Abstract
Background.
Glioblastoma (GBM) is the most common primary malignant brain tumor. Nomograms are often used for individualized estimation of prognosis. This study aimed to build and independently validate a nomogram to estimate individualized survival probabilities for patients with newly diagnosed GBM, using data from 2 independent NRG Oncology Radiation Therapy Oncology Group (RTOG) clinical trials.
Methods.
This analysis included information on 799 (RTOG 0525) and 555 (RTOG 0825) eligible and randomized patients with newly diagnosed GBM and contained the following variables: age at diagnosis, race, gender, Karnofsky performance status (KPS), extent of resection, O6-methylguanine-DNA methyltransferase (MGMT) methylation status, and survival (in months). Survival was assessed using Cox proportional hazards regression, random survival forests, and recursive partitioning analysis, with adjustment for known prognostic factors. The models were developed using the 0525 data and were independently validated using the 0825 data. Models were internally validated using 10-fold cross-validation, and individually predicted 6-, 12-, and 24-month survival probabilities were generated to measure the predictive accuracy and calibration against the actual survival status.
Results.
A final nomogram was built using the Cox proportional hazards model. Factors that increased the probability of shorter survival included greater age at diagnosis, male gender, lower KPS, not having total resection, and unmethylated MGMT status.
Conclusions.
A nomogram that assesses individualized survival probabilities (6-, 12-, and 24-mo) for patients with newly diagnosed GBM could be useful to health care providers for counseling patients regarding treatment decisions and optimizing therapeutic approaches. Free software for implementing this nomogram is provided: http://cancer4.case.edu/rCalculator/rCalculator.html.
Keywords: glioblastoma, nomogram, NRG Oncology, RTOG, survival.
Importance of the Study
GBM is the most common primary malignant brain tumor. A nomogram accounts for several prognostic factors and is an easily accessible tool for physicians to use on behalf of their patients for predicting survival, developing individualized cancer prognosis, and deciding the interval for follow-up and/or imaging. A nomogram for assessing survival estimates for patients with GBM has been developed and independently validated. To facilitate clinical use of this nomogram, free software for its implementation is provided (http://cancer4.case.edu/rCalculator/rCalculator.html). The nomogram provides an individualized estimate of survival rather than a group estimate and can be used as a companion to a recursive partitioning analysis class assignment. This tool should be useful to patients and health care providers for counseling patients and their families regarding treatment decisions, follow-up, and prognosis.
Glioblastoma (GBM), the most common primary malignant brain tumor, has one of the highest mortality rates, with median overall survival of 12 to 14 months.1 Although there have been modest increases in overall survival since the widespread acceptance of combination chemoradiotherapy utilizing temozolomide, most patients die within 2 years after diagnosis. Treatment benefit is particularly evident for those who have tumors characterized by hypermethylation of the promoter region of the O6-methylguanine-DNA methyltransferase gene (MGMT), referred to as methylated MGMT.2–4 The current standard therapy for GBM includes maximal resection followed by concomitant radiation therapy and temozolomide, then maintenance with 6–12 months of temozolomide as single agent therapy.2
A nomogram accounts for several prognostic factors and is an easily accessible tool for physicians to use on behalf of their patients for (i) predicting survival, (ii) developing an individualized cancer prognosis valuable for treatment decision making, and (iii) deciding the interval for follow-up and/or imaging.5 Such nomograms have been created for various diseases, including brain metastases6 and one previously for GBM using data from a European Organisation for Research and Treatment of Cancer‒National Cancer Institute of Canada (EORTC-NCIC) clinical trial (EORTC 26981/22981-NCIC), which enrolled patients between August 2000 and March 2002 (N = 573); however, both of these nomograms were internally validated only.7
The purpose of this study was to develop and then independently validate a nomogram for estimation of individualized survival probabilities for GBM patients treated with combination chemoradiation (temozolomide) that would be readily accessible for clinical use electronically. This was done using data from 2 independent, recent, and non-overlapping NRG Oncology Radiation Therapy Oncology Group (RTOG) clinical trials, 05258 and 0825.9
Methods
Data Collection and Study Population
De-identified data were provided by NRG Oncology RTOG for the clinical trials 0525 and 0825.8,9 In 0525, patients were enrolled from January 2006 through June 2008,8 and in 0825, patients were enrolled from April 2009 through May 2011.9 Protocol approval was received from the institutional review board at each study site, and informed consent was obtained from each patient prior to participation. The 2 trials included information on a total of 833 (0525) and 621 (0825) randomized and eligible patients with newly diagnosed GBM. For each patient, the following variables were obtained: survival/follow-up time in months, survival status (dead or alive), age at diagnosis (continuous), race (white, black, or other), gender (male or female), Karnofsky performance status (KPS) (70, 80, 90, or 100), extent of resection (total/gross, subtotal, or other), MGMT status (unmethylated or methylated), and recursive partitioning analysis (RPA) class (III, IV, or V).10,11 The analysis included only randomized patients who completed concurrent chemoradiation from both trials. After removing individuals with missing covariates (0525: 34 patients; 0825: 66 patients), 799 patients from 0525 and 555 from 0825 were used for this analysis.
Statistical Analyses
Overall survival was examined for each trial using the Kaplan–Meier method and was compared between the 2 trials using the log-rank test. Survival was assessed with adjustment for age at diagnosis, race, gender, KPS, extent of resection, and MGMT status using Cox proportional hazards (CPH) regression, random survival forests (RSF), and RPA. In RSF, an ensemble tree method for analyzing right-censored survival data was used to generate 1000 trees, and all possible split points for each variable were examined to find the optimal split solution. In RPA, a full recursive classification tree was generated and the least important splits were removed recursively to acquire the final subclassification tree with the minimal model deviance.
The models were trained using the data from 0525 and were independently validated using the data from 0825. Models were also internally validated using 10-fold cross-validation, and individual predicted 6-, 12-, and 24-month survival probabilities were generated to measure predictive accuracy compared with the actual survival as “ground truth.” The predictive accuracies for overall survival of the 3 statistical approaches (CPH regression, RSF, and RPA) were calculated using the concordance index, which is equivalent to the area under the receiver operating characteristic curve for censored data, ranging from 0.5 to 1.12 A final nomogram was built using the method with the greatest predictive accuracy for individualized estimation of survival. Calibration of the final model was visually evaluated by assigning all patients into quintiles of the nomogram-predicted 12-month survival probabilities and plotting the mean nomogram predicted 12-month survival probability against the Kaplan–Meier estimated 12-month survival for each quintile. Calibration curves were also drawn for each dataset for predicted 6-month survival. Finally, histograms were generated to visually display and evaluate predicted 12-month survival by RPA class (III, IV, and V), which was developed by NRG Oncology RTOG for categorizing glioblastoma based on patients’ age at diagnosis, KPS, neurological function, mental status, and extent of resection. All analyses were performed using R v3.1.2 (http://www.r-project.org/).
Results
Patient Selection and Patient Characteristics
Patients from both treatment arms from both trials were used for analysis, given that there was no difference in primary outcomes by treatment arm for these trials and there was no treatment interaction with prognostic variables of interest in the experimental arms of the trials. When all data from trials 0525 and 0825 were combined, P values for interaction with treatment arm were greater than .16, validating the lack of any substantial treatment effect/interaction with the baseline prognostic variables. For trial 0525 only, all the P values for interaction with treatment arm were greater than .16, and for trial 0825 only, all the P values were greater than .20. The average age at diagnosis for both trials was 56.5 years, with 0825 having a slightly older population than 0525 (0825 mean age: 57.70 y vs 0525 mean age: 55.70 y; P = .002). Similarly, the median age at diagnosis for both trials was 58.0 years, with 0825 having a slightly older population than 0525 (0825 median age: 59.0 y vs 0525 median age: 57.0 y; P = .004). The vast majority of the patients from both trials were white (96.2%), with no significant difference between the 2 trials (P = .488). The majority of patients had KPS of at least 90 (0525: 44.6%; 0825: 42.9%). Slightly more than half of the patients had surgeon-defined gross total resection (57.5%), but this varied between trials, with 54.3% for 0525 and 62.0% for 0825 (P = .016). Approximately two-thirds of the patients had tumors with unmethylated MGMT (69.5%), which did not differ significantly between the 2 trials (P = .265). Patient characteristics are presented in Table 1.
Table 1.
Patient characteristics
| Overall (N = 1354) |
Trial 0525 (training)8 (N = 799) |
Trial 0825 (independent validation9) (N = 555) |
P | |
|---|---|---|---|---|
| Age at diagnosis, mean (SD), median (interquartile range) |
56.52 (11.62) 58.0 (5.0, 64.0) |
55.70 (11.98) 57.0 (48.5, 64.0) |
57.70 (11.00) 59.0 (51.0, 65.0) |
.002 .004 |
| Race, N | .644 | |||
| White | 1145 (96.2%) | 617 (95.8%) | 528 (96.7%) | |
| Black or African American | 23 (1.9%) | 13 (2.0%) | 10 (1.8%) | |
| Other | 22 (1.8%) | 14 (2.2%) | 8 (1.5%) | |
| Gender, N | .488 | |||
| Male | 802 (59.2%) | 466 (58.3%) | 335 (6.4%) | |
| Female | 553 (4.8%) | 333 (41.7%) | 220 (39.6%) | |
| KPS, N | .010 | |||
| 70 | 161 (11.9%) | 91 (11.4%) | 70 (12.6%) | |
| 80 | 304 (22.5%) | 159 (19.9%) | 145 (26.1%) | |
| 90 | 594 (43.9%) | 356 (44.6%) | 238 (42.9%) | |
| 100 | 295 (21.8%) | 193 (24.2%) | 102 (18.4%) | |
| Resection, N | .016 | |||
| Subtotal | 538 (39.7%) | 339 (42.4%) | 199 (35.9%) | |
| Total (gross) | 778 (57.5%) | 434 (54.3%) | 344 (62.0%) | |
| Other | 38 (2.8%) | 26 (3.3%) | 12 (2.2%) | |
| RPA, N (10, 11) | .002 | |||
| III | 246 (18.3%) | 171 (21.4%) | 75 (13.7%) | |
| IV | 884 (65.7%) | 505 (63.2%) | 379 (69.4%) | |
| V | 215 (16.0%) | 123 (15.4%) | 92 (16.8%) | |
| MGMT, N | .265 | |||
| Methylated | 388 (3.5%) | 233 (31.8%) | 155 (28.8%) | |
| Unmethylated | 883 (69.5%) | 499 (68.2%) | 384 (71.2%) | |
| Overall survival vital status, N | <.001 | |||
| Alive | 359 (26.5%) | 174 (21.8%) | 185 (33.3%) | |
| Dead | 995 (73.5%) | 625 (78.2%) | 370 (66.7%) | |
Survival by the Kaplan–Meier Method
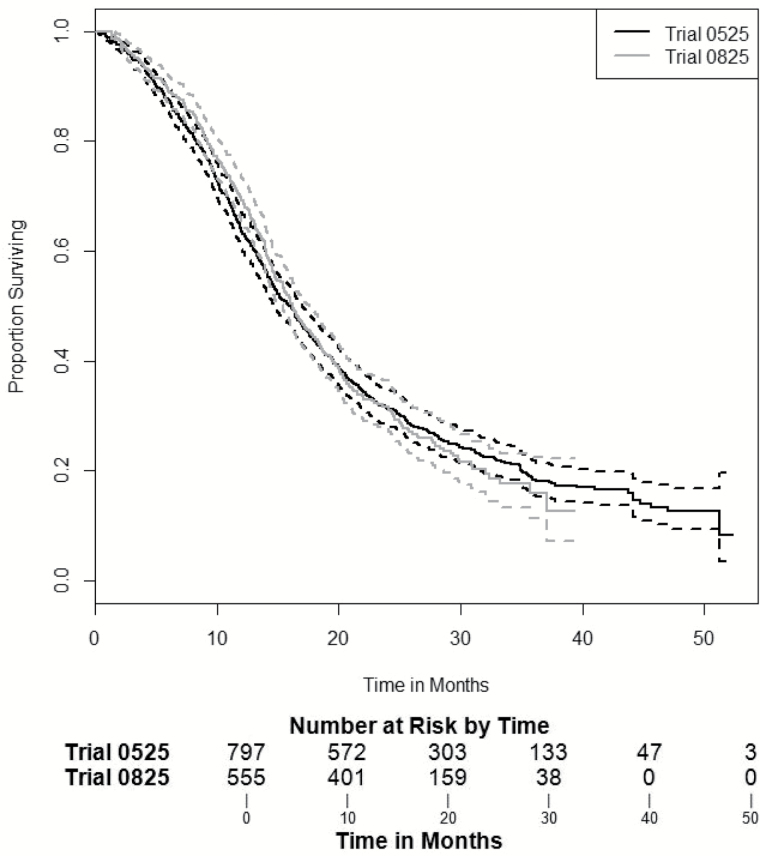
To assess differences in survival between the 2 trials, without adjusting for any covariates, Kaplan–Meier curves were generated for each trial with the pointwise 95% confidence intervals (Fig. 1). In 0525, there were 625 deaths out of the 799 patients, with a median survival time of 16.0 months (95% CI: [14.7, 17.0]). In 0825 there were 370 deaths out of the 555 patients, with a median survival time of 16.2 months (95% CI: [15.4, 17.6]). No significant difference in median survival time was found between these 2 trials (P = .996), with similar 1-, 2-, and 3-year survival rates (Fig. 1).
Fig. 1.
Kaplan–Meier survival results with 95% CI curves for median survival in months and 95% CI for 05258: 16.0 (14.7, 17.0) and 08259: 16.2 (15.4, 17.6); log-rank test by clinical trial P = .996).
Survival Analysis Results and Internal Cross-Validation
The training dataset (0525) included information on age at diagnosis, gender, KPS, extent of resection, and MGMT methylation status. After 10-fold cross-validation was performed on the training set, the concordance indices were calculated for each statistical method for predicting survival at 3 time points: 6, 12, and 24 months (Supplementary Table 1). For all 3 time points, the CPH analysis (6-, 12-, and 24-mo: 0.656, 0.657, 0.657) outperformed RSF (6-, 12-, and 24-mo: 0.562; 0.602; 0.597) and RPA (6-, 12-, and 24-mo: 0.617; 0.617; 0.617). The RSF ranked the covariates in order of importance, with age at diagnosis being the most important variable and extent of resection being least important relatively. The RPA showed that the most important split in predicting mortality was age at diagnosis (<52.5 y vs ≥52.5 y). Further detailed results from the RSF and RPA are shown in Supplementary Table 2 and Supplementary Figure 1, respectively.
Nomogram and Independent Validation
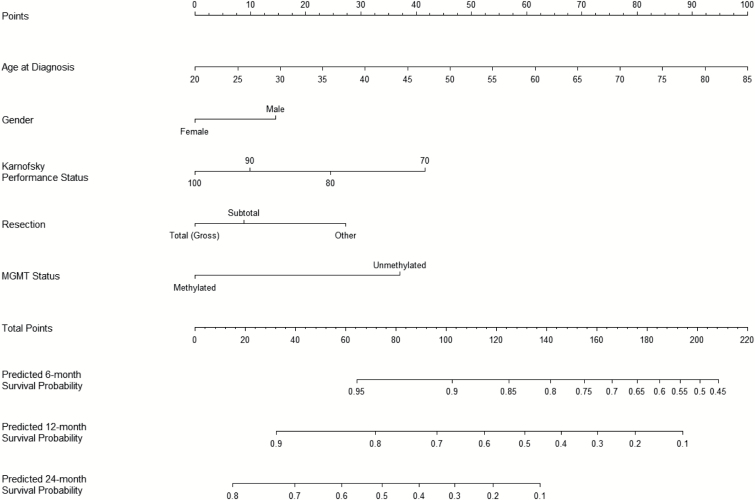
The nomogram to estimate 6-, 12-, and 24-month survival probabilities was built using the training dataset (0525) and validated on the independent dataset (0825, independent validation data) (Fig. 2) using the CPH model (Table 2). An online calculator for the final nomogram is available via an Internet interface at http://cancer4.case.edu/rCalculator/rCalculator.html.
Fig. 2.
Final nomogram built on training data 05258 and validated on 08259 (http://cancer4.case.edu/rCalculator/rCalculator.html).
Table 2.
Cox proportional hazards results on training dataset (0525)8
| Variable | Estimate | Standard Error | 95% CI | Wald Z | P |
|---|---|---|---|---|---|
| Age at diagnosis (continuous) |
1.030 | 0.0039 | (1.026, 1.034) | 7.55 | <.0001 |
| Gender (male vs female) |
1.319 | 0.0869 | (1.209, 1.439) | 3.19 | .0014 |
| KPS (80 vs 70) |
0.723 | 0.1503 | (0.622, 0.840) | −2.16 | .0307 |
| KPS (90 vs 70) |
0.547 | 0.1373 | (0.477, 0.628) | −4.40 | <.0001 |
| KPS (100 vs 70) |
0.453 | 0.1524 | (0.389, 0.528) | −5.20 | <.0001 |
| Resection (subtotal vs total) |
1.184 | 0.0881 | (1.084, 1.293) | 1.91 | .0557 |
| Resection (other vs total) |
1.680 | 0.2299 | (1.335, 2.114) | 2.26 | .0240 |
|
MGMT status (methylated vs unmethylated) |
0.493 | 0.0972 | (0.447, 0.543) | −7.27 | <.0001 |
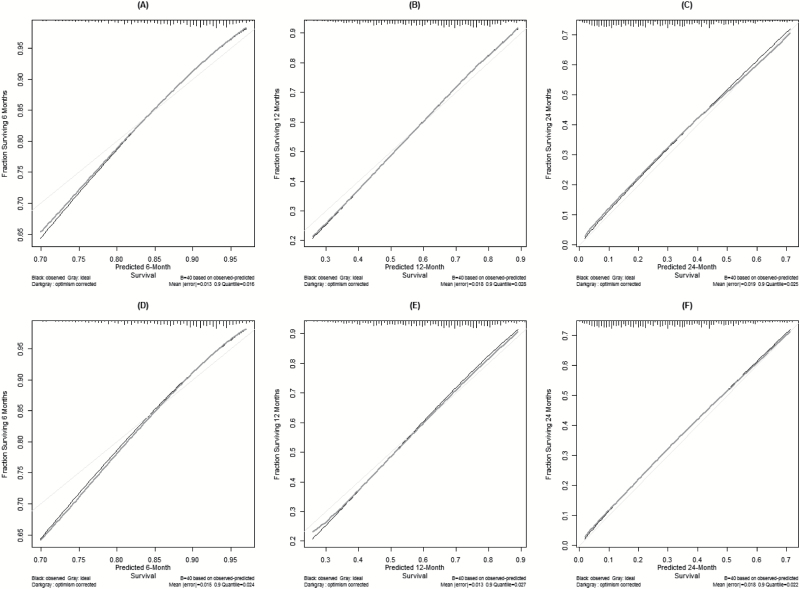
The final CPH model was well calibrated (Supplementary Figure 2), with a concordance index of 0.657. For each quintile group, the estimated versus observed 12-month survival probabilities intersected the 45-degree line, indicating that the predicted value approximated the observed value within a 95% confidence interval (Supplementary Figure 2).
Calibration curves were also drawn for the training dataset (0525) for predicted 6-, 12-, and 24-month survival, as well as for the independent validation dataset (0825) for a visual comparison. In Fig. 3 the black line shows the observed survival rates, the gray line shows the ideal survival rates, and the dark gray line shows the optimism-corrected survival rates. The optimism-corrected—also known as bias-corrected or overfitting-corrected—line is produced using a bootstrap approach to estimate predicted and observed values based on a nonparametric smoother applied to a sequence of predicted values. For the predicted 6-month survival plots (Figs. 3A and 3D), the observed and optimism-corrected lines are nearly identical, which suggests that the model’s predictions match expectations, although both of these lines stray from the ideal 45-degree line toward the low end of the outcome scale. The predicted 12-month survival plots (Figs. 3B and 3E) are similar. The observed and optimism-corrected lines are essentially overlapping, although these lines stray a little from the ideal line toward the low end of the outcome scale. For the predicted 24-month survival plots (Figs. 3C and 3F), all 3 lines closely align, demonstrating near perfect calibration.
Fig. 3.
(A-F) Calibration curves for the nomogram training data (0525)8 at 6 months (A), 12 months (B), and 24 months (C) and for independent validation data (0825)9 at 6 months (D), 12 months (E), and 24 months (F).
Survival by Recursive Partitioning Analysis Class
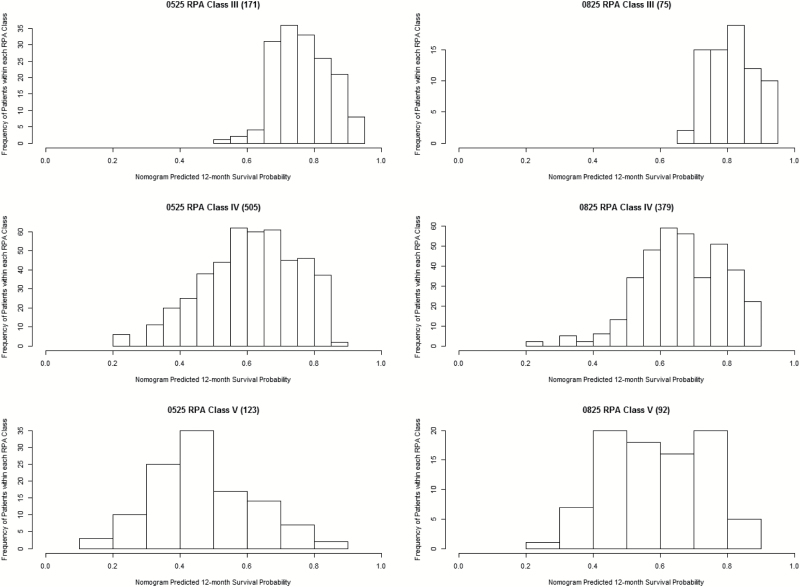
Finally, 12-month predicted survival by RPA class (III, IV, and V) (10, 11) was visually assessed through histograms for both the training and independent validation datasets (Fig. 4). The concordance index of RPA applied to the validation set was 0.582. The distributions between datasets are quite similar to each other. Overall, RPA class III had the best-predicted 12-month survival and RPA class IV had the worst-predicted 12-month survival.
Fig. 4.
12-month predicted survival by RPA class10,11 for training data (0525)8 and validation data (0825).9
Discussion
The objective of this analysis was to develop as well as validate, both internally and on an independent dataset, an individual prognostic nomogram for patients with newly diagnosed GBM. The CPH survival model was the best fitting and calibrated model, according to a 10-fold validation concordance index evaluating 3 survival approaches on the 0525 training data for 6-, 12-, and 24-month survival rates, and included age at diagnosis, gender, KPS, extent of tumor resection, and methylation status of the MGMT promoter. This model was then independently validated on the 0825 dataset.
Age at diagnosis, KPS, and extent of surgical resection have long been defined as important prognostic factors for GBM.13 A similar GBM nomogram-construction study has been published by Gorlia et al7 using data from an EORTC-NCIC clinical trial concluding that MGMT promoter methylation status is a significant prognostic factor for GBM (N = 573; P < .0001–.0003). Our results support their observation that MGMT promoter methylation status, age at diagnosis, KPS, and extent of surgical resection are significant; however, the EORTC nomogram did not include gender or KPS. In addition, their nomogram calculated median and 2-year survival only and did not include an independent validation of their final nomogram.7 The trials used for this analysis are the largest, most recent randomized trials available in the United States for GBM testing standard therapy versus an experimental treatment.
In the current literature, a higher incidence of GBM has been reported in men compared with women, with males having a GBM incidence rate 1.6 times greater than females. Moreover, men were found to have a significant survival advantage compared with women in the first year postdiagnosis, but afterward, the difference was not significant.14 Our results suggest that females have a slight survival advantage, though the difference is not significant.
Previous studies have identified biomarkers for GBM prognosis and prediction of treatment response. However, these markers are not consistently clinically utilized due to issues with their uniformity, complexity, applicability, and cost.15–17 Promoter methylation of MGMT is an accepted GBM biomarker, indicating sensitivity to the alkylating chemotherapeutic agent temozolomide.2–4 The utility of this biomarker is limited because it is present in only 20%–40% of patients; there is no consensus on optimal measurement methods, and no clear cutoff points between methylated and unmethylated MGMT have been prospectively established.18,19 However, Stupp et al4 found that methylation of the MGMT promoter was the strongest predictor for both outcome and clinical efficacy from temozolomide therapy.
This study had several limitations. In both the training (0525) and independent validation (0825) datasets, the vast majority of the patients were white. Therefore, race was not found to be a significant factor and was dropped from the CPH model, despite evidence for differences in survival by race in other studies.20,21 Our study represents a large group of patients with newly diagnosed GBM from 2 clinical trials conducted at many sites across the United States; however, our findings may or may not be generalizable to the general United States population with GBM. In both 0525 and 0825 all patients received current standard therapy for GBM—in 0525 one study arm also received dose-intensified temozolomide,8 and in 0825 one study arm also received bevacizumab.9 Trial 0525 only included patients who completed chemoradiation therapy. Neither clinical trial showed significant difference in clinical outcomes, therefore no further adjustments for treatment arm were performed.8,9 However, because all patients in this study satisfied the inclusion criteria for 0525 or 0825, this nomogram may not be applicable to GBM patients who would not satisfy these criteria. Caution should be used when applying this nomogram to patients with poor performance status (ie, KPS <70) and with biopsy-only patients. Future work could include assessment of the nomogram in a large group of consecutive GBM patients not treated on controlled clinical trials; in an extra-US population; or in elderly patients, some of whom have not been treated initially with radiotherapy. Additionally, further comparison of performance of other prognostic models for GBM could be tested. Recent randomized clinical trials in elderly patients with GBM suggest that with monotherapy, temozolomide results in superior outcomes in methylated tumors, whereas for unmethylated tumors, radiotherapy yields superior outcomes. In this context, MGMT methylation is viewed as both prognostic and predictive for the effect of temozolomide, but its exact prognostic and predictive value for radiotherapy in patients with GBM not receiving temozolomide still remains unclear. Our analysis did not include any radiotherapy-only patients and therefore this question remains unaddressed.
Despite the limitations, the CPH model was internally and independently validated with large contemporary clinical trial datasets including arms that received the current standard therapy.2 Furthermore, the same statistical analyses were performed switching the training and independent validation datasets for comparison (data not shown) and very limited variation was seen for the CPH model estimates, for the nomogram point assignments, or for the concordance indices. We also performed a sensitivity analysis using only patients from each trial on the standard therapy arms as training and independent validation sets (data not shown) and again very limited variation was seen for the CPH model estimates, for the nomogram point assignments, or for the concordance indices.
Conclusion
A nomogram for assessing survival estimates for patients with GBM has been developed. This tool provides an individualized estimate of survival, rather than a group estimate based on specific covariates. To facilitate clinical use of this nomogram, free software for its implementation is provided (http:// cancer4.case.edu/rCalculator/rCalculator.html). The nomogram provides an individualized estimate of survival rather than a group estimate and can be used as a companion to an RPA class assignment. This tool should be useful to patients and health care providers for counseling patients and their families regarding treatment decisions, follow-up, and prognosis.
Supplementary material
Supplementary material is available at Neuro-Oncology online.
Funding
This work was supported by grants U10CA21661 (RTOG-Ops-Stat), U10CA37422 (CCOP), U10CA180868 (NRG Oncology Operations), and U10CA180822 (NRG Oncology SDMC) from the National Cancer Institute (NCI), grants from Merck and Genentech, and a grant from the NCI to the Case Comprehensive Cancer Center (NCI P30-CA043703 to J.S.B.-S., H.G., D.L., S.S.L., A.E.S., M.M., and M.W.K).
Supplementary Material
Acknowledgements
We thank the NRG Oncology Brain and Ancillary Projects Committees for graciously providing the data needed for this work. This manuscript’s contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
Conflict of interest statement. H.G. reports that her father works for Genentech, which provided grants to support this work. D.L. reports research funding from the University of Iowa Hospitals and Clinics. A.B.L. reports consulting fees for Genentech, Bioclinica, VBI Vaccines, Sapience Therapeutics, Cortice Biosciences, Oxigene, and prIME Oncology. S.S.L. reports research support from Elekta AB, travel expenses and honoraria from Varian Medical Systems and Accuray Inc outside the submitted work. A.E.S. reports consultation agreements with Monteris Medical Inc, Medtronic Inc, Pharmakokenesis, and ownership options from Surgical Theatre Inc. E.P.S. reports personal fees from Merck and grants from AbbVie and from Novocure outside the submitted work. M.A.V. reports CMO, co-founder, options, and royalties from Infuseon Therapeutics, personal fees from Neuralstem serving as a DSMB member, and an honorarium from Pharmicokinesis outside the submitted work. T.J.C.W. reports travel expenses from Abbvie outside the submitted work. M.P.M. reports grants and personal fees from Novocure and Novelos, personal fees from Abbott, Phillips, BMS, Celldex, Roche, Elekta, Novartis, Cavion, Pharmacyclics, Monteris, and Varian outside the submitted work. J.S.B.-S.reports grants from NIH during the conduct of the study.
References
- 1. Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 4. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 5. Ogura K, Fujiwara T, Yasunaga H, et al. Development and external validation of nomograms predicting distant metastases and overall survival after neoadjuvant chemotherapy and surgery for patients with nonmetastatic osteosarcoma: a multi-institutional study. Cancer. 2015;121(21):3844–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barnholtz-Sloan JS, Yu C, Sloan AE, et al. A nomogram for individualized estimation of survival among patients with brain metastasis. Neuro Oncol. 2012;14(7):910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gorlia T, van den Bent MJ, Hegi ME, et al. Nomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981-22981/CE.3. Lancet Oncol. 2008;9(1):29–38. [DOI] [PubMed] [Google Scholar]
- 8. Gilbert MR, Wang M, Aldape KD, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. J Clin Oncol. 2013;31(32):4085–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Curran WJ, Jr, Scott CB, Horton J, et al. Recursive partitioning analysis of prognostic factors in three radiation therapy oncology group malignant glioma trials. J Natl Cancer Inst. 1993;85(9):704–710. [DOI] [PubMed] [Google Scholar]
- 11. Li J, Wang M, Won M, et al. Validation and simplification of the radiation therapy oncology group recursive partitioning analysis classification for glioblastoma. Int J Radiat Oncol. 2011;81(3):623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–387. [DOI] [PubMed] [Google Scholar]
- 13. Lacroix M, Abi-Said D, Fourney DR, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg. 2001;95(2):190–198. [DOI] [PubMed] [Google Scholar]
- 14. Thakkar JP, Dolecek TA, Horbinski C, et al. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomarkers Prev. 2014;23(10):1985–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. [DOI] [PubMed] [Google Scholar]
- 17. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hegi ME, Liu L, Herman JG, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26(25):4189–4199. [DOI] [PubMed] [Google Scholar]
- 19. Suri V, Jha P, Sharma MC, et al. O6-methylguanine DNA methyltransferase gene promoter methylation in high-grade gliomas: a review of current status. Neurol India. 2011;59(2):229–235. [DOI] [PubMed] [Google Scholar]
- 20. Chakrabarti I, Cockburn M, Cozen W, et al. A population-based description of glioblastoma multiforme in Los Angeles County, 1974–1999. Cancer. 2005;104(12):2798–2806. [DOI] [PubMed] [Google Scholar]
- 21. McLendon RE, Robinson JS, Jr, Chambers DB, et al. The glioblastoma multiforme in Georgia, 1977–1981. Cancer. 1985;56(4): 894–897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.