5 patients. In all cases, we found mutations or deletions that inactivate BAP1 coupled with chromosome 3 LOH (Fig. 1b; Supplementary Table 6–8). One case (RM-6) had 2 samples with both copies of BAP1 deleted—a single copy loss of part of chromosome 3p and an intrachromosomal fusion between the IQCF4 gene and the PBRM1 gene deleting ~700 kilobases encompassing BAP1 and the C-terminal 20 amino acids of polybromo-1 (PBRM1), including the stop codon (Fig. 1b). This event is predicted to inactivate PBRM1. Interestingly, 2 of these BAP1-mutant cases (RM-6 and RM-23) showed significant papillary morphology in addition to widespread rhabdoid cytomorphology (Fig. 3a), indicating a genetic connection between 2 WHO grade III subtypes of meningioma that to date have been largely considered distinct entities.
Fig. 3.
Characterization of genomic evolution of rhabdoid meningioma RM-23. (A) Co-occurrence of rhabdoid cytomorphology and papillary architecture in RM-23. (B) Phylogenetic tree inferred for familial rhabdoid meningioma RM-23 primary and recurrences. Branch thickness is proportional to the cancer-cell fraction (CCF) of mutations on that branch. Branch colors indicate types of tissue samples descended from each branch (gray, shared by all samples; blue, unique to primary sample; orange, present in recurrences; red, present in subclones of the recurrent tumor—eg, RM-23-c subclones 1, 2, and 3). Photomicrographs in circles show BAP1 immunohistochemistry for indicated tumors. Scale bar, 20µm. (C) Clinical information for patient RM-23. Timeline indicating relative sequence of major clinical events, including each surgery and other interventions. Representative images of preoperative MRI before each of the 4 surgical resections. While RM-23-a and RM-23-b were solitary masses, recurrences RM-23-c and RM-23-d comprised multiple spatially distinct masses along the falx. This pattern is consistent with the results from phylogenetic analysis showing that samples RM-23-c and RM-23-d comprised related but genetically distinct subclones. A list of mutation calls made for each of these 4 samples (RM-23-a to RM-23-d; noted as allelic fractions) is provided in Supplementary Table 7 (gene names and allelic fractions are in columns a–e).
Constitutional DNA was available for 3 of these 5 patients. In 2 cases (RM-6, RM-31), the BAP1 gene was wild-type in the constitutional DNA, whereas one case (RM-23) showed the same germline Y173X mutation that we had identified in the patient’s meningioma. Because our work is a retrospective analysis, our ability to access family history in RM-23 was limited, and a full pedigree analysis was not currently possible. Nonetheless, we found that that patient’s father and 2 paternal uncles had mesotheliomas, tumors known to arise as part of the BAP1 tumor predisposition syndrome.32–34
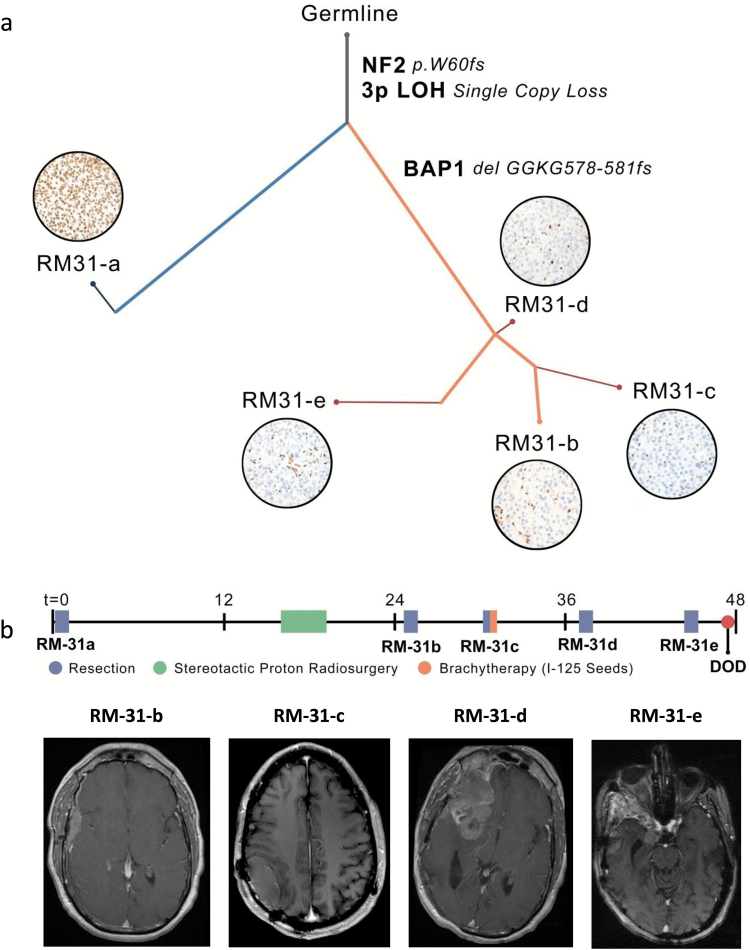
We next used the whole exome sequencing data to assess heterogeneity and evolutionary relationships24 in tumor samples from 2 patients with multiple recurrences—RM-23 with a germline Y173X BAP1 truncating mutation and RM-31 with a sporadic delGGKG (aa 578–581) BAP1 frameshift event (c.1733_1742delGTGGGAAGGG). This analysis demonstrated the clonal relationships between the primary and recurrent tumors within each patient (Fig. 3 and 4, Supplementary Figs. 7–11).
Fig. 4.
Characterization of genomic evolution of rhabdoid meningioma RM-31. (A) Phylogenetic tree inferred for sporadic rhabdoid meningioma RM-31 primary and recurrences. Branch thickness is proportional to the cancer-cell fraction (CCF) of mutations on that branch. Branch colors indicate types of tissue samples descended from each branch (gray, shared by all samples; blue, unique to primary sample; orange, present in recurrences; red, present in subclones of the recurrent tumor—eg, RM-23-c subclones 1, 2, and 3). Photomicrographs in circles show BAP1 immunohistochemistry for the indicated tumor. Scale bar, 20µm. (B) Clinical information for patient RM-31. Timeline of treatment indicating the relative sequence of major clinical events, including each surgery and other treatment interventions. Representative images of MRI before the indicated surgical resections. MRI from the patient prior to resection of primary tumor (RM-31-a) was performed at an outside facility and images were unavailable. A list of mutation calls made for each of these 5 samples (RM-31-a to RM-31-e; noted as allelic fractions) is provided in Supplementary Table 8 (gene names and allelic fractions are in columns a–f).
As predicted, RM-23 (Fig. 3) had BAP1 protein loss in the primary tumor (RM-23-a; Fig. 3b). We found extensive genomic heterogeneity with related but distinct subclones emerging in the last 2 recurrences (RM-23-c, RM-23-d; Fig. 3b; Supplementary Fig. 7; Supplementary Table 7). In those latter recurrences, the tumor was nodular, forming multiple spatially distinct masses, unlike the primary and first recurrence, which were distinct solitary masses (Fig. 3c). These anatomically distinct outgrowths may explain the extensive subclonal evolution we detected.
On the other hand, RM-31 (Fig. 4) had BAP1 protein intact in the primary tumor (RM-31-a; Fig 4a) and we found a branched evolutionary relationship between the primary and the recurrent tumors (Fig. 4a; Supplementary Fig. 10, Supplementary Table 8). In the primary tumor (RM-31-a), one BAP1 allele was inactivated due to monosomy of 3p, but the second BAP1 allele was intact (Supplementary Fig. 11), consistent with the retained BAP1 expression. This primary tumor also harbored a frameshift mutation in NF2 (W60fs; c.179delG). The clonally related recurrent tumor (RM-31-b) harbored this NF2 mutation but also was BAP1 negative due to inactivation of the second BAP1 allele because of a delGGKG (aa 578–581) frameshift event (Figs. 1b, 4a; Supplementary Fig. 10; Supplementary Table 8). Overt rhabdoid features were absent in the primary tumor (RM-31-a) but present in the recurrent tumors, following biallelic BAP1 inactivation. These findings suggest that meningiomas with monosomy 3p and intact BAP1 protein expression may recur as high-grade rhabdoid tumors if the second BAP1 allele is inactivated (Fig. 4b).
Discussion
Our work has practical implications for the care of patients with meningiomas, which comprise one-third of primary brain tumors. First, by demonstrating that rhabdoid meningiomas often harbor BAP1 mutations, we further sharpen the emerging molecular-genetic taxonomy of meningiomas that was described in the introductory section. Assessing the BAP1 expression status of suspected rhabdoid meningiomas—ones with either focal or widespread rhabdoid cytomorphology—will help remedy the nosologic dilemma that has muddled the diagnosis and prognosis of rhabdoid meningiomas. Such an assessment can be performed routinely using simple and inexpensive IHC testing for BAP1 protein expression, which has been validated in uveal melanoma to capture tumors with nonsynonymous mutations, in addition to those with epigenetic mechanisms of gene silencing.35 Of note, a caveat of our current study is that the number of rhabdoid meningioma cases we have analyzed is small, given the relative rarity of this meningioma subtype. Hence, the association of BAP1 mutations with rhabdoid meningiomas and further assessment of the clinical implications of BAP1 inactivation will require an even larger multi-institutional effort for future validation.
The straightforward IHC staining which we utilized will provide a molecular criterion that can now be assessed prospectively as a tool for risk-stratifying patients, identifying those requiring closer postoperative surveillance imaging or even adjuvant radiation therapy. Highlighting the need for an objective molecular marker that could potentially guide patient management, we found that 13 of 40 patients who had BAP1-intact WHO grades I–III meningiomas with rhabdoid features had received radiation therapy following the diagnosis. It is conceivable that a portion of these patients may have been overtreated. Additional multicenter prospective studies will be required to assess the value of BAP1 IHC in guiding adjuvant care. In our study, cases with inactivation of BAP1 had negative staining in essentially all tumor cells. In principle, IHC staining may help identify cases with loss of BAP1 in a subclonal population, and the clinical significance of such changes and the potential underlying genetic modifications resulting in focal BAP1 loss will need to be assessed.
Of additional clinical importance, our work links a distinct meningioma subtype with a tumor predisposition syndrome that principally gives rise to tumors that occur outside of the CNS, including uveal and cutaneous melanoma, mesothelioma, and renal cell carcinoma. Identification of BAP1-deficient meningiomas should elicit an evaluation for the BAP1 tumor predisposition syndrome, although a subset of these BAP1-negative tumors clearly arise due to sporadic somatic BAP1 mutations. Moreover, our work suggests that patients with known germline BAP1 mutations may warrant monitoring for the development of meningiomas.
Our work also suggests that meningiomas with one copy loss of chromosome 3p and retained BAP1 protein expression may have the propensity to evolve into BAP1-deficient high-grade rhabdoid meningiomas, as in the case of RM-31 (Fig. 4). In addition to characterizing traditional biomarkers,36 assessing the genomic aberrations of meningiomas is becoming an increasingly integral part of patient management.14,37–42 This increase in molecular testing will allow the identification of patients with “monosomy 3p meningiomas.” Additional multicenter studies with larger cohorts can now be used to assess whether such meningiomas indeed have a higher propensity to evolve into rhabdoid meningiomas.
We found that BAP1-mutant rhabdoid meningiomas also harbor mutations in genes that are altered in other BAP1-mutant tumors such as mesothelioma34 and clear cell renal cell carcinoma.43 For example, BAP1-mutant rhabdoid meningiomas also had genomic aberrations in the tumor suppressor genes NF2 (RM-15: p.A367fs, c.1100_1101CA>C and RM-31: p.W60fs, c.179delG) and FBXW7 (RM-6: p.N635fs, c.1905_1924CTTTGTAATTACCAGCTCAG>G and RM-15: p.G246X; c.736G>T), which have both been found to be altered in mesothelioma. In mesothelioma, alterations in BAP1 and NF2 tend to occur together in sarcomatoid subtypes, ones that have more aggressive behavior. We also describe a BAP1-mutant rhabdoid meningioma that also harbors an inactivating event in PBRM1 (RM-6). BAP1 and PBRM1 occur together in a subset of renal cell carcinomas that also display rhabdoid features.43 Such rhabdoid renal cell carcinomas are associated with poor outcome. The finding that genes altered in BAP1-mutant rhabdoid meningiomas are also altered in other BAP1-mutant tumors suggests that these tumors may share mechanisms of pathogenesis. Moreover, further studies will be able to assess whether patients with meningiomas that have BAP1 mutations co-occurring with NF2, FBXW7, or PBRM1 mutations have worse outcomes than those not harboring mutations in these additional genes.
Consistent with the poor prognosis seen in other BAP1-mutant tumors, our work shows that BAP1-deficient meningiomas appear to be highly aggressive and often lethal malignancies. All BAP1-deficient cases in our cohort had WHO grade II or III histology, and the ones with sufficient clinical follow-up demonstrated aggressive clinical behavior. Thus, novel treatment approaches are urgently needed for this molecular genetic subtype of meningioma. Recent work has shown that BAP1-deficient tumor cells are highly sensitive to genotoxic stressors43 and to inhibitors of enhancer of zeste homolog 2 (EZH2),44 providing promising therapeutic avenues to explore in pre-clinical models and clinical trial testing.
Supplementary material
Supplementary material is available online at Neuro-Oncology (http://neuro-oncology.oxfordjournals.org/).
Funding
This work was supported by the Brain Science Foundation (S.S., P.B.), the Jared Branfman Sunflowers for Life Fund for Pediatric Brain and Spinal Cancer Research (S.S.), grant 35–1041 from King Abdulaziz City for Science and Technology (KACST) no. 110968, Saudi Arabia (M.A.A.), and the Ludwig Center at Harvard (S.S.). PKB is funded by NIH K12 grant (2K12CA090354-11), Brain Science Foundation, Susan G. Komen for the Cure, Terri Brodeur Breast Cancer Foundation, Conquer Cancer Foundation, and American Brain Tumor Association (ABTA). G.M.S. is supported by the ABTA Basic Research Fellowship supported by the Humor to Fight the Tumor Committee.
Supplementary Material
Acknowledgments
We thank staff of the Center for Cancer Genome Discovery (CCGD) at the Dana Farber Cancer Institute and Center for Advanced Molecular Diagnostics (CAMD), specifically Matthew Ducar, Elizabeth Garcia, Michele Baltay, Yonghui Jia, and Bruce Wollison for DNA sequencing and analysis. We thank Marian Slaney, Sebastian Valentin, and Karen Bryan for assistance with histopathology, Terri Woo for immunohistochemistry, Marc Listewnik for assistance with aCGH analysis, and Amaro Taylor-Weiner for valuable guidance.
Conflict of interest statement. The authors do not have a conflict of interest to report.
References
- 1. Louis DN, Ohgaki H, Wiestler OD, et al. WHO Classification of Tumours of the Central Nervous System. Revised 4th ed. Lyon, France: International Agency for Research on Cancer; 2016:232–257. [Google Scholar]
- 2. Abedalthagafi MS, Merrill PH, Bi WL, et al. Angiomatous meningiomas have a distinct genetic profile with multiple chromosomal polysomies including polysomy of chromosome 5. Oncotarget. 2014;5(21):10596–10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brastianos PK, Horowitz PM, Santagata S, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45(3):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reuss DE, Piro RM, Jones DT, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013;125(3):351–358. [DOI] [PubMed] [Google Scholar]
- 6. Smith MJ, O’Sullivan J, Bhaskar SS, et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet. 2013;45(3):295–298. [DOI] [PubMed] [Google Scholar]
- 7. Abedalthagafi M, Bi WL, Aizer AA, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 2016;18(5):649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trofatter JA, MacCollin MM, Rutter JL, et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;72(5):791–800. [DOI] [PubMed] [Google Scholar]
- 9. Smith MJ, Wallace AJ, Bennett C, et al. Germline SMARCE1 mutations predispose to both spinal and cranial clear cell meningiomas. J Pathol. 2014;234(4):436–440. [DOI] [PubMed] [Google Scholar]
- 10. Kepes JJ, Moral LA, Wilkinson SB, et al. Rhabdoid transformation of tumor cells in meningiomas: a histologic indication of increased proliferative activity: report of four cases. Am J Surg Pathol. 1998;22(2):231–238. [DOI] [PubMed] [Google Scholar]
- 11. Perry A, Scheithauer BW, Stafford SL, et al. “Rhabdoid” meningioma: an aggressive variant. Am J Surg Pathol. 1998;22(12):1482–1490. [DOI] [PubMed] [Google Scholar]
- 12. Vaubel RA, Chen SG, Raleigh DR, et al. Meningiomas with rhabdoid features lacking other histologic features of malignancy: a study of 44 cases and review of the literature. J Neuropathol Exp Neurol. 2016;75(1):44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rogers CL, Perry A, Pugh S, et al. Pathology concordance levels for meningioma classification and grading in NRG Oncology RTOG Trial 0539. Neuro Oncol. 2016;18(4):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abedalthagafi MS, Bi WL, Merrill PH, et al. ARID1A and TERT promoter mutations in dedifferentiated meningioma. Cancer Genet. 2015;208(6):345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Altshuler DM, Gibbs RA, Peltonen L, et al. ; International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Browning BL, Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am J Hum Genet. 2009;85(6):847–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cibulskis K, McKenna A, Fennell T, et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27(18):2601–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saunders CT, Wong WS, Swamy S, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28(14):1811–1817. [DOI] [PubMed] [Google Scholar]
- 24. Brastianos PK, Carter SL, Santagata S, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5(11):1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Craig JM, Vena N, Ramkissoon S, et al. DNA fragmentation simulation method (FSM) and fragment size matching improve aCGH performance of FFPE tissues. PLoS One. 2012;7(6):e38881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carbone M, Yang H, Pass HI et al. BAP1 and cancer. Nat Rev Cancer. 2013;13(3):153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20(11):1242–1253. [DOI] [PubMed] [Google Scholar]
- 28. Lu C, Xie M, Wendl MC, et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun. 2015;6:10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48(12):856–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de la Fouchardiere A, Cabaret O, Petre J, et al. Primary leptomeningeal melanoma is part of the BAP1-related cancer syndrome. Acta Neuropathol. 2015;129(6):921–923. [DOI] [PubMed] [Google Scholar]
- 31. Popova T, Hebert L, Jacquemin V, et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am J Hum Genet. 2013;92(6):974–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43(10):1022–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nasu M, Emi M, Pastorino S, et al. High incidence of somatic BAP1 alterations in sporadic malignant mesothelioma. J Thorac Oncol. 2015;10(4):565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bueno R, Stawiski EW, Goldstein LD, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48(4):407–416. [DOI] [PubMed] [Google Scholar]
- 35. van de Nes JA, Nelles J, Kreis S, et al. Comparing the prognostic value of BAP1 mutation pattern, chromosome 3 status, and BAP1 immunohistochemistry in uveal melanoma. Am J Surg Pathol. 2016;40(6):796–805. [DOI] [PubMed] [Google Scholar]
- 36. Olar A, Wani KM, Sulman EP, et al. Mitotic index is an independent predictor of recurrence-free survival in meningioma. Brain Pathol. 2015;25(3):266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sahm F, Schrimpf D, Olar A, et al. TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst. 2016;108(5):. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goutagny S, Nault JC, Mallet M, et al. High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol. 2014;24(2):184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perry A, Banerjee R, Lohse CM, et al. A role for chromosome 9p21 deletions in the malignant progression of meningiomas and the prognosis of anaplastic meningiomas. Brain Pathol. 2002;12(2):183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boström J, Meyer-Puttlitz B, Wolter M, et al. Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. Am J Pathol. 2001;159(2):661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aizer AA, Abedalthagafi M, Bi WL, et al. A prognostic cytogenetic scoring system to guide the adjuvant management of patients with atypical meningioma. Neuro Oncol. 2016;18(2):269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Domingues PH, Sousa P, Otero Á, et al. Proposal for a new risk stratification classification for meningioma based on patient age, WHO tumor grade, size, localization, and karyotype. Neuro Oncol. 2014;16(5):735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peña-Llopis S, Vega-Rubín-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44(7):751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. LaFave LM, Béguelin W, Koche R, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21(11):1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.