Abstract
Background.
Human mesenchymal stem cells (hMSCs) have been shown to reside as stromal cells in human gliomas as glioma-associated hMSCs (GA-hMSCs), but their biological role remains unclear. Because recent evidence indicates that GA-hMSCs drive tumor cell proliferation and stemness, we hypothesized that a higher percentage of GA-hMSCs in tumors predicts poor patient prognosis.
Method.
We determined the percentage of cells coexpressing GA-hMSC markers CD105+/CD73+/CD90+ from patients with newly diagnosed high-grade glioma and analyzed the association between this percentage and overall survival (OS) in 3 independent cohorts: fresh surgical glioblastoma specimens (cohort 1, N = 9), cultured tumor specimens at passage 3 (cohort 2, N = 28), and The Cancer Genome Atlas (TCGA) database.
Results.
In all cohorts, patient OS correlated with the percentages of GA-hMSCs in tumors. For cohort 1, the median OS of patients with tumors with a low percentage of triple-positive cells was 46 months, and for tumors with a high percentage of triple-positive cells, it was 12 months (hazard ratio [HR] = 0.24; 95% CI: 0.02–0.5, P = .02). For cohort 2, the median OS of patients with tumors with a low percentage of GA-hMSCs was 66 months, and for tumors with a high percentage, it was 11 months (HR = 0.38; 95% CI: 0.13–0.9, P = .04). In the database of TCGA, the median OS times in patients with high and low coexpression levels of CD105/CD73/CD90 were 8.4 months and 13.1 months (HR = 0.4; 95% CI: 0.1–0.88; P = .04), respectively.
Conclusions.
The percentage of GA-MSCs inversely correlates with OS, suggesting a role for GA-MSCs in promoting aggressive behavior of gliomas.
Keywords: glioblastoma, mesenchymal stem cells, microenvironment, prognosis
Glioblastoma (GBM) is a recalcitrant disease, as patients with GBM survive on average only 14 months despite maximal treatment.1,2 The outcome of patients with GBM can vary depending on patient and tumor characteristics.3 Age, Karnofsky performance scale (KPS) score, and extent of resection are the most significant clinical predictors of survival for patients with GBM.4–6 Expression of epidermal growth factor receptor variant III, methylation of the O6-methylguanine DNA methyltransferase (MGMT) promoter, and mutations in isocitrate dehydrogenase genes (IDH1 and IDH2) have been identified as molecular prognostic factors.7–11
The tumor microenvironment is also critical in determining the biological behavior of solid tumors, including GBM.12 Although astrocytes, endothelia, and microglia are traditionally thought to make up the microenvironment of gliomas,13 recent evidence indicates that gliomas may also contain mesenchymal stem cells (MSCs), recruited either from local brain sources or from the bone marrow.14,15 MSCs are stem cells that were originally isolated from the bone marrow. Most recently, MSCs have been shown to reside in all organs, including the brain,16 as part of the perivascular niche.16 MSCs are characterized by their ability to grow as spindle-shaped cells in culture, their coexpression of CD105, CD73, and CD90, their lack of expression of CD45, CD34, and CD133, and their ability to undergo tri-mesenchymal differentiation into adipocytes, chondrocytes, and osteocytes.17
In solid tumors, MSCs have been shown to potentiate tumor progression or regression by releasing paracrine signals.18–22 Using the same methods developed for culturing bone marrow MSCs, we recently isolated and cultured human MSCs (hMSCs) from a large number of surgically resected high-grade gliomas, which we called glioma-associated hMSCs (GA-hMSCs). We showed that these GA-hMSCs were not tumorigenic but were capable of increasing the proliferation, stemness, and tumorigenicity of glioma-initiating cells (GICs), which are the cells responsible for glioma formation and recurrence after treatment. The effects of GA-hMSCs were mediated through the secretion of interleukin-6, which activates signal transducer and activator of transcription 3 in GICs.15 Similar results have been reported by others using glioma cell lines.23
Despite these laboratory findings, however, it remains unclear whether the presence of GA-MSCs in tumor samples alters the prognosis of patients with GBM. Given our translational studies, we hypothesized that tumors with higher percentages of GA-hMSCs would be more aggressive and would be associated with shorter overall survival (OS) compared with gliomas with lower percentages of GA-hMSCs. To test this hypothesis, we isolated GA-MSCs from surgical high-grade glioma specimens, based on coexpression of the MSC markers CD105 (Endoglin), CD73 (NT5E), and CD90 (THY1) and determined their fraction in the tumor mass (cohort 1) or in culture tumors (passage 3, cohort 2). In both of these cohorts, we found that higher percentages of GA-hMSCs correlated with poorer OS. We further validated this result using The Cancer Genome Atlas (TCGA) and found a similar association between high coexpression of MSC marker genes and poor patient outcome.
Patients and Methods
Patients
During the study period of September 2005 to May 2015, tumors from 48 consecutive patients operated on for the resection of glioma were analyzed for GA-hMSCs. Tumors from patients with a previous history of low-grade glioma and secondary transformation glioma were excluded (n = 5), as were patients with recurrent disease (n = 8) or those with insufficient follow-up (n = 3). The remaining 32 patients with newly diagnosed primary supratentorial high-grade glioma met the inclusion criteria and were included. Clinical data were obtained from the Department of Neurosurgery Prospective Database at The University of Texas M.D. Anderson Cancer Center (M.D. Anderson). All study participants provided informed consent according to an institutional review board–approved protocol (LAB04-0001).
Brain Tumor Specimens and Cohorts
Tumor specimens (N = 32) were obtained directly from the operating room and were assessed by a neuropathologist (Table 1). In 4 patients, the tumors were made into single cell suspensions and analyzed by flow cytometry analysis only. In 5 patients, the tumor was divided into 2 parts, with one part being made into a single cell suspension for flow cytometry analysis and the other part used to establish in vitro cultures within 4 h of tumor removal. In the remaining 23 patients, the specimen was used only to establish cultures within 4 h of tumor removal. Therefore, 2 cohorts were available for analysis—cohort 1 included tumor specimens that were directly assayed by flow cytometry for the percentage of MSCs without any culturing of the specimen (cohort 1, N = 9). The second cohort included tumor specimens that were cultured for 3 passages and then the cultured cells were assayed by flow cytometry for the percentage of MSCs (cohort 2, N = 28). Twelve of these specimens were reported previously (see Table 1 and Hossain et al15).
Table 1.
Characteristics of 32 patients with high-grade glioma
| Patient | Age (years) | Pathology | IDH1 Mutation Status |
KPS score on Admission | EOR (%) | Fraction of GA-MSCs in Tissue (%) | Fraction of GA-MSCs in Culture at Passage 3 (%) | Survival (mo) |
Status |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 58 | GBM | M | 90 | 100 | 1.1 | nd | 15 | D |
| 2 | 45 | GBM | WT | 90 | 100 | 3.7 | nd | 54 | D |
| 3 | 29 | GBM | M | 90 | 91 | 5.1 | nd | 38 | D |
| 4 | 53 | GBM | nd | 80 | 95 | 12.6 | nd | 17 | D |
| 5* | 45 | GBM | WT | 100 | 100 | 0.7 | 4.4 | 67 | A |
| 6 | 24 | GBM | M | 100 | 100 | 2.5 | 16.4 | 22 | D |
| 7 | 70 | GBM | nd | 80 | 95 | 8.9 | 65.9 | 5 | D |
| 8* | 59 | GBM | WT | 90 | 100 | 9.3 | 84.5 | 10 | D |
| 9 | 55 | GBM | nd | 90 | 95 | 19.5 | 92.3 | 14 | D |
| 10* | 41 | AA | nd | 100 | 100 | nd | 2.0 | 110 | A |
| 11* | 54 | AA | M | 100 | 100 | nd | 4.0 | 96 | A |
| 12* | 32 | AA | M | 90 | 91 | nd | 9.7 | 66 | D |
| 13 | 55 | GBM | WT | 60 | 54 | nd | 16.4 | 2 | D |
| 14 | 67 | GBM | WT | 90 | 99 | nd | 20.7 | 15 | D |
| 15* | 60 | GBM | WT | 90 | 100 | nd | 22.0 | 46 | D |
| 16* | 36 | AA | M | 100 | 100 | nd | 24.8 | 73 | A |
| 17 | 22 | AA | M | 100 | 100 | nd | 25.0 | 15 | A |
| 18 | 34 | GBM | M | 100 | 85 | nd | 25.4 | 27 | A |
| 19* | 30 | AA | WT | 100 | 96 | nd | 27.7 | 97 | A |
| 20 | 75 | GBM | WT | 90 | 100 | nd | 29.0 | 11 | D |
| 21 | 60 | GBM | WT | 90 | 100 | nd | 30.2 | 11 | D |
| 22 | 66 | GBM | WT | 100 | 100 | nd | 33.0 | 6 | D |
| 23 | 44 | GBM | nd | 80 | 100 | nd | 44.2 | 2 | D |
| 24 | 72 | GBM | WT | 100 | 100 | nd | 44.4 | 5 | D |
| 25 | 56 | GBM | WT | 80 | 51 | nd | 44.8 | 14 | D |
| 26* | 68 | GBM | nd | 100 | 100 | nd | 46.3 | 10 | D |
| 27 | 32 | GBM | nd | 90 | 100 | nd | 49.3 | 17 | D |
| 28* | 33 | AA | WT | 90 | 76 | nd | 53.0 | 83 | A |
| 29 | 48 | GBM | WT | 90 | 80 | nd | 55.1 | 10 | D |
| 30 | 61 | GBM | WT | 90 | 100 | nd | 64.2 | 30 | A |
| 31* | 49 | GBM | WT | 90 | 100 | nd | 71.9 | 25 | A |
| 32* | 63 | GBM | WT | 90 | 100 | nd | 78.9 | 47 | A |
Abbreviations: A, alive at the time of last follow-up; D, dead at the time of last follow-up; EOR, extent of resection; M, positive IDH1 mutation status; nd, not done; WT, wild type.
Cohort 1 = patients 1–9 (N = 9).
Cohort 2 = patients 5–32 (N = 28).
These specimens were reported previously by Hossain at el.15
Flow Cytometry Analysis of High-Grade Glioma Surgical Specimens
Tumor specimens were washed twice in serum-free minimal essential medium–alpha (MEM-α) (Mediatech), minced, dissociated, and passed through a series of cell strainers. Single cells were resuspended in phosphate-buffered saline (PBS) and counted in a Vi-Cell machine (Beckman Coulter); 5 × 105 cells were resuspended in 100µL of fluorescence activated cell sorting (FACS) buffer (PBS with 10% fetal bovine serum) and incubated at 4°C for 30 min with phycoerythrin-, fluorescein isothiocyanate-, and Alexa Fluor 647- (Molecular Probes), or allophycocyanin-conjugated antibodies against human CD105, CD90 (both from eBioscience), CD73, CD34, CD45 (BD Biosciences), and CD133 (Miltenyi Biotech). Cells were analyzed (20000 events/sample) using a FACSCalibur (BD Biosciences) flow cytometer equipped with BD CellQuest Pro software v5.1.1 (Apple).
In vitro Culture of Glioma-Associated Mesenchymal Stem Cells
Tumor specimens were cultured as described by Pittenger et al24 for isolation of bone marrow MSCs (BM-hMSCs), but modified for whole tissues. After dissociation, single cells were resuspended in “standard MSC medium,” composed of MEM-α plus 10% certified fetal bovine serum (Lonza), 2 mM L-glutamine (50 U/mL; Mediatech), and penicillin–streptomycin (50 U/mL–50 mg/mL; Flow Laboratories); 2 × 106 live cells were plated in 75 cm2 flasks. After 24 h, nonadherent cells were removed by 2 washes with PBS, and adherent cells were cultured to confluence. Cells were trypsinized (0.25% trypsin with 0.1% EDTA) and subcultured at a density of 5000 cells/cm2 through 3 passages. Cell cultures were observed/photographed using a Zeiss Axiovert 200 microscope with a digital camera (Zeiss AxioCam MRc) and XCAP-Plus software v2.1 (Epix).
Differentiation Protocols
GA-MSCs were differentiated into osteocytes, adipocytes, and chondrocytes using induction and maintenance media from Lonza, according to the manufacturer’s protocol. For detection of osteogenic or adipogenic differentiation, cells were stained with 40 mM Alizarin Red or with Oil Red O, respectively. For chondrogenic differentiation, pelleted specimens were formalin fixed/paraffin embedded, and sections were stained using Safranin O. BM-hMSCs were a positive control.
Immunohistochemistry
Immunohistochemistry for IDH1-R132H was done on 5-micron-thick formalin-fixed/paraffin-embedded tumor sections. Antigen retrieval was performed in citrate buffer (pH 6.0) in a microwave oven. Antibody specific for the mutant IDH1-R132H protein (dil 1:100, H09, Dianova) was used. Secondary antibody labeled with the streptavidin biotin kit (Universal) was used as a detection system (Dako). A senior pathologist (G.N.F.) blinded to the survival analysis evaluated the results of the immunostaining.
Statistical Analysis
M.D. Anderson Cohort Outcome Analysis. Data are presented as the mean±SD or the median (range). Patients’ characteristics were analyzed using Fisher’s exact test for determining their association with the fraction of cells coexpressing CD105, CD73, and CD90. The fraction of cells coexpressing CD105, CD73, and CD90 was determined for each surgical specimen (cohort 1, N = 9) and for each cultured specimen (cohort 2, N = 28). X-tile software v3.6.1 (Yale University) was used to identify the optimal cutoff of the fraction of triple-positive cells that generated 2 populations that were significantly different in OS. Based on this cutoff, the patients were separated into high and low coexpressing groups. OS was defined as the time from diagnosis to time of death or last day of follow-up. The estimated median for OS was determined by the Kaplan–Meier method, using the log-rank test to compare groups. The median survival as a function of the fraction of cells coexpressing CD105, CD73, and CD90 was determined using Somer’s Dyx rank correlation with censored data. The individual and joint association of predictor variables with OS was assessed using Cox proportional hazards regressions. Survival analyses for the M.D. Anderson patient cohort employed GraphPad Prism 6 software and Spotfire S+ 8.2 for Windows (TIBCO Software), while the remaining analyses employed SPSS software v21.
TCGA Cohort Outcome Analysis. Gene expression data (level 3) and the clinical data of the 507 glioblastoma patients in TCGA were downloaded from the data portal of TCGA (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm).25 Patients with excisional biopsy or undefined surgical treatment were excluded, leaving 414 patients whose tumors were resected and were included in the analysis. A gene-based Kaplan–Meier analysis was performed to assess OS among patients’ normalized expression values of Endoglin (CD105), NT5E (CD73), and THY (CD90). The normalized values were separated into quintiles. Expression values were analyzed separately and then the OS times of patients sharing expression values in the highest quintile of Endoglin, NT5E, and THY were compared with those of patients sharing expression values in the lowest quintile of these genes using the log-rank test.
Results
The M.D. Anderson Cohorts
Thirty-two tumor specimens from patients with newly diagnosed supratentorial high-grade glioma were included in the study (Table 1). The mean age of the patients was 49.9±15 years, and their median KPS score was 90 (range, 60–100). They all underwent tumor resection, with a mean extent of resection of 94 ± 12.5% (range, 51%–100%).
Analysis of GA-hMSCs in Specimens Before Culture (Cohort 1)
Fresh surgical tumor specimens from 9 patients were dissociated (see “Patients and Methods”), and single cells were analyzed by flow cytometry for the coexpression of the MSC markers CD105, CD73, and CD90 within 4 h of tumor removal (cohort 1, N = 9). Cells coexpressing all 3 markers (triple-positive), consistent with the definition of GA-hMSCs, were identified in all the specimens, indicating that GA-hMSCs exist in high-grade gliomas before culturing (Table 1, specimens 1–9). The average fraction of triple-positive cells was 7.04 ± 6.2%. Importantly, the fraction of triple-positive cells varied among specimens, with a range as low as 0.7% and as high as 19.5% (Table 1), indicating that the percentage of GA-hMSCs varies among tumors.
The median OS duration for the patients in this cohort, with surgical specimens that were analyzed before culture (N = 9), was 17 months (95% CI: 11.2–22.8 mo). There was an inverse correlation between patient survival and the fraction of cells coexpressing CD105, CD73, and CD90 (n = 9; Somer’s Dyx rank correlation coefficient = −0.39; 95% CI: −0.74 to −0.04), suggesting an association between a high fraction of GA-hMSCs and a poor survival outcome (Fig. 1A). An X-tile plot analysis (see “Patients and Methods”) revealed that patients with tumors harboring a low percentage of triple-positive cells (defined as ≤5.1%) had a statistically significant survival advantage compared with patients whose tumors harbored a high percentage of triple-positive cells (>5.1%). Specifically, the estimated median OS for patients with tumors containing a low-percentage of triple-positive cells was 46 months, whereas the estimated median OS for patients with tumors containing a high percentage of triple-positive cells was 12 months (hazard ratio [HR] = 0.24; 95% CI: 0.02–0.5, P = .02; Fig. 1B).
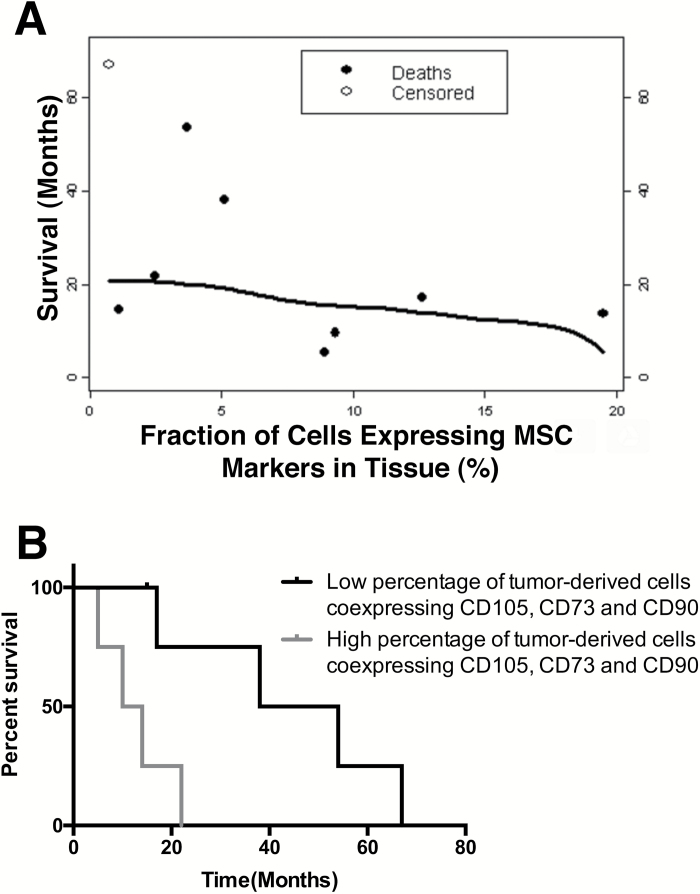
Fig. 1.
High percentage of GA-MSCs in fresh surgical specimens predicts poor overall survival of patients with high-grade glioma. (A) A correlation between the percentage of triple-positive (CD105+/CD73+/CD90+) cells identified in high-grade glioma fresh surgical specimens and patients OS (Somer’s Dyx rank correlation coefficient = −0.39; 95% CI: −0.74 to −0.04). (B) Kaplan–Meier survival curves in patients with low (≤5.1%) and high percentage of triple-positive (CD105+/CD73+/CD90+) cells (log-rank test, P = .02).
Analysis of GA-hMSCs in Glioma Specimens After Culturing (Cohort 2)
We aimed to demonstrate the correlation between the fraction of triple-positive cells and OS in a larger cohort of patients available from our institution. We previously reported that GA-hMSCs can be cultured from glioma specimens using the same protocols that have been used to culture bone marrow hMSCs (Hossain et al).15 We also previously reported that the percentage of triple-positive cells (ie, GA-hMSCs) varied from culture to culture when assayed at passage 3, a point in the culture processing where the cellular debride from the specimen was gone, but the culture still contained a mixture of phenotypically different cell types. We therefore hypothesized that we could use the percentage of triple-positive cells (GA-hMSCs) at passage 3 as a surrogate for the percentage of GA-hMSCs in tumor specimens before culturing. To test this hypothesis, we graphed the percentage of triple-positive cells in culture at passage 3 as a function of the percentage of triple-positive cells in the original uncultured specimen using 5 tumors for which we had both measurements of GA-hMSCs. Consistent with our previous report,15 we found a statistically significant positive correlation between the percentage of triple-positive cells in culture and the percentage of triple-positive cells in the original uncultured specimen (N = 5, r = 0.9; 95% CI: 0.07 to 0.99, P = .039; Fig. 2A, Table 1).
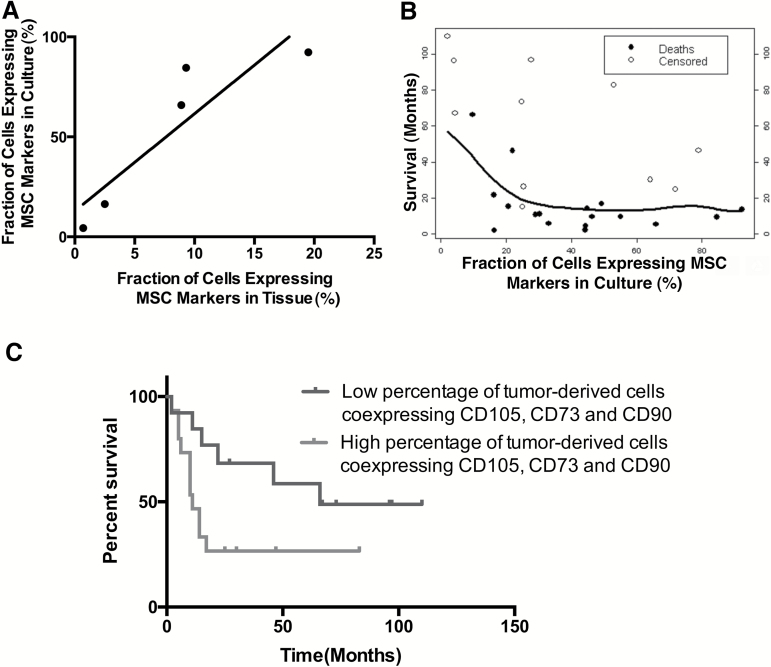
Fig. 2.
A high percentage of GA-MSCs in the culture of surgical specimens predicts poor OS of patients with high-grade glioma. (A) A correlation between the percentage of triple-positive (CD105+/CD73+/CD90+) cells identified in high-grade glioma fresh surgical specimens and the percentage of triple-positive cells identified in culture (assayed at passage 3) derived from the specimen (r = 0.9). (B) A correlation between the percentage of triple-positive (CD105+/CD73+/CD90+) cells identified in high-grade glioma in culture and patients’ OS (Somer’s Dyx rank correlation coefficient = −0.24; 95% CI: −0.54 to 0.07). (C) Kaplan–Meier survival curves in patients with low (≤29%) and high percentage of triple-positive (CD105+/CD73+/CD90+) cells (log-rank test, P = 0.04).
Based on this correlation, 28 specimens were cultured for GA-hMSCs using the protocol of Pittenger et al24 (including the 5 specimens that were also characterized before culture) (Table 1). Twelve of these glioma specimens were previously reported.15 In all cases, cells resembling MSCs were identified. Consistent with the definition of MSCs, these cells were spindle-shaped (Fig. 3A) and adherent to the plastic dish. Most samples were further cultured after passage 3, demonstrating that the cells could be subcultured multiple times. Flow cytometric analysis showed that they coexpressed CD105, CD73, and CD90 (Fig. 3B and 3C) and were negative for CD45, CD34, or CD133 (Fig. 3D). The median percentage of triple-positive cells in the cultures was 31.62%, with a range of 2%–92.3% (N = 28, Table 1, specimens 5–32). Finally, all cultured cells could be differentiated into at least 2 of the 3 mesenchymal cell types, namely osteocytes (Fig. 3E), adipocytes (Fig. 3F), and chondrocytes (Fig. 3G), when they were exposed to the appropriate differentiation media.
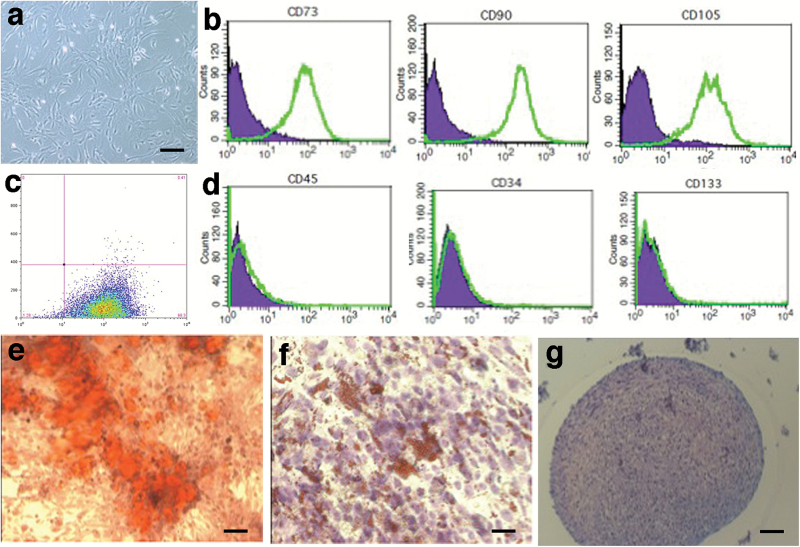
Fig. 3.
Characterization of tumor-associated mesenchymal stem cells (MSCs) isolated from glioblastoma surgical specimens. (A) In culture, spindle-shape appearance of GA-MSCs. Bar, 50 µm. (B) Representative flow cytometry analysis for positive markers: CD73 (left field), CD90 (middle field), and CD105 (right field). (C) Triple-positive (CD105+/CD73+/CD90+) cells are shown in Fig. 1C. (D) Representative flow cytometry analysis for negative markers: CD45 (left field), CD34 (middle field), and CD133 (right field). Differentiation of GA-MSCs into osteocytes (E, calcium deposits in red; bar, 200 µm), adipocytes (F, fat droplets in red; bar, 200 µm), and chondrocytes (G, cartilage in purple; bar, 100 µm).
Using this well-characterized cohort of tumors, we analyzed the correlation between patient survival and the fraction of cells coexpressing CD105, CD73, and CD90, as measured in culture (N = 28). An inverse correlation suggested an association between a high fraction of cells coexpressing CD105, CD73, and CD90 and a short survival time (Somer’s Dyx rank correlation coefficient = −0.24; 95% CI: −0.54 to 0.07), supporting our initial observation (Fig. 2B). Similar to cohort 1, in this second larger cohort (N = 28), an X-tile plot analysis revealed that patients with tumors harboring a low percentage of triple-positive cells (defined as ≤29% of cultured cells) had a statistically significant survival advantage compared with patients whose tumors harbored a high percentage of triple-positive cells (>29%). Specifically, the estimated median OS for patients with a low percentage of triple-positive cells in culture was 66 months and the estimated median OS for patients with a high percentage of triple-positive cells in culture was 11 months (N = 28; HR = 0.38; 95% CI: 0.13–0.9, P = 0.04; Fig. 2C).
In a univariate Cox regression analysis, a high fraction of cells coexpressing CD105, CD73, and CD90 (P = .05), older age (P = .01), KPS score ≤80 (P = .003), and a pathology of glioblastoma (P = 0.02) predicted short-term survival, whereas IDH1 mutation status (P = .18) did not. In a multivariate Cox proportional hazards model including the fraction of cells coexpressing CD105, CD73, and CD90, age, KPS score, and the type of pathology, a good performance score on admission was the only variable (KPS score >80; HR = 4.37; 95% CI: 1.21–15.8, P = .03) associated with long OS. The fraction of cells coexpressing CD105, CD73, and CD90 (HR = 1.23; 95% CI: .39–3.87, P = .73), age (HR = 1.03; 95% CI: .98–1.08, P = .24), and type of pathology (HR = 7.79; 95% CI: .79–76.69, P = .08) were not independent predictors of OS in this multivariate analysis. However, in a further analysis, we found that anaplastic astrocytoma (AA) as well as IDH1-mutated tumor samples were associated with a fraction of triple-positive cells that fell below the defined cutoff (Fisher’s exact test; P = .03 and P = .02, respectively), supporting our hypothesis that the fraction of triple-positive cells in high-grade glioma inversely correlates with OS. In fact, 6 of the 7 patients with AA and all patients with IDH1-mutated tumors (N = 6) were found in the group of patients with a low fraction of triple-positive cells (Table 1). This finding is what would be expected given that AA and IDH1 mutations are associated with favorable outcome.
TCGA Dataset Cohort
To further demonstrate our findings in a larger, independent cohort, we used the dataset of TCGA. Of 414 patients included in our analysis, 170 were male and 244 were female, with a mean age of 57.9 ± 14.53 years and a median KPS score of 80 at diagnosis. Initially, we performed a Cox regression analysis to determine whether there was an association between the expression levels of the individual GA-hMSC marker genes and survival outcomes. Gene expression of Endoglin (CD105; P = .29), NT5E (CD73; P = 0.4), and THY1 (CD90; P = .18), when evaluated independently, showed no significant correlation with patient survival. To determine whether coexpression levels of all 3 MSC markers—Endoglin (mean normalized expression level 5.8 ± 0.55; range, 4.67–8), NT5E (mean normalized expression level 7.42 ± 1.37; range, 4.09–11.96), and THY1 (mean normalized expression level 8.2 ± 1.14; range, 4.01–10.87)—correlated with survival, we compared the outcomes of patients with coexpression levels in the highest quintile with those in the lowest quintile. Of 414 patients, 18 coexpressed Endoglin, NT5E, and THY1 in the lower quintile of gene expression, and 10 coexpressed the 3 markers in the highest quintile (Fig. 4A, B). Kaplan–Meier survival plots for patients with high (highest quintile) and low (lowest quintile) levels of Endoglin, NT5E, and THY1 gene coexpression were significantly different (HR = 0.4; 95% CI: 0.1–0.88; P = .04). Patients with high gene coexpression levels had a median OS of 8.4 months compared with 13.1 months for patients with low gene coexpression (Fig. 4C). We evaluated the ages and KPS scores of patients with high and low levels of gene coexpression and found that those with high levels of triple gene expression were older (mean age, 64.4 ± 4.3 y) than those with low levels (mean age, 51.76 ± 3.5 y; P = .03). The median KPS scores at diagnosis did not differ between the groups (P = .25), nor did the distribution of IDH1 mutation (P = .5) or MGMT methylation status (P = .64).
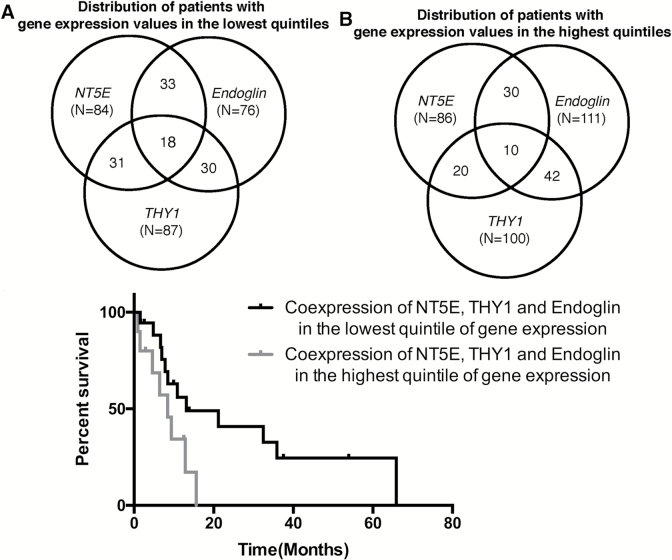
Fig. 4.
The distribution of patients represented in The Cancer Genome Atlas (TCGA) cohort, with gene expression levels at the lowest (A) or highest (B) quintile in Endoglin, NT5E, and THY1 genes separately and their overlaps. (C) Kaplan–Meier survival curves of patients having glioblastomas with high (upper quintile of gene expression) and low (lowest quintile of gene expression) levels of Endoglin, NT5E, and THY1 gene expression based on data from the database of TCGA.
In a Cox regression analysis, only the low quintile of Endoglin, NT5E, THY1 gene coexpression predicated long-term outcome (HR = 2.8; 95% CI: 0.1–7.5; P = .04). In contrast, age (HR = 1.0; 95% CI: 0.98–1.0; P = .37), KPS score (HR = 0.98; 95% CI: 0.96–1.0; P = .14), MGMT methylation status (HR = 0.96; 95% CI: 0.34–2.7; P = .94), and IDH1 mutation status (HR = 0.85; 95% CI: 1.9–3.8; P = .83) were not associated with survival in this selected cohort.
Discussion
We and others have shown previously that GA-hMSCs exist as stromal cells in human high-grade gliomas based on cell morphology, triple marker positivity (CD105/CD73/CD90), and tri-mesenchymal differentiation of cultured human specimens.14,15 We have also shown that GA-MSCs are not themselves tumorigenic but are stromal cells that enhance the proliferation and stemness of GICs.15 We now show, based on 2 cohorts of patients in which GA-hMSCs were identified either in the initial tumor before culture or in cultures of tumors at passage 3, and based on a third larger cohort from the dataset of TCGA, that the percentage of GA-hMSCs in gliomas varies from tumor to tumor and that patients with high percentages of GA-MSCs in their tumors have a worse OS than those with a low percentage, indicating that the fraction of GA-MSCs in the tumor mass is prognostic, and suggesting that the cellular composition of the microenvironment of gliomas influences patient outcome.
Tumor-associated MSCs (TA-MSCs) have been identified in other solid tumors.26–30 Along with endothelial cells and immune cells, TA-MSCs form the stroma of solid tumors, due to their well-documented natural tropism for developing tumors.30,31 Moreover, MSCs have been shown to influence tumor behavior.32,33 MSCs enhance tumor growth and tumor invasion in epithelial cancer subtypes such as colon,34 lung,35 breast,26,28 skin,36 and prostate.29 They may also contribute to establishment of distant metastases29,34,37 and may enhance resistance of cancer cells to chemotherapy. In contrast to this tumor-promoting role, MSCs were found to have a tumor-suppressive role in a Kaposi’s sarcoma model,22 adding to the complexity of tumor–MSC interactions. Despite these findings, little is known about the presence of MSCs in glioblastomas or their role in that microenvironment. Traditionally, the stroma of gliomas was thought to consist of vascular cells, microglia, immune cells, and neural progenitors.13 However, here and in a recent study,15 we demonstrated the presence of cells coexpressing CD105, CD73, and CD90, consistent with GA-hMSCs. The identification of such cells in fresh glioblastoma surgical specimens eliminates the possibility of their being artifacts resulting from culturing conditions and strengthens the observation that such populations exist within gliomas. Moreover, when cultured, GA-MSCs appeared as spindle-shaped cells adhering to the plastic dish, coexpressing MSC markers, as seen in the uncultured cells (see Fig. 3),17 and capable of tri-mesenchymal differentiation.24 These characteristics all agree with the International Society for Cellular Therapy's definition of MSCs,17 indicating that GA-hMSCs are part of the glioma microenvironment.
Because the fraction of GA-MSCs identified in the surgical high-grade glioma specimens varied among patients, we hypothesized that this fraction was associated with patient survival. This hypothesis is supported by our recent finding that GA-MSCs alter the biological behavior of GICs by increasing their proliferation and self-renewal capacity,15 suggesting a tumor-promoting role for GA-hMSCs in glioma. Consistent with this concept, we now show for the first time that a high percentage of GA-hMSCs in fresh glioma specimens (N = 9) and in cultured glioma specimens (N = 28) correlates with worse survival and marks gliomas with aggressive clinical behavior. Analyses of both cohorts identified a fraction of GA-MSCs above which there was a statistically significant worsening of survival. Importantly, the fraction of GA-MSCs was a predictor of survival in univariate analysis, along with age, KPS score, and the type of pathology. In multivariate analysis, only KPS score remained an independent predictor of survival. Nevertheless, in samples derived from patients with a less aggressive disease, namely those with AA pathology or IDH mutation, the fraction of GA-MSCs was statistically significantly lower than in patients with more aggressive disease, such as GBM or IDH wild-type glioma. Intriguingly, in contrast to previous reports that found that mutated IDH1 correlated with favorable prognosis in large populations of GBM patients, IDH1 mutation status was not associated with prognosis in our cohort, probably because of the small number of IDH-mutant GBM tumors in our cohort and possibly because of statistical interactions with the percentage of GA-hMSCs in this relatively small cohort.
To demonstrate our finding of an inverse relationship between the fraction of GA-MSCs in the tumor and the survival of glioma patients, in a larger and independent cohort, we performed a gene-based survival outcome analysis using the dataset of TCGA. When analyzed individually, expression of the 3 MSC surface markers did not correlate with survival. However, when all 3 markers were analyzed together (as a surrogate for coexpression on MSCs), high and low coexpression of triple markers predicted shorter and longer OS, respectively, thus supporting our initial observation. The analysis of TCGA also allowed us to examine the predictive role of known molecular factors such as IDH1 mutation and MGMT methylation. Neither of these covariates predicted survival outcome in our selected cohort, whereas the levels of Endoglin, NT5E, and THY1 gene coexpression did. These findings suggests that the coexpression level of the triple marker is an independent predictor of survival; however, this result should be evaluated with caution because of the small sample size. To our knowledge, this is the first time the database of TCGA has been used to study the glioblastoma microenvironment as a determinant for patient outcomes. However, because whole surgical specimens were used in this TCGA analysis, we cannot know what fraction of individual cells expressed the 3 surface markers simultaneously. Regardless, our data are compelling and provide impetus for further studies on GA-MSCs in malignant gliomas.
Although this study does not address the reason the fraction of GM-MSCs in high-grade glioma is associated with clinical outcome, in a recent paper15 we suggested that GA-MSCs increased proliferation and self-renewal of GICs at least partly by secreting the cytokine interleukin-6 and by activating signal transducer and activator of transcription 3. GA-MSCs may also promote angiogenesis by recruiting endothelial progenitor cells and by facilitating blood vessel formation.38 Both mechanisms provide a biological basis underlying our observation that increased percentages of GA-hMSCs are associated with worse clinical outcome.
Funding
We acknowledge the following for support of this study: the National Institutes of Health: 5R01 CA115729-05; National Cancer Institute, SPORE in Brain Cancer (Project 1, Core B, and Core C: 1P50 CA127001-06; the Broach Foundation for Brain Cancer Research; the Elias Family Fund; the Gene Pennebaker Brain Cancer Fund; the Anthony Bullock III Foundation; the Brian McCulloch Fund, the Uncle Kory Foundation, the Jason & Priscilla Hiley Fund, and philanthropic contributions to The University of Texas M.D. Anderson Moon Shots Program.
Supplementary Material
Acknowledgment
We thank David M. Wildrick, PhD, for editorial assistance.
Conflict of interest statement. No author declares a conflict of interest to disclose.
References
- 1. DeAngelis LM. Brain tumors. N Engl J Med. 2001;344(2):114–123. [DOI] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Gilbert H, Kagan AR, Cassidy F, et al. Glioblastoma multiforme is not a uniform disease! Cancer Clin Trials. 1981;4(1):87–89. [PubMed] [Google Scholar]
- 4. Curran WJ, Jr, Scott CB, Horton J, et al. Recursive partitioning analysis of prognostic factors in three Radiation Therapy Oncology Group malignant glioma trials. J Natl Cancer Inst. 1993;85(9):704–710. [DOI] [PubMed] [Google Scholar]
- 5. Lamborn KR, Chang SM, Prados MD. Prognostic factors for survival of patients with glioblastoma: recursive partitioning analysis. Neuro Oncol. 2004;6(3):227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lacroix M, Abi-Said D, Fourney DR, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg. 2001;95(2):190–198. [DOI] [PubMed] [Google Scholar]
- 7. Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. [DOI] [PubMed] [Google Scholar]
- 8. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 9. Rivera AL, Pelloski CE, Gilbert MR, et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12(2):116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. [DOI] [PubMed] [Google Scholar]
- 11. Heimberger AB, Suki D, Yang D, et al. The natural history of EGFR and EGFRvIII in glioblastoma patients. J Transl Med. 2005;3:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell. 2010;18(6):884–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Charles NA, Holland EC, Gilbertson R, et al. The brain tumor microenvironment. Glia. 2012;60(3):502–514. [DOI] [PubMed] [Google Scholar]
- 14. Kim YG, Jeon S, Sin GY, et al. Existence of glioma stroma mesenchymal stemlike cells in Korean glioma specimens. Childs Nerv Syst. 2013;29(4):549–563. [DOI] [PubMed] [Google Scholar]
- 15. Hossain A, Gumin J, Gao F, et al. Mesenchymal stem cells isolated from human gliomas increase proliferation and maintain stemness of glioma stem cells through the IL-6/gp130/STAT3 pathway. Stem Cells. 2015;33(8):2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang SG, Shinojima N, Hossain A, et al. Isolation and perivascular localization of mesenchymal stem cells from mouse brain. Neurosurgery. 2010;67(3):711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2(4):313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kidd S, Spaeth E, Klopp A, et al. The (in) auspicious role of mesenchymal stromal cells in cancer: be it friend or foe. Cytotherapy. 2008;10(7):657–667. [DOI] [PubMed] [Google Scholar]
- 19. Spaeth EL, Dembinski JL, Sasser AK, et al. Mesenchymal stem cell transition to tumor-associated fibroblasts contributes to fibrovascular network expansion and tumor progression. PLoS One. 2009;4(4):e4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mishra PJ, Humeniuk R, Medina DJ, et al. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 2008;68(11):4331–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu W, Xu W, Jiang R, et al. Mesenchymal stem cells derived from bone marrow favor tumor cell growth in vivo. Exp Mol Pathol. 2006;80(3):267–274. [DOI] [PubMed] [Google Scholar]
- 22. Khakoo AY, Pati S, Anderson SA, et al. Human mesenchymal stem cells exert potent antitumorigenic effects in a model of Kaposi’s sarcoma. J Exp Med. 2006;203(5):1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Behnan J, Isakson P, Joel M, et al. Recruited brain tumor-derived mesenchymal stem cells contribute to brain tumor progression. Stem Cells. 2014;32(5):1110–1123. [DOI] [PubMed] [Google Scholar]
- 24. Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. [DOI] [PubMed] [Google Scholar]
- 25. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu S, Ginestier C, Ou SJ, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011;71(2):614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McLean K, Gong Y, Choi Y, et al. Human ovarian carcinoma–associated mesenchymal stem cells regulate cancer stem cells and tumorigenesis via altered BMP production. J Clin Invest. 2011;121(8):3206–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan XL, Fu CJ, Chen L, et al. Mesenchymal stem cells from primary breast cancer tissue promote cancer proliferation and enhance mammosphere formation partially via EGF/EGFR/Akt pathway. Breast Cancer Res Treat. 2012;132(1):153–164. [DOI] [PubMed] [Google Scholar]
- 29. Luo J, Ok Lee S, Liang L, et al. Infiltrating bone marrow mesenchymal stem cells increase prostate cancer stem cell population and metastatic ability via secreting cytokines to suppress androgen receptor signaling. Oncogene. 2014;33(21):2768–2778. [DOI] [PubMed] [Google Scholar]
- 30. Coffelt SB, Marini FC, Watson K, et al. The pro-inflammatory peptide LL-37 promotes ovarian tumor progression through recruitment of multipotent mesenchymal stromal cells. Proc Natl Acad Sci U S A. 2009;106(10):3806–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–1659. [DOI] [PubMed] [Google Scholar]
- 32. Bergfeld SA, DeClerck YA. Bone marrow-derived mesenchymal stem cells and the tumor microenvironment. Cancer Metastasis Rev. 2010;29(2):249–261. [DOI] [PubMed] [Google Scholar]
- 33. Cuiffo BG, Karnoub AE. Mesenchymal stem cells in tumor development: emerging roles and concepts. Cell Adh Migr. 2012;6(3):220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shinagawa K, Kitadai Y, Tanaka M, et al. Mesenchymal stem cells enhance growth and metastasis of colon cancer. Int J Cancer. 2010;127(10):2323–2333. [DOI] [PubMed] [Google Scholar]
- 35. Suzuki K, Sun R, Origuchi M, et al. Mesenchymal stromal cells promote tumor growth through the enhancement of neovascularization. Mol Med. 2011;17(7-8):579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kucerova L, Matuskova M, Hlubinova K, et al. Tumor cell behaviour modulation by mesenchymal stromal cells. Mol Cancer. 2010;9:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goldstein RH, Reagan MR, Anderson K, et al. Human bone marrow-derived MSCs can home to orthotopic breast cancer tumors and promote bone metastasis. Cancer Res. 2010;70(24):10044–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Au P, Tam J, Fukumura D, et al. Bone marrow-derived mesenchymal stem cells facilitate engineering of long-lasting functional vasculature. Blood. 2008;111(9):4551–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.