Abstract
Chromosomal translocations joining in-frame members of the fibroblast growth factor receptor–transforming acidic coiled-coil gene families (the FGFR-TACC gene fusions) were first discovered in human glioblastoma multiforme (GBM) and later in many other cancer types. Here, we review this rapidly expanding field of research and discuss the unique biological and clinical features conferred to isocitrate dehydrogenase wild-type glioma cells by FGFR-TACC fusions. FGFR-TACC fusions generate powerful oncogenes that combine growth-promoting effects with aneuploidy through the activation of as yet unclear intracellular signaling mechanisms. FGFR-TACC fusions appear to be clonal tumor-initiating events that confer strong sensitivity to FGFR tyrosine kinase inhibitors. Screening assays have recently been reported for the accurate identification of FGFR-TACC fusion variants in human cancer, and early clinical data have shown promising effects in cancer patients harboring FGFR-TACC fusions and treated with FGFR inhibitors. Thus, FGFR-TACC gene fusions provide a “low-hanging fruit” model for the validation of precision medicine paradigms in human GBM.
Keywords: chromosomal translocations, FGFR-TACC, glioma, personalized therapy
Recurrent chromosomal rearrangements in cancer have been described for more than 50 years and identification of chromosomal rearrangements has been an important effort in cancer genetics, especially after initial studies showed that in many cases the fusion of 2 genes carries oncogenic functions.1,2 In general, fusion oncogenes are not common in human tumors, but their importance for the understanding of tumor biology has been extremely high and they represent powerful examples of success in targeted therapies for selected tumor types.3 Mechanistic studies of the BCR-ABL1 and PML-RARA oncogenes more than a decade ago have translated into successful therapies for 2 previously deadly types of hematological cancer: imatinib for the treatment of chronic myelogenous leukemia and combined arsenic trioxide and retinoic acid for the treatment of acute pro-myelocytic leukemia.4,5 More recently, EML4–ALK fusion was identified in non–small cell lung cancer, leading to an exceptionally rapid translation into clinical benefit for patients treated with the anaplastic lymphoma kinase (ALK) inhibitor crizotinib.6–9 Thus, the finding that a subset of human glioblastoma multiforme (GBM) harbors oncogenic fusions that join the members of the fibroblast growth factor receptor 3 (FGFR3) and FGFR1 tyrosine kinases (TKs) to the transforming acidic coiled-coil (TACC) proteins TACC3 and TACC1, respectively, has raised hope that inhibition of FGFR could be a valuable therapeutic option for this subgroup of deadly type of brain cancer.10
Architecture of FGFR-TACC Rearrangement
The 3′ partners of the FGFR-TACC chromosomal translocations code for members of the TACC protein family, which includes the proteins TACC1, TACC2, and TACC3.11 The distinctive feature of TACC proteins is a coiled-coil domain at the C-terminus, known as the TACC domain, which mediates localization to the centrosome and mitotic spindle.11,12 TACC proteins are hypothesized to be oncogenic in several human tumors, including GBM.13,14 The most frequent configuration of FGFR-TACC chromosomal translocations in malignant glioma implicates the genes coding for FGFR3 and TACC3, which are located 48 kb apart on human chromosome 4p16 (Fig. 1).10,15,16 Other FGFR-TACC fusions (FGFR1-TACC1 and FGFR2-TACC2) join the remaining members of the FGFR and TACC families.10,15,17,18 They retain the close chromosomal location, with FGFR1 and TACC1 paired on chromosome 8p11 and FGFR2 and TACC2 paired on chromosome 10q26.19
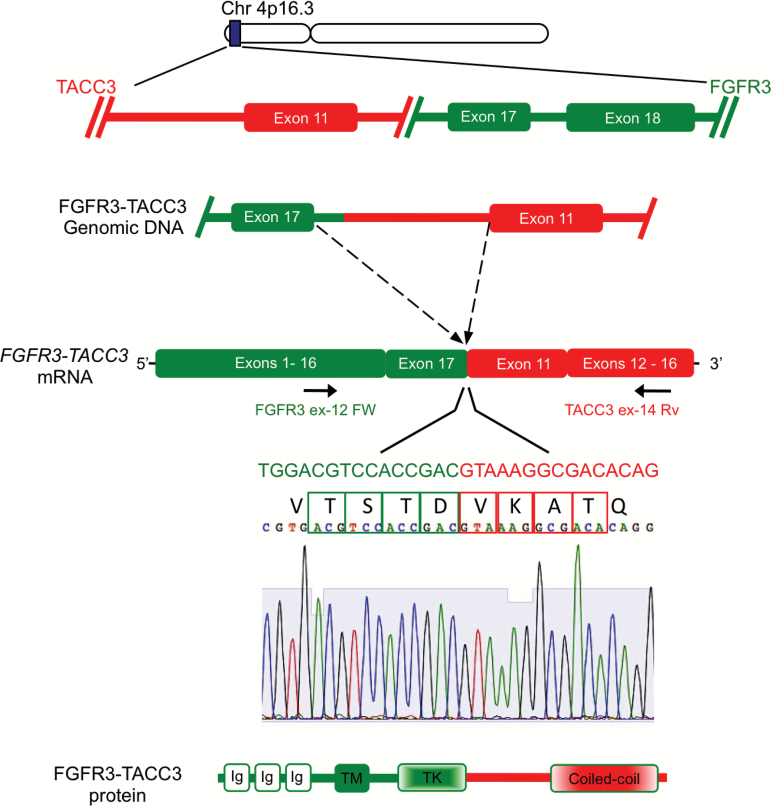
Fig. 1.
Structure of FGFR3-TACC3 rearrangement in GBM. Genomic organization of the FGFR3 and TACC3 loci (top). In an FGFR3-TACC3 variant reported in GBM, the genomic rearrangement causes the juxtaposition of exon 17 and a small portion of intron 17 of the FGFR3 gene with intron 10 of the TACC3 gene, leading to in-frame fusion of exon 17 of FGFR3 and exon 11 of TACC3 as indicated by the Sanger sequence of the joint mRNA. This fusion structure is one of the most frequent mRNA fusion variants identified in GBM. Arrows indicate the position of the diagnostic primers used in the RT-PCR screening assay for FGFR3-TACC3. The structure of the FGFR3-TACC3 fusion protein is shown in the bottom panel and invariably includes the TK domain of FGFR3 and the coiled-coil domain of TACC3.
In our initial report, we showed that ~3% of human GBM harbors rearrangements involving FGFR3 and TACC3 and FGFR1 and TACC1, respectively.10 The cDNA of the first identified FGFR3-TACC3 fusion contained an open reading frame coding for a protein of 1048 amino acids resulting from the in-frame fusion of the FGFR3 N-terminus (residues 1 to 758) with the TACC3 C-terminus (residues 549 to 838). The genomic breakpoint on chromosome 4 (1808966 for FGFR3 and 1737080 for TACC3, genome build GRCh37/hg19) falls within FGFR3 exon 17 and TACC3 intron 7, which gives rise to a transcript in which the 5′ FGFR3 exon 16 is spliced to the 3′ TACC3 exon 8.10 The architecture of FGFR-TACC intra-chromosomal rearrangement is duplication with inversion. The observation of micro-homology sequences within a 10-base region at the DNA junctions of FGFR3 and TACC3 was consistent with results reported for other intra-chromosomal rearrangements in human cancer.20,21 The analysis of high-density single nucleotide polymorphism (SNP) 6.0 arrays of 158 GBM samples from The Cancer Genome Atlas revealed focal amplification events involving only the exons of the FGFR3 gene that are included in the fusion breakpoint in the 5 FGFR3-TACC3–positive samples in the dataset. However, none of 10 samples that displayed different degrees of copy number gains encompassing the entire FGFR3 and TACC3 loci harbored FGFR3-TACC3 fusions.10,15 Duplications or low levels of amplification have been overlooked in the past because they involve small chromosomal segments beyond the resolution of cytogenetics, and any copy number variation survey with a lower resolution than high-density SNP arrays will likely fail to identify the genomic marks of FGFR3-TACC3–positive cases.
After the initial discovery of FGFR-TACC fusions in human GBM, recurrent FGFR-TACC gene fusions have been identified in many tumor types with a frequency typically between 1% and 4%.10,15,16,18,22–32 When considered in aggregate, FGFR-TACC fusions have now emerged as one of the most recurrent chromosomal translocations across multiple types of human cancer.17,33 However, when all cancer types are considered, FGFR3-TACC3 fusions are by far the most frequent gene fusion variant (Table 1). As larger data have become available, a notable variability among FGFR3–TACC3 fusion isoforms has been documented, with rearrangements that join FGFR3 exon 17 with TACC3 (different exons) being more frequent and many fusion variants occurring in individual cases.15 In Fig. 1 we show the structure of the fusion mRNA that joins FGFR3 exon 17 to TACC3 exon 11, which was reported as a recurrent FGFR3-TACC3 fusion mRNA variant in human GBM.15 While the high degree of heterogeneity of the FGFR3 and TACC3 genomic breakpoints poses a significant challenge toward the design of an accurate and sensitive assay for the screening of FGFR3-TACC3 fusions in human cancer, the amplicons of each FGFR-TACC fusion cDNA invariably join in-frame the entire FGFR-TK domain upstream of TACC-coding sequences, which always include the intact coiled-coil TACC domain. The loss of the 3′ untranslated region of FGFR3 would eliminate gene regulation by miR-99a, thus leading to uncontrolled expression of the fusion gene.16
Table 1.
Frequency of FGFR-TACC fusions in human cancer
| Cancer Type | Frequency (%) | Reference(s) |
|---|---|---|
| Glioblastoma | 3 | 10, 15 |
| Pediatric low-grade glioma | 6–7 | 18 |
| Glioblastoma | 8.3 | 16 |
| Bladder cancer | 3 | 25, 30 |
| Head & neck | 3 | 31, 32 |
| Non–small cell lung carcinoma | 1.3–3 | 31 |
| Lung squamous cell carcinoma | 3.5–4.2 | 27, 29 |
| Lung adenocarcinoma | 0.5 | 23 |
| Sporadic intrahepatic cholangiocarcinoma | Undefined | 22 |
| Nasopharyngeal carcinoma, esophageal squamous cell carcinoma | 2.1–2.5 | 32 |
| Cervical squamous cell carcinoma | 3 | 24 |
| Grades II–III glioma | 3.5 | 15 |
| Triple negative breast cancer | 1.85 | 28 |
The incidence of chromosomal translocations fusing in-frame the FGFR3 and TACC3 genes is considerably higher than the frequency of FGFR1-TACC1 rearrangements. However, based on available data, the biological and oncogenic functions of FGFR1-TACC1 are similar to those assigned to FGFR3-TACC310. As in previous reports, in this review we refer to FGFR-TACC to define the combination of FGFR1-TACC1 and FGFR3-TACC3 variants. However, it is important to note that most studies addressed the functional properties of FGFR3-TACC3.
Signaling and Biological Activities of FGFR-TACC Fusion Proteins
Fibroblast growth factor (FGF) signaling is a highly complex growth factor signaling pathway that regulates a multitude of fundamental pathways in development and adult organism and controls key cell functions, such as proliferation, differentiation, and survival.34,35 FGFRs signal as dimers, and ligand-dependent dimerization leads to conformational changes of the receptor structure that activates the intracellular kinase domain, resulting in intermolecular transphosphorylation of the TK domains and intracellular tails. Phosphorylated tyrosine residues on the receptor function as docking sites for adaptor proteins, which themselves may be directly phosphorylated by FGFR. FGF stimulation leads to tyrosine phosphorylation of the docking protein fibroblast growth factor receptor substrate 2 alpha (FRS2α) and FRS2β, followed by recruitment of multiple Grb2/Sos complexes and Grb2/Gab1 complexes resulting in activation of the Ras/mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3 kinase (PI3K) signaling pathways, respectively.36,37 In addition to FGFR signaling activation, FRS2α recruits negative regulators and is central to a negative feedback mechanism, whereby threonine phosphorylation of FRS2α by MAPK reduces tyrosine phosphorylation of FRS2α, recruitment of Grb2, and attenuation of the MAPK response.38 Downstream FGFR signaling can also be attenuated through the induction of MAPK phosphatases, Sprouty proteins, and Sef (“similar expression to FGF”) family members that modulate receptor signaling at several points in the signal transduction cascade.39 In contrast to wild-type FGFR, FGFR-TACC fusion proteins are constitutively active receptor TKs (RTKs).10,16,30,31 The inclusion of the TACC coiled-coil domain with its ability to form dimers promotes fusion protein dimerization, auto-phosphorylation, and FGFR tyrosine kinase activation. Recently, several constitutive phosphorylated tyrosine residues in the key FGFR3-TK have been identified in FGFR3-TACC3 by mass spectrometry.40 However, the intracellular signaling events that operate downstream of FGFR-TACC and execute its oncogenic functions are not completely understood and it is unknown how negative regulatory mechanisms of FGFR signaling are countered. The experimental evidence that has been gathered so far indicates that FRS2 is phosphorylated in cells expressing FGFR-TACC fusions.10 This finding suggests that the major mediator of FGFR signaling remains constitutively bound to the juxta-membrane domain of the FGFR3 moiety of the fusion, and this is likely independent of FGFR-TACC cellular compartmentalization. The question that is still unresolved is whether FRS2 recruits the signaling components that are activated by FGFR-TACC. It was reported that the ectopic expression of FGFR3-TACC3 in certain cell types activates extracellular signal-regulated kinase ERK/MAPK/MAPK and possibly also PI3K/Akt and signal transducer and activator of transcription 3 (STAT3) pathways.16,30 However, activation of the canonical FGFR pathways engaged by cell membrane-bound FGFR (MAPK, PI3K/Akt, and STAT3) was absent when FGFR-TACC fusions were expressed in mouse and human astrocytes and other cell types.10,31,40 Furthermore, pharmacologic inhibition of FGFR-TK in astrocytes ectopically expressing FGFR3-TACC3 and human GBM-derived gliomaspheres that express endogenous FGFR3-TACC3 fusion did not affect MAPK or PI3K/Akt (A.L. and A.I., personal communication). These findings could be explained with the reduced localization of FGFR3-TACC3 to the cell membrane, in contrast to its predominant intracellular compartmentalization and/or with a more effective negative feedback on canonical FGFR signaling in certain cellular contexts.10,40 Clearly, the weak activation of canonical cell membrane-bound FGFR signals by FGFR-TACC fusions has to be complemented by the ability of FGFR-TACC to engage other transforming, “noncanonical” signaling events that remain to be charted. The aberrant FGFR-TACC signaling is likely to be dictated by the TACC-guided intracellular localization of the fusion protein. Indeed, the TACC domain is absolutely essential for constitutive RTK signaling and the oncogenic activities of FGFR-TACC fusion proteins.10,16,31 It is known that TACC proteins localize to centrosomes and mitotic spindles, where they stabilize spindle microtubules during mitosis, thus controlling the process of chromosome segregation.11,12 In interphase, TACC proteins are primarily nuclear and it was suggested that they function as transcriptional coactivators.41 Consistent with a role of the TACC domain of FGFR-TACC in directing the intracellular compartmentalization of FGFR-TACC fusions are the findings that FGFR3-TACC3 tends to accumulate at the spindle poles of cells undergoing mitosis and displays substantial nuclear localization during interphase.10,40 It is logical to hypothesize that the aberrant localization of a constitutively active FGFR-TACC kinase in these compartments may cause direct phosphorylation events on atypical substrates, some of which may be essential for the biological and oncogenic functions associated with FGFR-TACC translocations. It is noteworthy that, beside the potent transforming activity displayed by FGFR-TACC fusions in vitro and in vivo, expression of FGFR-TACC proteins generates profound mitotic aberrations that result in chromosome mis-segregation and rampant aneuploidy (Fig. 2).10 The reported association between acute induction of aneuploidy and reduced fitness of normal cells42 is probably the reason for the detrimental effects that were observed in primary human astrocytes after acute expression of FGFR3-TACC3.10 However, the concurrent activation of as yet unknown growth-promoting pathways allows cells transduced with FGFR3-TACC3 to overcome the loss of fitness, resume growth, and ultimately develop a hyperproliferative and aneuploid state in long-term cultures. This series of events is also the likely explanation for the long latency of glioblastoma development (>6 mo) in mice in which ectopic expression of FGFR3-TACC3 was induced by lentiviral transduction of neural progenitor cells in the dentate gyrus.10 The elucidation of the signaling cascade activated by FGFR-TACC proteins in neural cells is the first step that can reveal which intracellular networks are affected and which control mechanisms of signal duration, signal amplitude, and spatial localization are disrupted by FGFR-TACC fusions leading to cellular transformation.
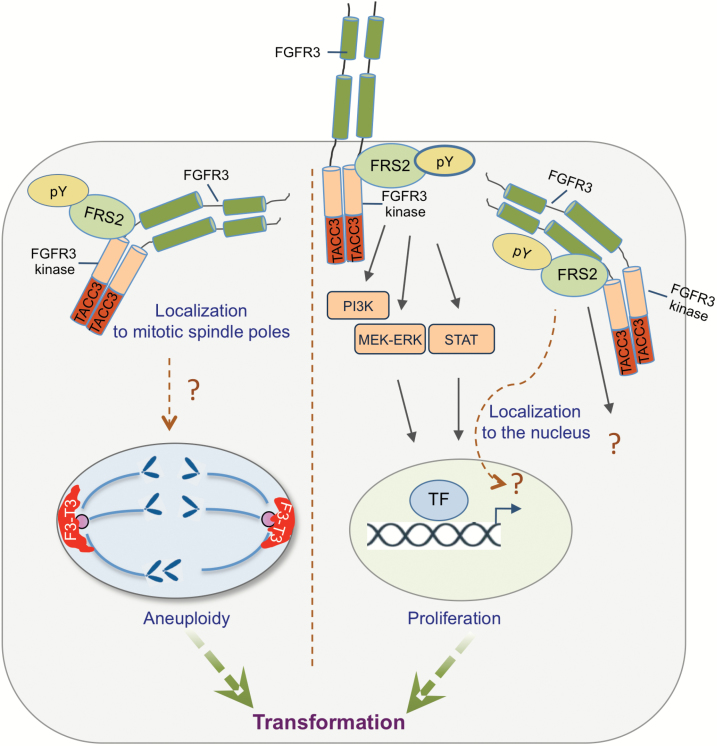
Fig. 2.
Mechanisms of cellular transformation by FGFR-TACC fusions. FGFR-TACC fusion proteins form dimers through the coiled-coil domain of TACC leading to auto-phosphorylation and constitutive FGFR tyrosine kinase activation. The active kinase acts weakly through canonical FGFR signaling (right). FGFR-TACC fusions engage unknown, noncanonical substrates in cellular compartments that may be dictated by the TACC moiety (eg, mitotic spindle, left; nuclear compartment, right), leading to hyperproliferation and aneuploidy. Phosphorylation of aberrant substrates in the cytosol may also promote other, yet undefined oncogenic activities.
FGFR-TACC Fusions as Actionable Targets in Glioma
Challenges and Opportunities
Recent findings reported that, beside GBM, FGFR-TACC fusions occur in the subgroup of isocitrate dehydrogenase (IDH) wild-type lower-grade glioma (World Health Organization grades II–III) with prevalence similar to GBM (~3%).15 Interestingly, FGFR-TACC fusions are invariably excluded in the larger lower-grade glioma IDH-mutant group, which has a relatively better prognosis in comparison with the IDH wild-type counterpart that manifests molecular and clinical features of GBM (“molecular GBM”).43,44 Thus, the finding that FGFR-TACC fusions occur in IDH wild-type but not in IDH-mutant glioma provides a clue to the molecular characterization of this glioma subtype.
The identification of FGFR-TACC fusions in IDH wild-type glioma, followed by the validation of these chromosomal rearrangements as powerful tumor-initiating events and addicting cancer alterations in mouse and human glioma, offered an unprecedented opportunity for testing the value of FGFR-TACC fusions as actionable lesions in neuro-oncology. The encouraging outcome of FGFR inhibition in preclinical studies in the subcutaneous and orthotopic setting in the mouse10,15 reinforced the theoretical rationale for therapeutic inhibition of FGFR-TACC in patients with FGFR-TACC–positive GBM. Preliminary data from a phase I trial showed clear antitumor activity in 2 patients with recurrent FGFR3-TACC3–positive GBM15,45 and prompted us to evaluate FGFR inhibition with AZD4547 in recurrent gliomas with FGFR-TACC fusions (Table 2).
Table 2.
Current clinical development of FGFR targeting anticancer drugs in GBM and other cancers
| Drug | Company | Target | Clinical Development |
|---|---|---|---|
| Glioblastoma | |||
| ADZ4547 | Astra Zeneca | FGFR1-3 | Phase II (EudraCT 2014-005428-81) |
| BGJ398 | Novartis | FGFR1-3 | Phase II (NCT01975701) |
| Solid tumors/lymphoma | |||
| JNJ-42756493 | Janssen R&D LLC | Pan-FGFR | Phase I (NCT01703481) |
| Debio 1347-101 | Debiopharm International SA | FGFR1-3 | Phase I (NCT01948297) |
| INCB054828 | Incyte Corporation | FGFR1-3 | Phase I (NCT02393248) |
| LY2874455 | Eli Lilly | FGFR1-4 | Phase I (NCT01212107) completed |
| TAS-120 | Taiho Pharma Co. Ltd. | FGFR1-4 | Phase I (NCT02052778) |
In this setting, the low frequency of FGFR-TACC fusions in malignant glioma poses the significant challenge of having to screen a large number of tumors to identify a considerable cohort of patients to enroll in clinical trials targeting FGFR-TACC fusions. However, the selection of FGFR-TACC–positive gliomas is the prerequisite for ideal and long overdue studies in neuro-oncology, involving trial population preselected on the basis of a tumor marker central to tumor biology and validated as a predictor of clinical response from preclinical studies.
A Screening Assay for Detection of FGFR-TACC Fusions
The accurate and sensitive identification of FGFR-TACC fusions in human glioma is challenging. Although recurrent FGFR3-TACC3 fusion transcripts have been reported, there is a remarkable variability of FGFR3-TACC3 variant mRNAs in human cancer.10,15,16,18,22–32 This notion underscores the difficulty of designing screening assays for the sensitive detection of all possible FGFR-TACC variants. Even more pronounced is the structural heterogeneity of FGFR-TACC fusions at the genomic level with distinct breakpoints within the FGFR3 and TACC3 genomic loci generating identical transcripts. To address such complexity, an unbiased real-time (RT)-PCR–based screening assay was proposed and validated to detect all possible FGFR3-TACC3 variants.15 The amplicons targeted by this assay include the key functional domains of FGFR3-TACC3, namely the TK-coding domain of FGFR3 and the TACC-coding domain of TACC3 (see arrows indicating the position of the primers used in the diagnostic RT-PCR assay in Fig. 1). Moreover, as FGFR3-TACC3 fusion proteins accumulate at high levels in FGFR3-TACC3–positive tumors, immunostaining of formalin-fixed paraffin embedded GBM samples using antibodies against the N-terminal region of FGFR3, which is invariably retained in all fusion variants, can be used for efficient and sensitive preselection of tumors for subsequent RT-PCR analysis. Specifically, strong and diffuse FGFR3 N-terminal expression was found in 31/32 FGFR3-TACC3–positive gliomas (M.S., personal communication) (Fig. 3). Thus, the standard molecular characterization of IDH wild-type glioma performed by diagnostic pathology laboratories should include this simple and reliable immunodetection of the FGFR3 N-terminal epitope.
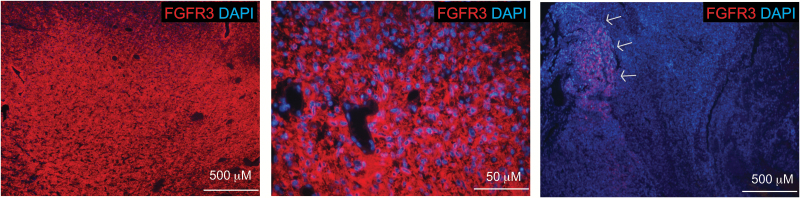
Fig. 3.
Immunostaining for FGFR3 N-terminus in formalin-fixed paraffin embedded GBM. Left panel shows elevated and widespread expression of FGFR3 (red). Middle panel shows the higher magnification microphotograph of FGFR3 immunostaining (red) and nuclear staining with 4′,6′-diamidino-2-phenylindole (blue) for the sample presented in the left panel. RT-PCR confirmed that the sample harbored the FGFR3-TACC3 rearrangement. Right panel shows weak FGFR3 positivity only in a limited area (arrows) in a GBM tumor that tested negative for FGFR3-TACC3 gene fusion when analyzed by RT-PCR. Values of the scale bars are indicated for each microphotograph.
FGFR-TACC as Proof of Principle for Personalized Clinical Trials in Neuro-oncology
A formidable limitation for a successful or at least interpretable clinical study in neuro-oncology remains the ability of the potentially effective compounds to efficiently reach the target across the blood–brain barrier and the validation of the target after treatment.46 As for most anticancer drugs, FGFR inhibitors were not developed specifically for tumors of the CNS, and the biodistribution of drugs within intracranial tumors is largely unknown because of the lack of pharmacokinetic studies in patients with malignant glioma.45,47–49 This remains a notable shortcoming that prevents clinical success, and therefore major efforts are needed to learn whether the different FGFR inhibitors reach therapeutic concentrations in CNS tumors through preclinical and clinical phase 0 studies.50–53 Two critical parameters deserve consideration to ensure functionally the success of FGFR targeted therapy in FGFR-TACC–positive glioma. The first aspect is the extent of target representation within the tumor and the ability to measure the target. It is well established that GBM tumors are highly heterogeneous, being characterized by coexistence of tumor clones harboring different genetic alterations (such as those involving RTK-coding genes) even within neighboring cells.54,55 When combined with the additional layer of heterogeneity emerging from the analysis of geographically distant biopsies and residual tumor cells from the surgical resection margin of the same tumor mass,56–58 it is clear that FGFR inhibition in GBM will have to address the notable degree of heterogeneity that characterizes the majority of human GBM.59 Thus, the question is whether FGFR-TACC fusions are clonal genetic alterations that can be clearly evaluated for their widespread occurrence within the tumor sample. The only study that analyzed a cohort of 12 GBM samples harboring FGFR3-TACC3 fusions by immunohistochemistry suggested that FGFR3-TACC3 fusion proteins are homogeneously expressed within the tumor mass and thus represent an optimal target for inhibition (31 of 32 FGFR3-TACC3–positive samples scored positive in follow-up analysis; M.S., personal communication).15 Considering the sensitivity and specificity of the FGFR3 antibody that recognizes the FGFR3 component of FGFR3-TACC3 fusions, the analysis at the single cell level of the distribution of the fusion protein within the tumor mass is a criterion that should always be considered when enrolling molecularly positive GBM in targeted studies. The strong tumor initiating capacity manifested by FGFR-TACC is consistent with the recent findings that FGFR-TACC fusions were reported as clonal events in clinical GBM specimens.10,15,60 Ultimately, the therapeutic value of single-agent clinical trials relies on the presence of the target in the vast majority of tumor cells, whereas only little benefit—if any at all—is to be expected when targeting biomarkers expressed by subclonal fractions of cells, regardless of the functional role of such biomarkers.
The second challenge for successful FGFR-TACC targeting is the longitudinal path of evolution of FGFR-TACC–positive GBM following the standard of care treatments (radiotherapy plus temozolomide) that is administered to most GBM patients at diagnosis. As early trials targeting FGFR-TACC will have to be focused primarily on patients harboring recurrent tumors, the evidence that FGFR-TACC fusions persist as clonal events in recurrent GBM is an essential notion that will have to be validated to ensure a rational enrollment of recurrent patients. At the moment, there is a dearth of information on whether and how GBM evolution affects the presence or even the clonality of FGFR-TACC fusions. Encouraging data from a limited number of matched primary/recurrent GBM pairs have suggested that, if detected at diagnosis, FGFR-TACC fusions are retained at recurrence following treatment with radiotherapy plus temozolomide with similarly widespread expression as observed at diagnosis.15,60
The strong oncogenic effects associated with FGFR-TACC fusions and the preclinical data showing acquired sensitivity toward FGFR inhibitors by tumors expressing FGFR-TACC are encouraging elements linking FGFR-TACC positivity with tumor sensitivity to FGFR inhibition in human GBM. Conversely, there are variable oncogenic effects associated with FGFR mutations in human cancer, including GBM. While modeling of certain mutations (FGFR2, FGFR3, etc) is linked to clear oncogenic transformation, the introduction of other tumor-specific mutations lacks any measurable oncogenic effect and fails to trigger sensitivity to FGFR inhibitors.61 Considering the heterogeneous spectrum of FGFR mutations in human cancer and glioma, rather than proceeding directly with attempts to clinically target FGFR mutations in GBM patients, the discovery of new and functionally untested mutations of FGFR genes should be followed by modeling the mutant kinases for oncogenic impact and testing the response of the corresponding patient-derived cells to FGFR inhibitory compounds. Similarly, overexpression of wild-type FGFR genes, such as that resulting from gene amplification, typically fails to drive addiction to FGFR signaling.10,62 Therefore, when detected in the GBM context, FGFR amplification cannot be considered a definite “druggable” event, at least in the absence of personalized preclinical testing.
Development of Resistance to FGFR Inhibitors
The encouraging preclinical and clinical responses recorded in human tumors harboring FGFR-TACC fusions should instigate our thinking on how to overcome the emergence of acquired mechanisms of resistance to FGFR inhibitors, an event that invariably limits the efficacy of inhibitors targeting RTK cancer drivers.63 It is currently unknown whether resistance of FGFR-TACC–positive gliomas to FGFR-TK inhibitors will be driven by FGFR gatekeeper mutations and/or the activation of bypass signaling pathways, 2 prominent but independent mechanisms conferring resistance to other genetic alterations of FGFR genes in different cancer types.62 In particular, epidermal growth factor receptor (EGFR) activation has been identified as a mechanism of resistance in bladder cancer cells with FGFR3 mutations after treatment with FGFR inhibitors.64 Whereas alterations of EGFR (amplification, EGFR variant III, etc) are excluded from untreated malignant glioma harboring FGFR-TACC fusions,15 it will be interesting to determine whether mechanisms of EGFR activation are selected for the induction of resistance to FGFR inhibitors. In other cancer cell lines, MET proto-oncogene, receptor tyrosine kinase activation has also been implicated in resistance to FGFR inhibition.65 As for EGFR, MET is recurrently activated in GBM and might therefore play a role in conferring resistance to FGFR inhibition in FGFR-TACC–positive GBM.66
Conclusions
Targeting the genetic alterations that activate FGFR-TK in cancer is a relatively new field of research. The compounds initially developed as inhibitors of FGFR kinases exhibited multi-kinase inhibitor capacity, displayed only limited potency against FGFR, and were characterized by considerable toxicity. During the last decade, highly potent and specific FGFR inhibitory molecules have been released, and several of them are now being tested in clinical cancer trials (Table 2). The results of the recently published clinical studies testing FGFR inhibitors in FGFR-TACC–positive tumors, including GBM, raise cautious optimism for future studies.15,45
Funding
This work was supported by National Institutes of Health grants to A.L. (R01CA101644, R01CA185486, and U54CA193313), and A.I. (R01CA178546, R01CA190891, U54CA193313, and a grant from The Chemotherapy Foundation).
Conflict of interest statement. No authors have a conflict of interest with the manuscript.
References
- 1. Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7(4):233–245. [DOI] [PubMed] [Google Scholar]
- 2. Prensner JR, Chinnaiyan AM. Oncogenic gene fusions in epithelial carcinomas. Curr Opin Genet Dev. 2009;19(1):82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ali MA. Chronic myeloid leukemia in the era of tyrosine kinase inhibitors: an evolving paradigm of molecularly targeted therapy. Mol Diagn Ther. 2016;20(4):315–333. [DOI] [PubMed] [Google Scholar]
- 4. Mahon FX, Etienne G. Deep molecular response in chronic myeloid leukemia: the new goal of therapy? Clin Cancer Res. 2014;20(2):310–322. [DOI] [PubMed] [Google Scholar]
- 5. Zhu G, Mische SE, Seigneres B. Novel treatment of acute promyelocytic leukemia: As2O3, retinoic acid and retinoid pharmacology. Curr Pharm Biotechnol. 2013;14(9):849–858. [DOI] [PubMed] [Google Scholar]
- 6. Gerber DE, Minna JD. ALK inhibition for non-small cell lung cancer: from discovery to therapy in record time. Cancer Cell. 2010;18(6):548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Husain H, Rudin CM. ALK-targeted therapy for lung cancer: ready for prime time. Oncology (Williston Park). 2011;25(7):597–601. [PubMed] [Google Scholar]
- 8. Perner S, Wagner PL, Demichelis F, et al. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia. 2008;10(3):298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368(25):2385–2394. [DOI] [PubMed] [Google Scholar]
- 10. Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peset I, Vernos I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 2008;18(8):379–388. [DOI] [PubMed] [Google Scholar]
- 12. Hood FE, Royle SJ. Pulling it together: the mitotic function of TACC3. Bioarchitecture. 2011;1(3):105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duncan CG, Killela PJ, Payne CA, et al. Integrated genomic analyses identify ERRFI1 and TACC3 as glioblastoma-targeted genes. Oncotarget. 2010;1(4):265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yao R, Natsume Y, Saiki Y, et al. Disruption of Tacc3 function leads to in vivo tumor regression. Oncogene. 2012;31(2):135–148. [DOI] [PubMed] [Google Scholar]
- 15. Di Stefano AL, Fucci A, Frattini V, et al. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res. 2015;21(14):3307–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parker BC, Annala MJ, Cogdell DE, et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013;123(2):855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang J, Wu G, Miller CP, et al. ;St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45(6):602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Still IH, Vince P, Cowell JK. The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics. 1999;58(2):165–170. [DOI] [PubMed] [Google Scholar]
- 20. Bass AJ, Lawrence MS, Brace LE, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43(10):964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stephens PJ, McBride DJ, Lin ML, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462(7276):1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Borad MJ, Champion MD, Egan JB, et al. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet. 2014;10(2):e1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Capelletti M, Dodge ME, Ercan D, et al. Identification of recurrent FGFR3-TACC3 fusion oncogenes from lung adenocarcinoma. Clin Cancer Res. 2014;20(24):6551–6558. [DOI] [PubMed] [Google Scholar]
- 24. Carneiro BA, Elvin JA, Kamath SD, et al. FGFR3-TACC3: a novel gene fusion in cervical cancer. Gynecol Oncol Rep. 2015;13:53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo G, Sun X, Chen C, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45(12):1459–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Helsten T, Elkin S, Arthur E, et al. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res. 2016;22(1):259–267. [DOI] [PubMed] [Google Scholar]
- 27. Majewski IJ, Mittempergher L, Davidson NM, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol. 2013;230(3):270–276. [DOI] [PubMed] [Google Scholar]
- 28. Shaver TM, Lehmann BD, Beeler JS, et al. Diverse, biologically relevant, and targetable gene rearrangements in triple-negative breast cancer and other malignancies. Cancer Res. 2016;76(16):4850–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res. 2014;20(15):4107–4114. [DOI] [PubMed] [Google Scholar]
- 30. Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22(4):795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3(6):636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yuan L, Liu ZH, Lin ZR, et al. Recurrent FGFR3-TACC3 fusion gene in nasopharyngeal carcinoma. Cancer Biol Ther. 2014;15(12):1613–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoshihara K, Wang Q, Torres-Garcia W, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34(37):4845–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16(2):139–149. [DOI] [PubMed] [Google Scholar]
- 35. Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10(2):116–129. [DOI] [PubMed] [Google Scholar]
- 36. Hadari YR, Gotoh N, Kouhara H, et al. Critical role for the docking-protein FRS2 alpha in FGF receptor-mediated signal transduction pathways. Proc Natl Acad Sci U S A. 2001;98(15):8578–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kouhara H, Hadari YR, Spivak-Kroizman T, et al. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89(5):693–702. [DOI] [PubMed] [Google Scholar]
- 38. Lax I, Wong A, Lamothe B, et al. The docking protein FRS2alpha controls a MAP kinase-mediated negative feedback mechanism for signaling by FGF receptors. Mol Cell. 2002;10(4):709–719. [DOI] [PubMed] [Google Scholar]
- 39. Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. 2005;287(2):390–402. [DOI] [PubMed] [Google Scholar]
- 40. Nelson KN, Meyer AN, Siari A, et al. Oncogenic gene fusion FGFR3-TACC3 is regulated by tyrosine phosphorylation. Mol Cancer Res. 2016;14(5):458–469. [DOI] [PubMed] [Google Scholar]
- 41. Guo Y, Scheuermann TH, Partch CL, et al. Coiled-coil coactivators play a structural role mediating interactions in hypoxia-inducible factor heterodimerization. J Biol Chem. 2015;290(12):7707–7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179(2):737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cancer Genome Atlas Research Network et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26), 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ceccarelli M, Barthel FP, Malta TM, et al. ;TCGA Research Network. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tabernero J, Bahleda R, Dienstmann R, et al. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2015;33(30):3401–3408. [DOI] [PubMed] [Google Scholar]
- 46. Levin VA, Tonge PJ, Gallo JM, et al. CNS anticancer drug discovery and development conference white paper. Neuro Oncol. 2015;17Suppl 6, vi1–vi26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dieci MV, Arnedos M, Andre F, Soria JC. Fibroblast growth factor receptor inhibitors as a cancer treatment: from a biologic rationale to medical perspectives. Cancer Discov. 2013;3(3):264–279. [DOI] [PubMed] [Google Scholar]
- 48. Gavine PR, Mooney L, Kilgour E, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72(8):2045–2056. [DOI] [PubMed] [Google Scholar]
- 49. Guagnano V, Furet P, Spanka C, et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J Med Chem. 2011;54(20):7066–7083. [DOI] [PubMed] [Google Scholar]
- 50. Calvert AH, Plummer R. The development of phase I cancer trial methodologies: the use of pharmacokinetic and pharmacodynamic end points sets the scene for phase 0 cancer clinical trials. Clin Cancer Res. 2008;14(12):3664–3669. [DOI] [PubMed] [Google Scholar]
- 51. Kummar S, Kinders R, Rubinstein L, et al. Compressing drug development timelines in oncology using phase ‘0’ trials. Nat Rev Cancer. 2007;7(2):131–139. [DOI] [PubMed] [Google Scholar]
- 52. Kummar S, Rubinstein L, Kinders R, et al. Phase 0 clinical trials: conceptions and misconceptions. Cancer J. 2008;14(3):133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murgo AJ, Kummar S, Rubinstein L, et al. Designing phase 0 cancer clinical trials. Clin Cancer Res. 2008;14(12):3675–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Snuderl M, Fazlollahi L, Le LP, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. [DOI] [PubMed] [Google Scholar]
- 55. Szerlip NJ, Pedraza A, Chakravarty D, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109(8):3041–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Glas M, Rath BH, Simon M, et al. Residual tumor cells are unique cellular targets in glioblastoma. Ann Neurol. 2010;68(2):264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aum DJ, Kim DH, Beaumont TL, et al. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg Focus. 2014;37(6):E11. [DOI] [PubMed] [Google Scholar]
- 60. Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liao RG, Jung J, Tchaicha J, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73(16):5195–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hallinan N, Finn S, Cuffe S, et al. Targeting the fibroblast growth factor receptor family in cancer. Cancer Treat Rev. 2016;46:51–62. [DOI] [PubMed] [Google Scholar]
- 63. Sierra JR, Cepero V, Giordano S. Molecular mechanisms of acquired resistance to tyrosine kinase targeted therapy. Mol Cancer. 2010;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Herrera-Abreu MT, Pearson A, Campbell J, et al. Parallel RNA interference screens identify EGFR activation as an escape mechanism in FGFR3-mutant cancer. Cancer Discov. 2013;3(9):1058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Harbinski F, Craig VJ, Sanghavi S, et al. Rescue screens with secreted proteins reveal compensatory potential of receptor tyrosine kinases in driving cancer growth. Cancer Discov. 2012;2(10):948–959. [DOI] [PubMed] [Google Scholar]
- 66. Petterson SA, Dahlrot RH, Hermansen SK, et al. High levels of c-Met is associated with poor prognosis in glioblastoma. J Neurooncol. 2015;122(3):517–527. [DOI] [PubMed] [Google Scholar]