Abstract
Background.
Previous studies have shown that MR imaging features can be used to predict survival and molecular profile of glioblastoma. However, no study of a similar type has been performed on lower-grade gliomas (LGGs).
Methods.
Presurgical MRIs of 165 patients with diffuse low- and intermediate-grade gliomas (histological grades II and III) were scored according to the Visually Accessible Rembrandt Images (VASARI) annotations. Radiomic models using automated texture analysis and VASARI features were built to predict isocitrate dehydrogenase 1 (IDH1) mutation, 1p/19q codeletion status, histological grade, and tumor progression.
Results.
Interrater analysis showed significant agreement in all imaging features scored (k = 0.703–1.000). On multivariate Cox regression analysis, no enhancement and a smooth non-enhancing margin were associated with longer progression-free survival (PFS), while a smooth non-enhancing margin was associated with longer overall survival (OS) after taking into account age, grade, tumor location, histology, extent of resection, and IDH1 1p/19q subtype. Using logistic regression and bootstrap testing evaluations, texture models were found to possess higher prediction potential for IDH1 mutation, 1p/19q codeletion status, histological grade, and progression of LGGs than VASARI features, with areas under the receiver-operating characteristic curves of 0.86 ± 0.01, 0.96 ± 0.01, 0.86 ± 0.01, and 0.80 ± 0.01, respectively.
Conclusion.
No enhancement and a smooth non-enhancing margin on MRI were predictive of longer PFS, while a smooth non-enhancing margin was a significant predictor of longer OS in LGGs. Textural analyses of MR imaging data predicted IDH1 mutation, 1p/19q codeletion, histological grade, and tumor progression with high accuracy.
Keywords: 1p/19q codeletion, IDH1 mutation, lower-grade gliomas, MR imaging, texture analysis
Importance of the study
Previous studies have shown that MRI features scored according to VASARI annotations can be used to predict survival and molecular profile of glioblastoma. However, no study of similar type has been performed on LGGs. On multivariate analysis, we showed that no enhancement on MRI was associated with longer PFS, while a smooth non-enhancing margin was associated with longer PFS and OS compared with an irregular non-enhancing margin. Our results demonstrated that imaging features scored using a standardized vocabulary had prognostic value in addition to traditionally recognized clinical and molecular markers in LGGs. In addition, multivariable texture models extracted from baseline MRI scans have the potential to accurately classify LGGs in terms of IDH1 mutation, 1p/19q codeletion, histological grade, and tumor progression. With proven accuracy, textural analysis will complement invasive tissue sampling, guiding patient management at an earlier stage of disease and in follow-up. Textural analysis may even serve as the standard in cases where invasive procedure is not available or appropriate.
Diffuse lower-grade gliomas (LGGs) are infiltrative neoplasms that generally include diffuse low- and intermediate-grade gliomas (World Health Organization [WHO] grade II or III).1 The outcomes of these tumors are variable—some recur after treatment within months and even progress to glioblastoma (WHO grade IV gliomas), while others remain indolent for years.2 Traditionally, the natural progression of LGGs is thought to be dependent on their histological class (astrocytomas vs oligoastrocytomas vs oligodendrogliomas). However, a recent study from The Cancer Genome Atlas (TCGA) Research Network classified LGGs into 3 categories based on isocitrate dehydrogenase (IDH) mutation and 1p/19q codeletion status: gliomas with an IDH mutation and 1p/19q codeletion, gliomas with an IDH mutation but no 1p/19q codeletion, and gliomas with wild-type IDH.1 This new classification scheme has been shown to capture the biologic characteristics of LGGs with greater fidelity than does histological class.1
MRI has served as an important noninvasive method for diagnosing gliomas and monitoring their treatment response. Previous radiogenomic analyses of glioblastoma have shown that the proportion of contrast enhancement (CE) and longest axis length of tumor on MR were significantly associated with poor survival.3 In a recent study of 120 patients with primary grades III (n = 35) and IV (n = 85) gliomas, Zhang et al. built a model using nonredundant preoperative MRIs and clinical data with a random forest algorithm that achieved accuracies of 86% in the training cohort and 89% in the validation cohort in predicting IDH genotype.4 However, no similar study has been performed to determine the association of MR imaging features with survival and molecular markers in LGGs.
To make the assessment of MR imaging features in gliomas more accurate and reproducible, a comprehensive feature set known as the Visually Accessible Rembrandt Images (VASARI) was developed in 2008. The VASARI annotations include 30 distinct imaging features with corresponding criteria clustered by categories related to lesion location, morphology of the lesion substance, morphology of the lesion margin, alterations in the vicinity of the lesion, and remote alterations (https://wiki.nci.nih.gov/display/CIP/VASARI).5 Previous studies have shown that measurements of these features by VASARI were highly reproducible, clinically meaningful, and biologically relevant in glioblastoma.3,6
Radiomics extracts and mines a large number of medical imaging features.7 These approaches based on machine learning have been increasingly used to quantify tumor phenotypic characteristics and to predict clinical outcomes.7 In 2014, Aerts et al.8 demonstrated that imaging features quantifying tumor image intensity, shape, and texture extracted from computed tomography data of 1019 patients with lung or head-and-neck cancer not only had prognostic power in the independent datasets of lung and head-and-neck cancer patients, but also had associations with gene-expression patterns. In 2015, Vallières et al.9 built a radiomics model from joint 2-fluoro-2-deoxy-d-glucose PET and MRI texture features using bootstrapping evaluations that predicted lung metastases in soft tissue sarcomas of the extremities with high sensitivity and specificity.
The purpose of our study was to perform a comprehensive analysis of MR imaging features of LGGs by using the VASARI feature set, as they relate to patient survival and molecular markers. In addition, we explored the possibility of using textural features extracted from MR imaging to make binary predictions of wild-type IDH versus IDH1 mutation; IDH1 mutation with 1p/19q codeletion versus IDH1 mutation without 1p/19q codeletion; grade II versus grade III LGGs; and progression versus nonprogression of LGGs.
Materials and Methods
Study Data
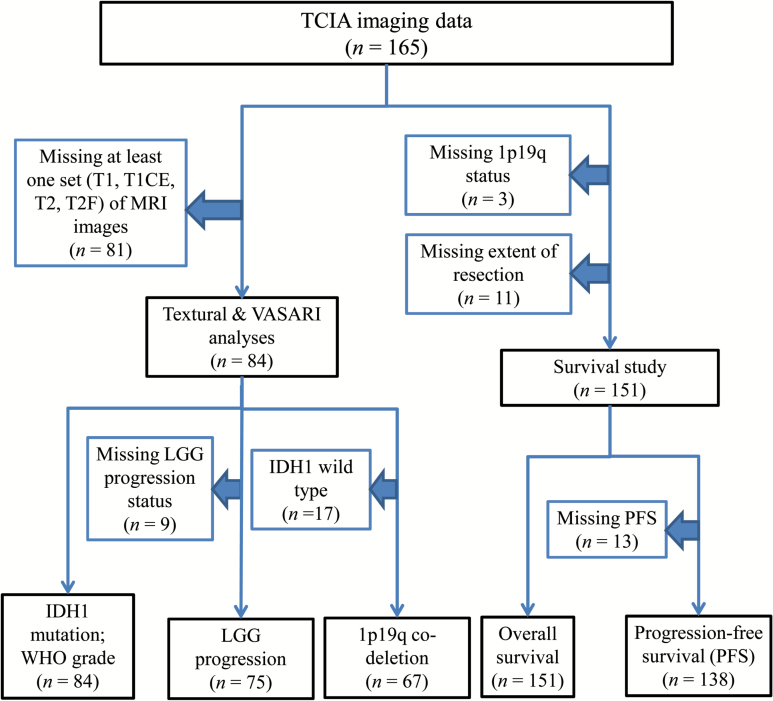
We identified 165 patients with diffuse low- and intermediate-grade gliomas (histological grades II and III) from TCGA who have overlapping presurgical MRI data from The Cancer Imaging Archive (TCIA),10,11 an imaging sharing resource that houses images corresponding to TCGA patients. A flowchart of the number of patients included for each analysis, along with the number of and reason why patients were excluded from each analysis, is shown in Fig. 1. As the patients had been previously de-identified by TCGA/TCIA, and their relevant information was available for public download, no institutional review board or Health Insurance Portability and Accountability Act approval was required for our study.
Fig. 1.
Flowchart of patients included and excluded for each analysis.
Imaging Review: VASARI Scores and Computation of Textures
For each patient, 2 neuroradiologists (H.T. and W.L.) with 7 and 20 years of experience, respectively, independently reviewed axial T1-weighted MR images before (T1W) and after gadolinium-based CE material administration (T1CE) as well as axial T2-weighted (T2W) and axial T2-weighted fluid attenuated inversion recovery (FLAIR) (T2F) images of the 165 LGG patients. The readers were blinded to the clinical data. Clear Canvas workstation, which allows visualization as well as annotation and markup of Digital Imaging and Communications in Medicine (DICOM) images, was used for imaging review. Each tumor was independently scored by the readers using the 30 imaging features defined according to the VASARI scoring system as previously described. The reader is referred to the Supplementary material for a complete description of the VASARI feature set. The interreader agreement for the imaging features was assessed using the kappa consistency test. Kappa values >0.81, in the range of 0.61–0.80, and <0.60 were considered to reflect excellent, good, and poor agreement, respectively. Final disagreement was resolved in a panel format including 2 additional coauthors (H.X.B. and L.Y.). For texture analysis, 3D regions of interest (ROIs) for each MR imaging set of each patient were manually drawn slice-by-slice in the axial plane for each of the available sequences (T1W, T2W, T1CE, T2F) by an expert radiologist. A total of 42 texture features were then extracted using 3D analysis from the T1W, T2W, T1CE, and T2F scans: 3 histogram-based textures, 8 texture features from the Gray-Level Co-occurrence Matrix (GLCM), 13 texture features from the Gray-Level Run-Length Matrix (GLRLM), 13 texture features from the Gray-Level Size Zone Matrix (GLSZM), and 5 texture features from the Neighborhood Gray-Tone Difference Matrix (NGTDM). The reader is referred to the Supplementary material for a complete description of all texture features, acronyms, and references.
Association of VASARI Imaging Features with Survival
Progression-free survival (PFS) was defined as the time that passes from the day on which a patient is enrolled and the date on which tumor progresses. Overall survival (OS) was defined as the time between initial diagnosis and death or last follow-up. We examined the association between each VASARI imaging feature with PFS and OS using the Kaplan–Meier survival curves and log-rank analyses. Features that were significant on the univariate analysis (P < .05) were entered into multivariate survival analysis based on the Cox proportional hazards ratio model, after incorporating clinical and pathological variables, including age, gender, extent of resection, histological type, histological grade, pretreatment Karnofsky performance scale (KPS), and IDH1 1p/19q codeletion status.
Prediction of IDH1 Mutation, 1p/19q Codeletion Status, Histological Grade, and LGG Progression with Textural Imaging Features
For this part of the work, the patient imaging data and outcome availability for each modeled outcome were as follows: (i) IDH1 mutation: 63 mutations, 21 wild type; (ii) 1p/19q codeletion status: 17 codeletion, 50 non-codeletion; (iii) histological grade: 35 grade II gliomas, 49 grade III gliomas; and (iv) LGG progression: 28 progressions, 47 nonprogressions. Binary clinical outcomes were modeled by predicting the class with the lowest number of instances.
Forty-two texture features were computed as described by Vallières et al.9 using the 40 possible combinations of the following extraction parameters: (i) 3D isotropic scales of 0.5 mm, 1 mm, 2 mm, 3 mm, and 4 mm; (ii) “uniform” and “equal-probability” quantization; (iii) number of gray-levels of 8, 16, 32, and 64. The 4 initial feature sets that were tested comprised one T1-based and one T2-based MR imaging sequence (each containing 2 × 42 × 40 = 3360 scan-texture-parameters): (i) T1W-T2W, (ii) T1W-T2F, (iii) T1CE-T2W, and (iv) T1CE-T2F.
Multivariate models were constructed for each initial feature set and modeled outcome using imbalanced-adjusted logistic regression, an adaptation of the method based on Schiller et al9 following the general methodology developed by Vallières et al.12 All initial feature sets first underwent feature set reduction using 100 bootstrap training samples to yield reduced feature sets of 25 different scan-texture features. Then, feature selection was performed by maximizing the area under the receiver-operating characteristic curve (AUC)632+ metric in 100 bootstrap training and testing samples to obtain texture models combining 1 to 10 variables (model order).13 For each outcome, the feature set and model order providing the combination of texture variables with the best parsimonious properties (ie, the simplest model with the best predictive properties) were chosen. The model was built using the feature set that provided the highest prediction curve while using the lowest model order before the AUC632+ metric started reaching a plateau or decreasing. Finally, the prediction performance of the 4 chosen texture models was estimated using average AUCs, sensitivities, and specificities obtained in 100 bootstrap testing samples.
Prediction of IDH1 Mutation, 1p/19q Codeletion Status, Histological Grade, and LGG Progression with VASARI Features
In order to directly compare the prognostic potential of VASARI features and texture features, we built VASARI multivariable models to predict IDH1 mutation, 1p/19q codeletion status, histological grade, and LGG progression. From the full set of VASARI features, the feature selection, model choice, and prediction estimation methods were the same as described in the section above for texture analysis.
Comparison of the Predictive Potential of Models Using Clinical Variables, VASARI Features, and Texture Features with Random Forest Analysis
Logistic or Cox regression analysis is best suited for modeling continuous variables only (eg, texture and VASARI features). These types of analyses provide a fast way to mine the best features for that category of inputs. On the other hand, random forests are more complex classifiers and are designed to implement any type of inputs, either categorical, as is often the case for clinical information, or continuous for radiomics data. In order to directly compare their predictive potential, clinical variables (categorical data), VASARI features, and texture features (continuous data) were thus included in a random forest analysis to predict IDH1 mutation, 1p/19q codeletion status, histological grade, and LGG progression. The clinical variables included age, KPS, histological type, grade (removed for histological grade outcome), laterality, location, gender, radiotherapy (yes vs no; removed for all outcomes except LGG progression), chemotherapy (yes vs no; removed for all outcomes except LGG progression), and IDH1 1p/19q subtype (removed for IDH1, 1p/19q outcomes). The VASARI and texture features included in the random forest analysis were the variables forming the best multivariable models as determined using the methods described in the 2 previous sections above. Random forest classifiers were trained (inherently using bootstrapping) on the whole cohort using 500 trees. Prediction performance was estimated using out-of-bag observations. Prediction balance between sensitivity and specificity was achieved by finding the optimal cost (ie, emphasis) on the classification of positive instances, and by maximizing 0.5*AUC + 0.5*(1 − |Sensitivity − Specificity|) on out-of-bag estimates. Random forest analysis was performed using clinical variables only; VASARI features only; texture features only; a combination of VASARI and texture features; and a combination of clinical variables, VASARI, and texture features.
Results
Interreader Agreement
Interrater analysis showed significant agreement in all VASARI imaging features scored. Interreader agreements for all the imaging features were good to excellent (kappa value = 0.703–1.000) (Supplementary Table 1).
Correlation between VASARI Imaging Features and Progression-Free Survival
Kaplan–Meier analysis showed that 7 VASARI features were significantly associated with PFS (P < .1): enhancement quality, proportion of enhancing tumor, proportion of non-enhancing tumor, definition of the non-enhancing margin, diffusion, satellites, and lesion size. On multivariate Cox regression analysis, enhancement quality, definition of the non-enhancing margin, the presurgical KPS score, and IDH1 1p/19q subtype were significantly associated with PFS after taking into account gender, tumor location, and histology (Table 1). Specifically, tumors with no enhancement had longer PFS than tumors with either mild or marked enhancement (P = .048 by log-rank test; Supplementary Fig. 1A). Tumors with a smooth margin were associated with improved PFS compared with those with an irregular margin (P = .02 by log-rank test; Supplementary Fig. 1B).
Table 1.
Correlation between VASARI imaging features and PFS on multivariate Cox analysis
| Variable | Hazard Ratio | P-value |
|---|---|---|
| Enhancement quality | 1.485 (1.051, 2.099) | .025 |
| Definition of the non- enhancing margin | 2.056 (1.168, 3.518) | .012 |
| KPS | 0.969 (0.942, 0.996) | .023 |
| IDH1 1p/19q subtype | 3.035 (1.937, 4.756) | <.001 |
Note: Data are hazard ratio estimates, with 95% CIs in parentheses, for variables included in the Cox regression model involving imaging features plus clinical variables, for the analysis of the association between the imaging features and PFS after adjusted for standard clinical variables.
Correlation between VASARI Imaging Features and Overall Survival
Kaplan–Meier analysis showed that 10 VASARI features were significantly associated with OS: eloquent brain involved, proportion of enhancing tumor, proportion of non-enhancing tumor, cysts, multifocal or multicentric, definition of the non-enhancing margin, proportion of edema, diffusion, enhancing tumor crosses midline, and satellites. On multivariate Cox regression analysis, definition of the non-enhancing margin, age, tumor grade, and IDH1 1p/19q subtype were significantly associated with OS after taking into account gender, tumor location, and histology (Table 2). Specifically, tumors with a smooth margin had longer OS than those with an irregular margin (P = .002 by log-rank test; Supplementary Fig. 1C).
Table 2.
Correlation between VASARI imaging features and OS on multivariate Cox analysis
| Variable | Hazard Ratio | P-value |
|---|---|---|
| Definition of the non-enhancing margin | 1.088 (1.047, 1.131) | .017 |
| Age | 1.088 (1.047, 1.131) | <.001 |
| WHO grade | 5.298 (2.027, 13.849) | .001 |
| IDH1 1p/19q subtype | 2.655 (1.489, 4.732) | .001 |
Note: Data are hazard ratio estimates, with 95% CIs in parentheses, for variables included in the Cox regression model involving imaging features plus clinical variables, for the analysis of the association between the imaging features and OS after adjusted for standard clinical variables.
Prediction of IDH1 Mutation, 1p/19q Codeletion Status, Histological Grade, and LGG Progression with Textural Imaging Features
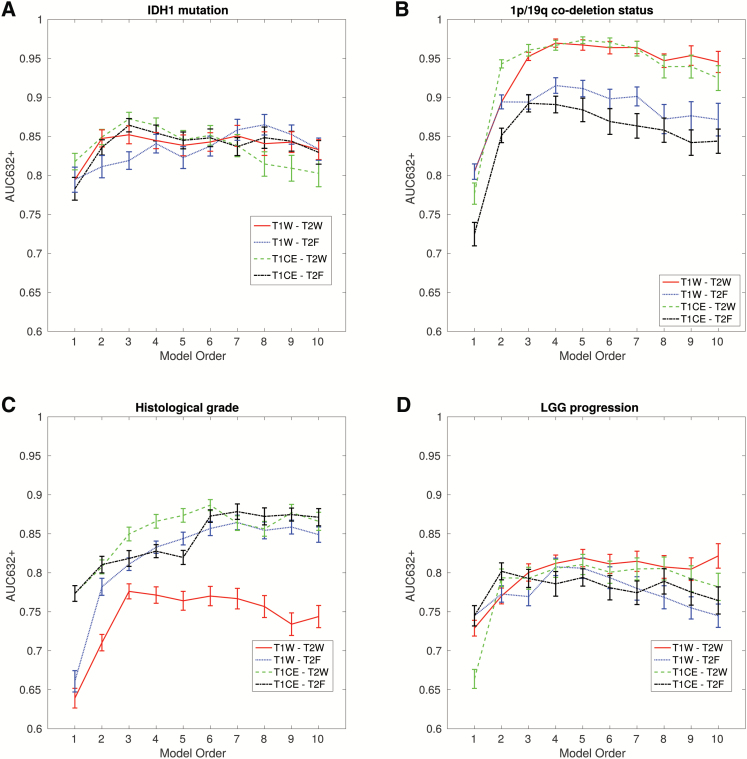
Following feature set reduction and feature selection using imbalanced-adjusted logistic regression and bootstrap resampling, texture models with orders 1 to 10 that maximized the AUC632+ metric were computed for each of the 4 feature sets to model the 4 outcomes (Fig. 2A–D). By inspecting the curves in Fig. 1, we determined that the combinations of 3 textures from the T1CE-T2W set, 3 textures from the T1CE-T2W set, 4 textures from the T1CE-T2W set, and 4 textures from the T1W-T2W set provided the best predictive properties for IDH1 mutation, 1p/19q codeletion status, histological grade, and LGG progression outcomes, respectively. For the IDH1 mutation outcome, the optimal set of features included global-skewness (T2W), GLRLM run-length variance (T2W), and GLRLM short run low Gray-level emphasis (T2W), which reached an AUC of 0.86 ± 0.01, a sensitivity of 0.75 ± 0.03, and a specificity of 0.78 ± 0.02. For 1p/19q codeletion status, the optimal set of features included GLRLM low Gray-level run emphasis (LGRE) (T1CE), GLSZM short zone low Gray-level emphasis (SZHGE) (T2W), and GLRLM long run high Gray-level emphasis (T2W), which reached an AUC of 0.96 ± 0.01, a sensitivity of 0.90 ± 0.02, and a specificity of 0.89 ± 0.02. For the histological grade outcome, the optimal set of features included GLCM-homogeneity (T1CE), GLSZM short-zone emphasis (SZE) (T2W), GLSZM-SZE (T1CE), and global-kurtosis (T1CE), which reached an AUC of 0.86 ± 0.01, a sensitivity of 0.74 ± 0.02, and a specificity of 0.79 ± 0.02. For the LGG progression outcome, the optimal set of features included GLRLM long run low Gray-level emphasis (T1W), GLRLM-LGRE (T2W), GLSZM-SZHGE (T1W),and GLSZM zone size variance (T2W), which reached an AUC of 0.80 ± 0.01, a sensitivity of 0.76 ± 0.03, and a specificity of 0.72 ± 0.03. The direction of correlation between each significant texture feature included in the final model and outcome is shown in Supplementary Table 2.
Fig. 2.
Inspection of predictive properties of multivariable texture models constructed from 4 feature sets: (i) T1W-T2W, (ii) T1W-T2F, (iii) T2CE-T2W, and (iv) T1CE-T2F. Estimation of prediction performance is shown for combinations of 1 to 10 texture features (model orders) in terms of the AUC632+ metric for: (A) IDH1 mutation, (B) 1p/19q codeletion status, (C) histological grade, and (D) LGG progression.
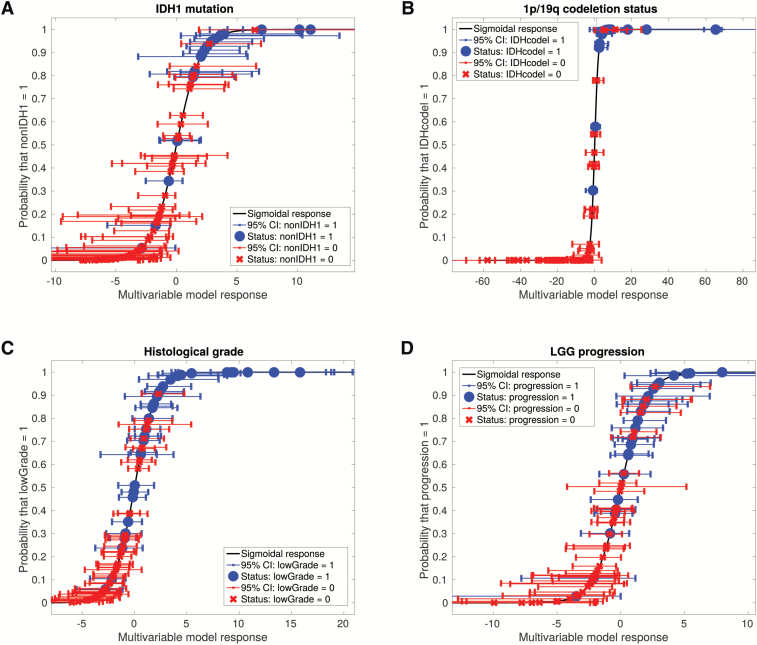
To demonstrate the classification capability of the optimal models shown in Supplementary Table 2, final logistic regression coefficients were computed for all models using 100 bootstrap training samples. The posterior probability of observing a given outcome as a function of the logistic regression response of the models was calculated along with the associated 95% CIs of the model responses in the bootstrap samples (Fig. 3).
Fig. 3.
Probability of observing a given outcome as a function of the response of optimal multivariable texture models developed in this work, for all patients of the cohort: (A) IDH1 mutation (nonIDH1), (B) 1p/19q codeletion status (IDHcodel), (C) histological grade (lowGrade), and (D) LGG progression (progression). It can be seen that the optimal texture models can significantly separate the patients of the 2 classes for each outcome, especially in the case of the 1p/19q codeletion status.
Prediction of IDH1 Mutation, 1p/19q Codeletion Status, Histological Grade, and LGG Progression with VASARI Features
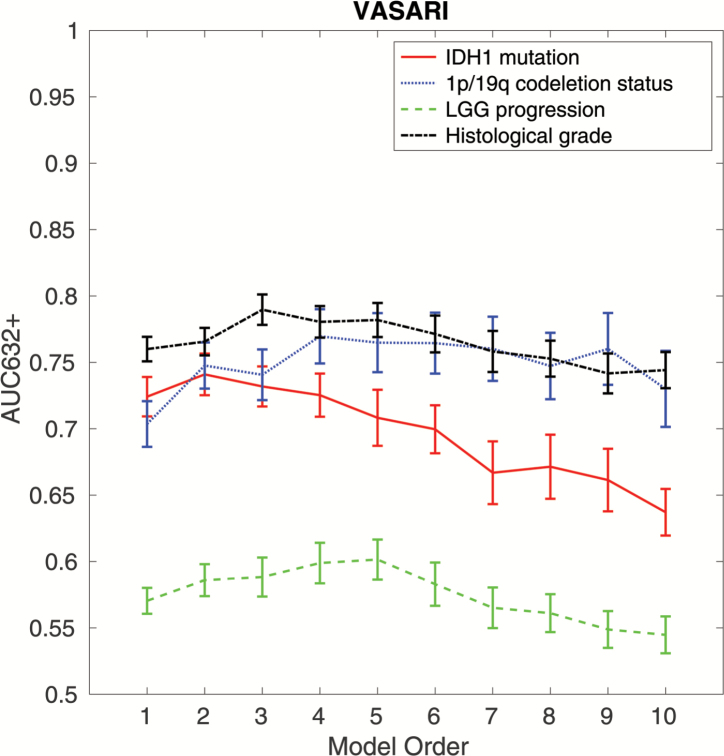
For the IDH1 mutation outcome, the optimal set of features included proportion necrosis and lesion size, which reached an AUC of 0.73 ± 0.02, a sensitivity of 0.69 + 0.03, and a specificity of 0.69 ± 0.02. For the 1p/19q codeletion outcome, the optimal set of features included multifocal or multicentric, edema proportion, tumor location, and enhancing proportion, which reached an AUC of 0.78 ± 0.01, a sensitivity of 0.72 ± 0.03, and a specificity of 0.67 ± 0.03. For the histological grade outcome, the optimal set of features included enhancing proportion, definition of the non-enhancing margin, and diffusion, which reached an AUC of 0.78 ± 0.01, a sensitivity of 0.72 ± 0.03, and a specificity of 0.67 ± 0.03. For the LGG progression outcome, the optimal set of features included tumor location, enhancement quality, necrosis proportion, T1/FLAIR ratio, and thickness of enhancing margin, which reached an AUC of 0.58 ± 0.02, a sensitivity of 0.54 ± 0.04, and a specificity of 0.58 ± 0.03. The results are shown in Fig. 4. The direction of correlation between each significant VASARI feature included in the final model and outcome is shown in Supplementary Table 3.
Fig. 4.
Inspection of predictive properties of VASARI models constructed for 4 different outcomes: (i) IDH1 mutation, (ii) 1p/19q codeletion status, (iii) histological grade, and (iv) LGG progression. Estimation of prediction performance is shown for combinations of 1 to 10 texture features (model orders) in terms of the AUC632+ metric.
Comparison of the Predictive Potential of Models Using Clinical Variables, VASARI Features, and Texture Features with Random Forest Analysis
Random forest analysis was used to directly compare the predictive potential of clinical, VASARI, and texture features (Supplementary Table 4). For IDH1 mutation, 1p/19q codeletion, and LGG progression, we found that the best category of predictor was texture features, with AUCs of 0.79, 0.88, and 0.70, respectively. For histological grade, the best category of predictor was VASARI features, with an AUC of 0.73. However, the combination of clinical, VASARI, and texture variables showed that clinical variables can successfully complement imaging features for the prediction of IDH1 mutation and histological grade, with respective AUCs of 0.86 and 0.78. On the other hand, 1p/19q codeletion was best modeled with a combination of texture and VASARI features only (AUC = 0.89), and LGG progression was best modeled with texture features only (AUC = 0.70). The detailed results are shown in Supplementary Tables 5, 6, 7, 8, and 9.
Discussion
We found a significant association between enhancement quality (none vs mild vs marked) and PFS. Specifically, no enhancement was associated with longer PFS than either mild or marked enhancement. In addition, a smooth definition of the non-enhancing margin was associated with longer PFS and OS than an irregular non-enhancing margin. Of note, these imaging features were still strongly associated with patients’ survival after incorporating age, KPS, extent of resection, tumor grade, and IDH 1p/19q subtype.
In glioblastoma, previous studies have shown that the proportion of CE on MRI was associated with survival.3,14 However, in LGG, the prognostic significance of CE on survival remains less well understood, and differing results have been reported depending on LGG and mutation status.15–17 In our study, the proportion of CE was not a significant predictor of either PFS or OS, but the lack of CE was a positive predictor of PFS. There was no significant difference in survival between patients who had tumors with mild enhancement and those who had tumors with marked enhancement. Grade III tumors were more likely to enhance than grade II tumors (84% vs 44%, respectively), but a significant proportion of the grade II tumors demonstrated CE as well.
We found that a smooth non-enhancing margin was associated with longer PFS and OS compared with an irregular non-enhancing margin. This is in contrast to a prior study in glioblastoma which found that a smooth edge of CE predicted poor prognosis, while a sharp edge was a positive factor.6 However, a previous study of 43 grade III gliomas found no significant association of non-enhancing margin with survival.18 To our knowledge, our study was the first to demonstrate that the definition of the non-enhancing margin on MRI can predict survival in a combined cohort of grade II and grade III gliomas.
In this study, we constructed a multivariable texture model extracted from baseline MRI scans that achieved an AUC of 0.86, a sensitivity of 0.75, and a specificity of 0.78 in predicting IDH1 mutation in LGGs. This logistic regression model performed better than a model using VASARI features via the same methods and comparable to the models built with random forest analysis incorporating clinical variables, VASARI, and texture features. In comparison to previous studies that relied on complete clinical data19 or advanced imaging techniques,20 our results demonstrated the ability of machine-learning algorithms to achieve accurate prediction of IDH mutation in LGGs with very few preoperative MRI texture features alone. These results, if validated with other datasets, may affect clinical management in the future, since these imaging studies were obtained routinely before surgery. They can also be useful in a research setting where a large amount of samples have to be genotyped for IDH mutation.
The codeletion of chromosomal arms 1p and 19q is a characteristic and early genetic event in oligodendroglial tumors.15 Tumors with 1p/19q codeletion are associated with a better prognosis and enhanced response to therapy.16 Moreover, grades II and III gliomas with 1p/19q codeletion are also mutated in IDH1/2.21 In a smaller study of 55 patients with oligodendrogliomas, Brown et al.17 used S-transform-based texture analysis of preoperative MR images to predict 1p/19q codeletion with an AUC of 0.94, a sensitivity of 0.93, and a specificity of 0.96, versus a sensitivity of 0.70 and a specificity of 0.63 for genotype prediction by blinded experts. We corroborated these results in a larger cohort of patients that included both astrocytomas and oligodendrogliomas, and our optimal texture model achieved an average bootstrap testing AUC of 0.96, a sensitivity of 0.90, and a specificity of 0.89 in predicting 1p/19q codeletion status in LGGs. This logistic regression multivariable model was superior to that built using either VASARI features or random forest analysis. Our results afforded a method for predicting 1p/19q codeletion that is both noninvasive and uses data from routinely acquired MR images.
The current standard for glioma grading is based on histopathological assessment, which has 2 major limitations. First, it is an invasive procedure. Second, it has an inherent sampling error, especially with stereotactic biopsy. Our multivariable texture model achieved an AUC of 0.86, a sensitivity of 0.74, and a specificity of 0.79 in distinguishing grade II from grade III gliomas, a performance which compared favorably with previous studies.20–22 However, these prior studies relied on carefully selected ROIs drawn by neuroradiologists and multiple advanced MR imaging modalities, including diffusion-weighted imaging, diffusion-tensor imaging, MR spectroscopy, and perfusion-weighted imaging. These resources may not be available in non-academic institutions. Our algorithm demonstrated the potential of achieving high predictive accuracy using only conventional MRI sequences.
One texture feature in our final model that predicted histological grade was GLCM-homogeneity on contrast-enhanced T1-weighted images. We found that high tumor “homogeneity” was associated with higher grade. This result may be counterintuitive, since previous studies in breast cancer23 and soft tissue sarcomas24 have demonstrated that tumor heterogeneity is associated with higher tumor grade and more aggressive pathological features. We have 2 possible explanations for our results. First, MR CE in grade II tumors is more heterogeneous than CE in grade III tumors. This characteristic of CE in grade II gliomas was not captured by the corresponding VASARI feature key, but was demonstrated by Pallud et al.,25 where among the 143 cases of grade II tumor with CE, CE was characterized as “patchy and faint” in 93 and “nodular-like” in 50. Second, for those tumors without CE, our previous observation in soft tissue sarcoma was that high texture homogeneity was mostly due to large homogeneous necrotic centers.9 This suggests that the tumor has a fast growing rate, thus compatible with higher grade.
The ultimate goal of using machine-learning algorithm to study glioma is to predict patient outcome. Our multivariable texture model constructed using logistic regression achieved an AUC of 0.80, a sensitivity of 0.76, and a specificity of 0.72 in predicting LGG progression. These results were superior to those obtained using either VASARI features (AUC of 0.58) or random forest analysis (AUC of 0.69). Few studies have investigated the use of radiomics, specifically texture analysis based on baseline MR imaging, to predict outcomes in patients with gliomas, and existing studies focused exclusively on glioblastoma.21,26 Our study was the first in the literature to demonstrate the potential of using textural analysis from baseline MR imaging to predict glioma progression.
We acknowledge several limitations to our study. First, some patients could not be included due to incompleteness of MR sequence data from TCIA. In addition, the heterogeneity of different imaging parameters used by different investigators contributing to TCGA/TCIA data could not be controlled. Second, clinical variables, such as PFS and extent of resection, are not available for all patients from TCGA. Third, this study was partly based on the radiologist-scored imaging features. Although a good to excellent interreader agreement based on kappa consistency test has been achieved, the scores can be subject to interreader variability and random errors during manual contour tracing. Lastly, collection and assessment of new LGGs is still ongoing by our group, and we need more cases to validate our initial conclusions made in this paper.
In conclusion, tumor enhancement and an irregular non-enhancing margin on MR imaging were associated with shorter PFS, while a smooth non-enhancing margin was a positive predictor of OS. Multivariable texture models extracted from baseline MRI scans were able to classify LGGs in terms of IDH1 mutation, 1p/19q codeletion, histological grade, and tumor progression with high sensitivity and specificity.
Online Resources
The ROI masks used for texture analysis are made available in DICOM format on the TCIA website: http://www.cancerimagingarchive.net/. All MATLAB software code used to compute the texture features and multivariable model results is freely shared under the GNU General Public License at: https://github.com/mvallieres/radiomics.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This work was supported by the Natural Science Foundation of China (81301988 to L.Y., 2014-1 to 2016-12), and China Ministry of Education Doctoral Program Spot Foundation (20130162120061 to L.Y.), Shenghua Yuying Project of Central South University to L.Y., and Natural Science Foundation of Hunan Province of China (14JJ2042, 2014-1 to 2016-12) to J.Z.
Conflict of interest statement. None declared.
Supplementary Material
References
- 1. Brat DJ, Verhaak RG, Aldape KD, et al. Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van den Bent MJ, Wefel JS, Schiff D, et al. Response assessment in neuro-oncology (a report of the RANO group): assessment of outcome in trials of diffuse low-grade gliomas. Lancet Oncol. 2011;12(6):583–593. [DOI] [PubMed] [Google Scholar]
- 3. Gutman DA, Cooper LA, Hwang SN, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267(2):560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang B, Chang K, Ramkissoon S, et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol. 2016;0(0):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiki for the VASARI feature set The National Cancer Institute Web site. Available at https://wikicancerimagingarchivenet/display/Public/VASARI+Research+Project Updated July 25, 2016. [Google Scholar]
- 6. Gevaert O, Mitchell LA, Achrol AS, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273(1):168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bai HX, Lee AM, Yang L, et al. Imaging genomics in cancer research: limitations and promises. Br J Radiol. 2016;89(1061):20151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vallières M, Freeman CR, Skamene SR, et al. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60(14):5471–5496. [DOI] [PubMed] [Google Scholar]
- 10. Pedano N, Flanders AE, Scarpace L. et al. Radiology data from The Cancer Genome Atlas Low Grade Glioma [TCGA-LGG] collection. Cancer Imaging Arch. 2016. doi:10.7937/K9/TCIA.2016.L4LTD3TK. [Google Scholar]
- 11. Clark K, Vendt B, Smith K. et al. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schiller TW, Chen Y, Naqa IE, et al. Modeling radiation-induced lung injury risk with an ensemble of support vector machines. Neurocomputing. 2010;73(10–12):1861–1867. [Google Scholar]
- 13. Sahiner B, Chan HP, Hadjiiski L. Classifier performance prediction for computer-aided diagnosis using a limited dataset. Med Phys. 2008;35(4):1559–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nicolasjilwan M, Hu Y, Yan C, et al. TCGA Glioma Phenotype Research Group. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. J Neuroradiol. 2015;42(4):212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pope WB, Sayre J, Perlina A, et al. MR imaging correlates of survival in patients with high-grade gliomas. AJNR Am J Neuroradiol. 2005;26(10):2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 16. Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66(20):9852–9861. [DOI] [PubMed] [Google Scholar]
- 17. Woehrer A, Sander P, Haberler C, et al. Research Committee of the European Confederation of Neuropathological Societies. FISH-based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice—a publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies (Euro-CNS). Clin Neuropathol. 2011;30(2):47–55. [DOI] [PubMed] [Google Scholar]
- 18. Brown R, Zlatescu M, Sijben A, et al. The use of magnetic resonance imaging to noninvasively detect genetic signatures in oligodendroglioma. Clin Cancer Res. 2008;14(8):2357–2362. [DOI] [PubMed] [Google Scholar]
- 19. Law M, Yang S, Wang H, et al. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am J Neuroradiol 2003;24(10):1989-1998. [PMC free article] [PubMed] [Google Scholar]
- 20. Zacharaki EI, Wang S, Chawla S, et al. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med. 2009;62(6):1609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caulo M, Panara V, Tortora D, et al. Data-driven grading of brain gliomas: a multiparametric MR imaging study. Radiology 2014;272(2):494-503. [DOI] [PubMed] [Google Scholar]
- 22. Soussan M, Orlhac F, Boubaya M, et al. Relationship between tumor heterogeneity measured on FDG-PET/CT and pathological prognostic factors in invasive breast cancer. PLoS One. 2014;9(4):e94017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao F, Ahlawat S, Farahani SJ, et al. Can MR imaging be used to predict tumor grade in soft-tissue sarcoma? Radiology. 2014;272(1):192–201. [DOI] [PubMed] [Google Scholar]
- 24. Pallud J, Capelle L, Taillandier L, et al. Prognostic significance of imaging contrast enhancement for WHO grade II gliomas. Neuro Oncol. 2009;11(2):176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee J, Jain R, Khalil K, et al. Texture feature ratios from relative CBV maps of perfusion MRI are associated with patient survival in glioblastoma. AJNR Am J Neuroradiol. 2016;37(1):37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang D, Rao G, Martinez J, et al. Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma. Med Phys. 2015;42(11):6725–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.