Abstract
Background.
Activating mutations or structural rearrangements in BRAF are identified in roughly 75% of all pediatric low-grade astrocytomas (PLGAs). However, first-generation RAF inhibitors approved for adult melanoma have poor blood–brain penetrance and are only effective on tumors that express the canonical BRAFV600E oncoprotein, which functions as a monomer. These drugs (type I antagonists that target the “DFG-in” conformation of the kinase) fail to block signaling via KIAA1549:BRAF, a truncation/fusion BRAF oncoprotein which functions as a dimer and is found in the most common form of PLGA.
Methods.
A panel of small molecule RAF inhibitors (including type II inhibitors, targeting the “DFG-out” conformation of the kinase) was screened for drugs showing efficacy on murine models of PLGA and on authentic human PLGA cells expressing KIAA1549:BRAF.
Results.
We identify a type II RAF inhibitor that serves as an equipotent antagonist of BRAFV600E, KIAA1549:BRAF, and other noncanonical BRAF oncoproteins that function as dimers. This drug (MLN2480, also known as TAK-580) has good brain penetrance and is active on authentic human PLGA cells in brain organotypic cultures.
Conclusion.
MLN2480 may be an effective therapeutic for BRAF mutant pediatric astrocytomas.
Keywords: MLN2480, pediatric low-grade astrocytoma, RAF dimers
Importance of the study
Primary cancers of the brain have surpassed leukemia to become the number one cause of cancer-related death in children. We identified a small molecule RAF inhibitor with therapeutic potential for low-grade astrocytomas, the most common type of pediatric brain cancer.
Activating mutants of BRAF are genetic drivers for many adult tumors, including the melanomas, papillary carcinomas of thyroid, and significant numbers of lung and colon carcinomas.1 Recent studies have extended the oncogenic repertoire of BRAF to pediatric low-grade astrocytomas (PLGAs), the most common brain tumor of childhood. Roughly 85% of juvenile pilocytic astrocytomas (World Health Organization [WHO] grade I; the most common type of PLGA) express constitutively active truncation/fusion forms of BRAF, the most common of which is known as KIAA1549:BRAF. All of these BRAF truncation/fusions involve deletion of an amino terminal autoinhibitory domain and result in the formation of constitutively active BRAF dimers.2–4 The prevalence of truncation/fusion mutations in juvenile pilocytic astrocytoma contrasts with adult cancers, where the most common oncogenic form of BRAF is a V600E substitution mutant which functions as a constitutively active monomer.5 The BRAFV600E mutation is rare in juvenile pilocytic astrocytomas but is seen in some fibrillary astrocytomas, gangliogliomas, and pleomorphic xanthoastrocytoma where its occurrence is mutually exclusive with BRAF truncation/fusion mutations.6–9
Small molecule RAF inhibitors have shown efficacy in adult patients with BRAFV600E mutant cancers. Unfortunately, the RAF inhibitors currently approved for these adult tumors (eg, vemurafenib, dabrafenib) are active only on monomeric BRAF oncoproteins with substitution mutations at position V600.1,10,11 The blood–brain barrier is another confounding issue for treating PLGAs with RAF inhibitors. Neither vemurafenib nor dabrafenib have good blood–brain penetrance. Juvenile pilocytic astrocytomas frequently show local breakdown of the blood–brain barrier, as shown by contrast enhancing MRI.12 However, as noted above, almost all juvenile pilocytic astrocytomas express truncation/fusion variants of BRAF, which function as dimers and do not respond to vemurafenib or dabrafenib.11 As noted above, a subset of the fibrillary astrocytomas, gangliogliomas, and pleomorphic xanthoastrocytomas express the BRAFV600E oncoprotein. Contrast enhancing examples of these tumors could, in principle, respond to vemurafenib or dabrafenib. However, these tumors tend to be more infiltrative than juvenile pilocytic astrocytomas, and many of them do not show evidence of local blood–brain barrier breakdown. A final concern with RAF inhibitors as therapeutics for pediatric patients are the skin rashes and secondary skin tumors (squamous cell carcinomas) observed in adult patients treated with vemurafenib or dabrafenib.1 These dermatologic complications reflect a combination of (i) time-delayed “rebound” signaling activity via drug-induced loss of extracellular signal-regulated kinase (ERK) feedback on the RAS/RAF/mitogen extracellular signal-regulated kinase signaling axis and (ii) “paradoxical activation” of wild-type RAF kinase dimers in cells with moderate RAS activity.5,10,13–15
The monomer-specific RAF inhibitors, vemurafenib and dabrafenib, are both type I antagonists that target the active or “DFG-in” conformation of the BRAF catalytic domain.16 Recent studies show that other types of RAF inhibitors, including type II antagonists that target the inactive (DFG-out) conformation of the kinase, can target both monomeric and dimeric forms of the BRAF oncoprotein. These same agents eliminate the rebound signaling and paradoxical activation that are observed with vemurafenib and dabrafenib.17–20 In studies summarized here, we describe a brain-penetrant type II RAF inhibitor that suppresses both monomeric and dimeric forms of the BRAF oncoprotein in human PLGA cells and has therapeutic potential for PLGAs.
Materials and Methods
Animal Procedures, Tissue Dissociation, and Cell Culture
Animal husbandry was performed according to Dana-Farber Cancer Institute guidelines under the Institutional Animal Care and Use Committee—approved protocols. The strains used have been described previously.21 Neuroprogenitor cells were isolated from embryonic day 14 medial ganglionic eminences, dispersed by trituration, grown under neurosphere suspension conditions, and passaged with Dulbecco’s modified Eagle’s medium (DMEM)/F12 supplemented with B27 and N2 (Invitrogen) in the presence of basic fibroblast growth factor (FGF) and epidermal growth factor (EGF) (20 ng/mL).22 Tumor/brain organotypic cultures employed murine E14 vibratome sections as a substrate for PLGA cells as detailed by Chadwick et al.23 PLGA cells were derived as previously described24 from fresh PLGA tumors acquired from patients undergoing neurosurgery at the Boston Children’s Hospital on an institutional review board—approved protocol as part of the Dana-Farber/Harvard Cancer Center Living Tissue Bank program.
In vitro Drug Screening Assay, In vivo Tumorigenesis Assay, and Drug Treatment
For in vitro drug screening studies, cells were plated at 1000–4000 cells per well and then treated with different drugs the next day at the indicated concentrations. Triplicate wells were used for each dose or time point. After drug treatment for 4 days, cell cultures were subjected to CellTiter-Glo luminescent cell viability assay (Promega) according to the standard protocol. For each drug at different doses, the luminescence signal was normalized to dimethyl sulfoxide (DMSO) control group and analyzed by GraphPad Prism. For the in vivo tumorigenesis study, 2×105 p53 null neuroprogenitor cells bearing BRAFV600E or KIAA1549:BRAF were injected into the right striatum of Icr-SCID or NCr nude mice (Taconic Farms) at the coordinates: A, 0 mm, L, 2.0 mm, and D, 2.0 mm relative to the bregma. Animals were euthanized at the onset of neurological/clinical symptoms. For the drug in vivo treatment experiment, cells stably expressing the luciferase gene were injected intracranially, and tumor growth was monitored using bioluminescence imaging. Bioluminescence was measured by injecting anesthetized animals with D-luciferin (100 mg/kg) and imaging using an IVIS (in vivo imaging system) camera (Caliper Life Sciences). Quantification of signal was performed using Living Image software (Caliper Life Sciences). Dosing was determined by the maximum amount of MLN2480 (also known as TAK-580) that could be reliably suspended in vehicle (0.5% carboxymethyl cellulose, 0.1% Tween-20) 30 mg/kg. This dose is far below the calculated maximum tolerated dose based on human trials for MLN2480 (2500 mg/kg).
In vivo Pharmacokinetic Study
Male Swiss albino mice were dosed via oral gavage (suspensions in 0.5% w/v Na carboxymethyl cellulose with 0.1% v/v Tween-80 in water). Blood and brain samples were collected at 0, 0.083 (for i.v. only), 0.25, 0.5, 1, 2, 4, 6 (for p.o. only), 8, 12, and 24 h for the i.v. and p.o. groups. The blood samples were collected from sets of 3 mice at each time point. Plasma samples were separated by centrifugation and stored below −70°C until bioanalysis. Brain samples were homogenized using ice-cold phosphate buffered saline (pH 7.4), and homogenates were stored below –70°C until analysis. All samples were processed for analysis and analyzed by liquid chromatography–tandem mass spectrometry (lower limit of quantification, 2.03 ng/mL for plasma and 10.16 ng/mL for brain). Pharmacokinetic parameters were calculated using the noncompartmental analysis tool of Phoenix WinNonlin (version 5.3).
Mass Spectrometry Imaging
Mouse brains harboring KIAA1549:BRAF fusion tumors (p53-/-) or wild type were flash frozen following treatment, stored at −80°C, and placed at −25°C one hour before use. Twelve-micron coronal tissue sections were prepared using a Microm HM550 cryostat, thaw mounted onto indium tin oxide–coated microscopic slides (Bruker Daltonics) for matrix-assisted laser desorption/ionization—mass spectrometry imaging (MALDI-MSI) and optical slides for hematoxylin and eosin staining. Samples were dried for 15 minutes in a desiccator. 2,5-Dihydroxybenzoic acid (40 mg/mL solution in methanol OH/0.2% trifluoroacetic acid 70:30 v/v) was deposited using the TM-Sprayer system (HTX Technologies) as previously described.25 Mass spectra were acquired using a 9.4 Tesla SolariX XR Fourier transform ion cyclotron resonance mass spectrometer (Bruker Daltonics). MALDI-MSI settings are in the Supplementary material.
KiNativ and Association Assays
BRAFV600E or KIAA1549:BRAF oncogenes containing neurospheres were incubated with the indicated compound (1 μM) for 1 h. Cell pellets were collected via centrifugation and stored at −80°C. Lysates were generated and incubated with an ATP biotin probe to afford labeling of unbound kinases. Labeled kinases were separated from the total lysate via streptavidin pull-down, removed from the streptavidin, digested, and identified by mass spectrometry.
To analyze the direct interaction between BRAF mutants and inhibitors, a kinase enrichment kit (Pierce) was used with ATP probe according to the instructions. Indicated variants of BRAF were cotransfected into human embryonic kidney (HEK)293 cells. Forty-eight hours after transfection, lysates were collected and desalted, then 1 mg lysate was preincubated with MLN2480 or vemurafenib at different doses for 10 min at room temperature, then labeled with desthiobiotin-ATP for another 10 min. Each sample was denatured by adding urea to 4 M then incubated with 50 μL 50% streptavidin agarose slurry for 1 h. The amounts of BRAF mutants that were labeled with biotinylated ATP were visualized and compared by western blot.
Co-immunoprecipitation Assay
One microgram of KIAA1549:BRAF with C-terminal tag (HA or V5) was either singly or doubly transfected into HEK293/Cos-7 cells; 24–48 h later, cells were starved with serum-free DMEM overnight followed by drug treatment for 90 min. Then cells were harvested in the following buffer: 20 mM Tris pH 7.5, 150 mM NaCl, 0.2% NP-40, 0.5% sodium deoxycholate, 10% glycerol with protease inhibitor cocktail (Roche) and phosphatase inhibitors. Immunoprecipitation of KIAA1549:BRAF-V5 was performed with anti-V5 affinity agarose (Sigma). Western blotting was performed using the electrochemiluminescence procedure according to the manufacturer’s instructions (Amersham Bioscience), with a rabbit polyclonal anti-HA antibody (1:2000 HA-probe [Y-11], Santa-Cruz) to detect the co-immunoprecipitated KIAA1549:BRAF-HA protein.
Additional details on plasmids and antibodies used, generation and use of retroviruses, transfection methodology, western blot analyses, and immunohistochemistry can be found in the Supplementary material.
Results
Type II RAF Antagonists Target Multiple Forms of BRAF Oncoprotein in a “Pathway Relevant” PLGA Murine Model
Patient-derived PLGA cells cannot be serially passaged in animals or in culture. For entry-level drug screens, we developed murine models of BRAF mutant PLGAs by transducing TP53 null neural progenitors with either KIAA1549:BRAF fusion or the BRAFV600E expression vectors. It should be stressed that our 2 models do not emulate the pediatric tumors at the genetic or histopathologic levels. In particular, the tumor protein (TP)53 gene is structurally intact in authentic PLGAs.26 However, rapid proliferations in culture and in vivo are enabled by ablation of TP53 and are desirable qualities for drug screening exercises, and as shown in Supplementary Fig. S1 our 2 models are “pathway relevant” to BRAF mutant PLGAs. In particular, the 2 BRAF-transformed cell lines proliferate in factor-free culture medium and form intracranial tumors in immune-suppressed mice. In contrast, TP53 null neural progenitors transduced with wild-type BRAF or with vector controls are not tumorigenic and require EGF and FGF to proliferate in culture.
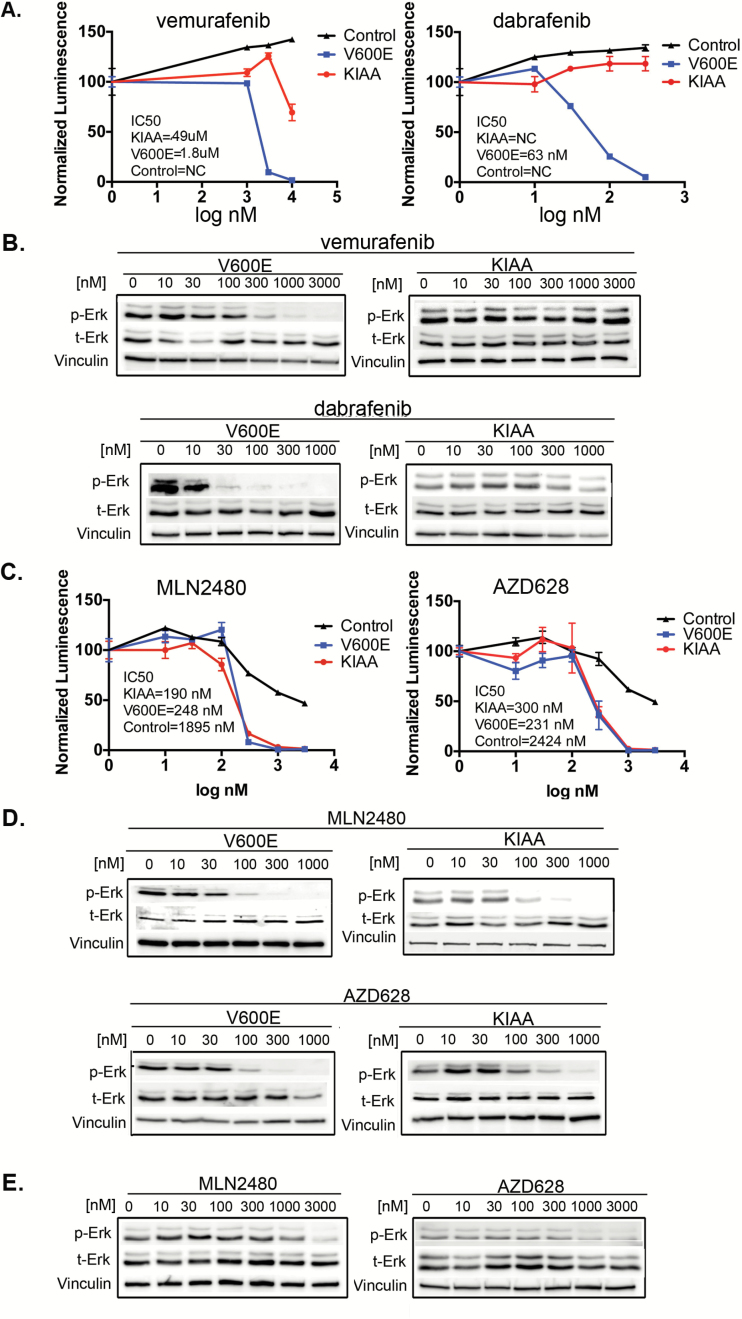
In agreement with others, the type I RAF inhibitors vemurafenib and dabrafenib are active on the V600E variant of our PLGA model10,15 but are much less effective on the KIAA1549:BRAF cells11 (Figs 1A, B, Supplementary Fig. S2). In contrast, a pair of type II RAF inhibitors, MLN248027 and AZD628,28 are equally active on both of the BRAF oncoproteins (Fig. 1C, D, Supplementary Fig. S2). Also, in accord with recent studies,17–19 these type II RAF inhibitors are active on other mutants of BRAF that are far less sensitive to the type I drugs (Supplementary Fig. S3). The effect of the type II drugs on TP53 null neural progenitors transduced with vector controls and cultured in the presence of EGF and bFGF is shown in Fig. 1E and Supplementary Fig. S2C. As indicated, the nontransformed cells responding to growth factors are ~10-fold more resistant to the type II drugs than their malignant counterparts driven by BRAF oncogenes, suggesting a potentially useful therapeutic window.
Fig. 1.
Type I and type II RAF inhibitors have differential effects on BRAF activating mutants. (A) p53-/- neuroprogenitor cells were transduced with the indicated constructs. Control cells (with EGF and basic FGF) and oncogene-transformed cells (in factor-free medium) were treated with vemurafenib or dabrafenib for 4 days and viability at day 4 was monitored by CellTiter Glo. (B) As in (A) above, except that treatment with vemurafenib or dabrafenib was for 1 hour and phospho-ERK signaling was monitored by immunoblot. (C, D) Proliferation and signal transduction were monitored as in panels (A) and (B) above. (E) Wild-type BRAF cells were treated as in (D) and phospho-ERK assayed by immunoblot.
MLN2480 and AZD628 Act Directly upon BRAF
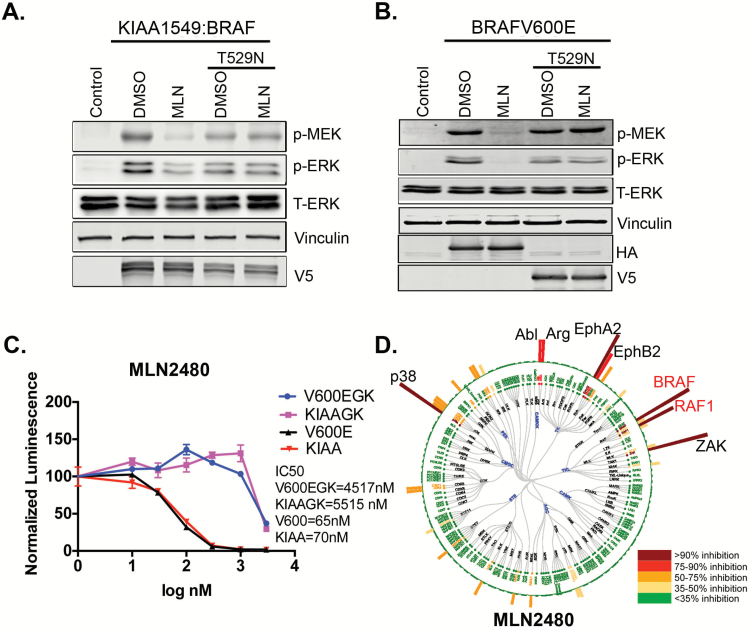
As shown by KiNativ assay,29 a chemical proteomics technique that displays competition between kinase inhibitors and biotinylated ATP, the type II inhibitors MLN2480 and AZD628 interact directly with BRAF in both BRAFV600E and KIAA1549:BRAF expressing neurospheres (Fig. 2D, Supplementary Fig. S4). However, several other protein kinases also appear to interact with MLN2480 and AZD628. To validate target specificity, we inserted the T529N gatekeeper mutation, which blocks drug binding,30 into both of the BRAF oncogene expression vectors. As shown, the gatekeeper mutation abolishes effects of the type II inhibitor MLN2480 on both proliferation and ERK signaling (Fig. 2A–C) in cells with either the V600E or the KIAA1549 oncoproteins. Similar results were obtained for V600E with AZD628 (Supplementary Fig. S5A). Thus type II inhibitors engage with BRAF (Fig. 2D, Supplementary Fig. S4), and their impact on proliferation and downstream RAF signaling is contingent upon this engagement. The inhibitory effects of a type I inhibitor (vemurafenib) on BRAFV600E are also eliminated when the gatekeeper mutation is introduced into BRAFV600E protein (Supplementary Fig. S5B).
Fig. 2.
The response of KIAA1549:BRAF expressing cells to type II RAF inhibitors requires drug interaction with the KIAA1549:BRAF oncoprotein. (A, B) Mouse neuroprogenitor cells transduced with the indicated constructs were treated with MLN2480 (MLN) at 1 µM or DMSO and examined by immunoblot. (C) p53-/- mouse neuroprogenitor cells stably transduced with the indicated constructs, then treated with MLN2480 and assayed for cell proliferation. V600EGK, BRAFV600E with gatekeeper mutation; KIAAGK, BRAF fusion with gatekeeper mutation (D) KiNativ assay for kinase specificity was performed after 1 hour of drug treatment in KIAA1549:BRAF expressing p53 null neural progenitor cells. The circle represents the kinase families tested in the assay and the bars extending outward from the circle represent relative inhibition of these kinases by MLN2480. MLN2480 binds native BRAF and RAF1 (aka CRAF) as indicated in the dendrogram.
Therapeutic Potential: MLN2480 Has Good Blood—Brain Barrier Penetrance
As an initial assessment of the potential ability of MLN2480 and AZD628 to access brain tumors, we performed standard pharmacokinetic experiments to quantify the drug levels in plasma and brain of mice following oral (10 mg/kg) drug administration. MLN2480 had a plasma:brain ratio of 24% and area under the curve (brain) of 41.6 h•µM following the oral dose, suggesting significant accumulation in the CNS, and was thus singled out for closer scrutiny as a potential therapeutic for PLGAs (Supplementary Table S1).
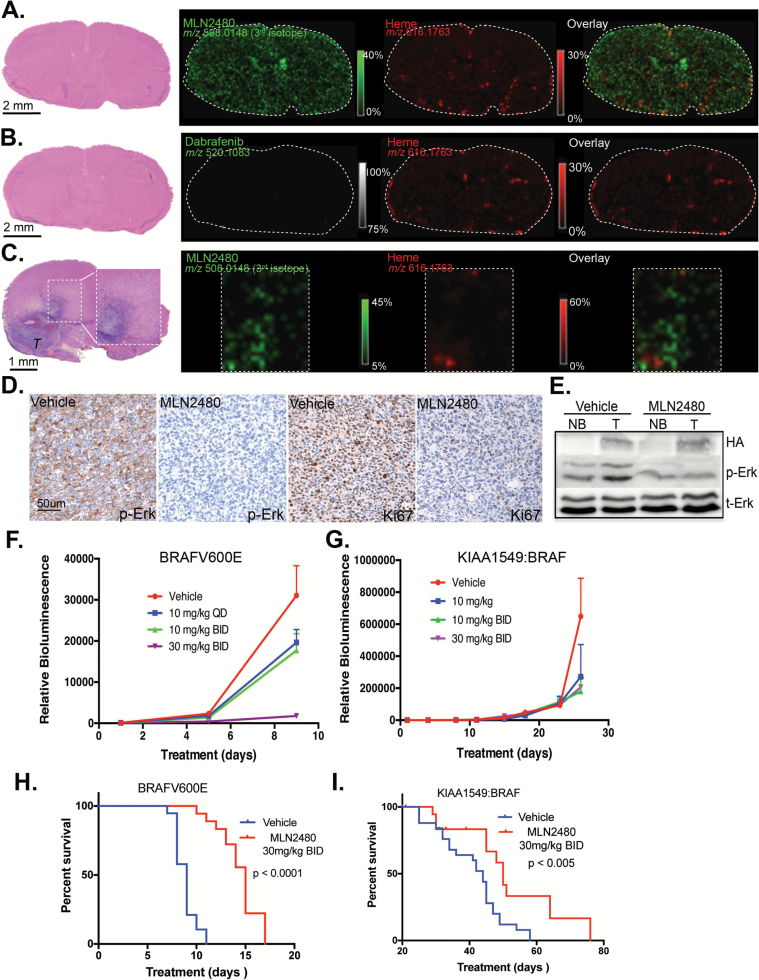
MALDI-MSI can be used to localize drug molecule and metabolite distributions in tissue sections. By co-imaging a marker that cannot escape the lumen of brain capillaries (eg, heme) together with the drug of interest, the ability of drugs to transit the blood—brain or blood—tumor barriers can be directly visualized.31 As indicated, MLN2480 readily segregates from heme to access the interstitial regions of healthy brain (Fig. 3A) as well as intracranial tumors generated by stereotactic injection of the KIAA1549:BRAF PLGA model cells (Fig. 3C). As a negative control, Fig. 3B shows dabrafenib, a type I RAF inhibitor has poor blood—brain penetrance.
Fig. 3.
MLN2480 can cross the blood–brain barrier and retard growth of BRAFV600E and KIAA1549:BRAF intracranial tumors in NCr-NU mice. (A) Hematoxylin and eosin (H&E) staining of a normal brain section following a single oral dose of 30 mg/kg MLN2480. Tissue was collected after 4 hours. Green and red colors indicate the distributions of MLN2480 (third isotope, m/z 508.0148) and heme (m/z 616.1763). (B) H&E staining of a normal brain section following a single oral dose of 100 mg/kg dabrafenib as in (A). White dotted area delineates normal brain regions, which were subjected to MALDI-MS imaging at 75 µm spatial resolution. Green and red colors indicate the distributions of dabrafenib and heme. (C) H&E staining of a KIAA1549:BRAF tumor bearing brain section following 41 days of twice daily oral MLN2480 treatment at 30 mg/kg. Brain tissue was collected 4 hours after the final dose. The representative images of MLN2480 and heme distribution in tumor region (“T”) highlighted in rectangle are indicated. (D) Decreased phospho-ERK signaling in KIAA1549:BRAF brain tumors treated with MLN2480 as shown by immunohistochemistry. Ki67-positive cells in MLN2480-treated tumors are also decreased. (E) Immunoblot analysis of phospho-ERK signaling in MLN2480 treated brain tumors (T) and from contralateral normal brain (NB). (F, G) p53-/- neuroprogenitor cells were stably transduced with BRAFV600E or KIAA1549:BRAF, then intracranially injected into NCr-NU mice. Mice harboring tumors were dosed orally with MLN2480 each day (days 1–9 for the BRAFV600E and 1–28 for KIAA1549:BRAF), as indicated, and subjected to bioluminescent imaging (mean ± SEM; P < .05. One way ANOVA, n = 10). (H, I) Kaplan–Meier analysis of mice with intracranial tumors as in F, G treated (or not) with MLN2480.
In accord with the MALDI-MSI images, immunohistochemical and immunoblot analyses show that MLN2480 suppresses phospho-ERK in both tumor tissue and healthy brain (Fig. 3D, E). Moreover, Ki67-positive tumor cells also decreased upon MLN2480 treatment (Fig. 3D). In separate experiments, the impact of MLN2480 on intracranial growth of both BRAFV600E and KIAA1549:BRAF-driven tumors was assessed. MLN2480 retarded the growth of both of these tumor types (Fig. 3F, G) and increased survival of the treated mice (Fig. 3H, I).
MLN2480 Is Active on BRAF Mutant Human PLGAs
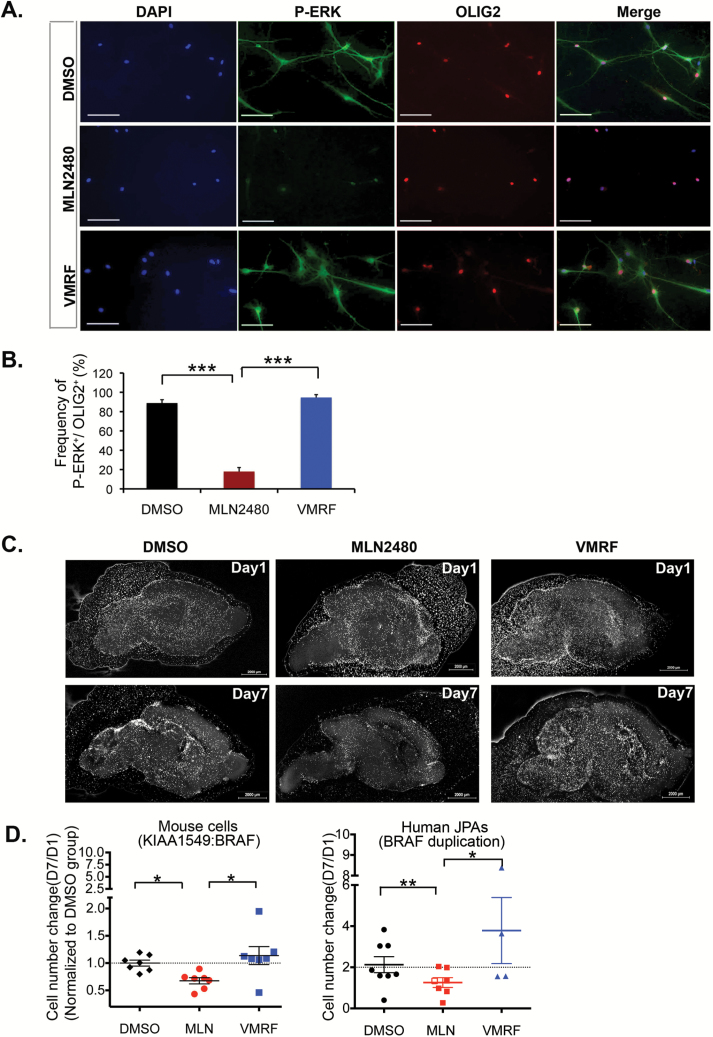
Signaling responses of freshly isolated PLGA cells (Supplementary Table S2) to MLN2480 were monitored in short-term (~24 h) experiments using cells plated on laminin-coated coverslips. Enzymatically disaggregated tumor samples contain some normal stromal cells. Signaling responses of OLIG2-positive tumor cells32–34 from a WHO grade I juvenile pilocytic astrocytoma with the KIAA1549 mutation to type I and type II RAF inhibitors are shown in Fig. 4A, B. As indicated, MLN2480 suppresses phospho-ERK in the OLIG2-positive cells while vemurafenib does not.
Fig. 4.
A type II RAF inhibitor is active on BRAF mutant human PLGAs. (A) Freshly isolated human PLGA cells expressing the BRAF truncation/fusion mutation (BT728, see Supplementary Table S2) were seeded onto laminin-coated microscope coverslips and incubated overnight in growth factor-free medium. The cells were then treated with 1 µM MLN2480 or vemurafenib (VMRF) for 1 hr, fixed and processed for immunofluorescence of phospho-ERK (P-ERK) or OLIG2. (B) Quantification of the P-ERK and OLIG2 images (mean ± SEM; ***P < .001; one way ANOVA with Bonferroni test; n = 441 cells). (C) Example of organotypic slice overlay assay using p53-/- murine neural progenitor cells transduced with KIAA1549:BRAF. Cells were labeled with Cm-Dil, seeded onto brain slices (see “Materials and Methods”) and then incubated with vehicle control (DMSO) or with 1 µM MLN2480 or vemurafenib (VMRF) as indicated for 7 days. Slices were imaged for Cm-Dil at days 1 and 7. (D) Quantification of drug responses on slice overlay cultures. KIAA1549:BRAF-transformed mouse cells or, alternatively, cells from 8 different juvenile pilocytic astrocytomas bearing the truncation/fusion BRAF mutation were processed as in panel (C) above. The overlay cultures were incubated with DMSO or with drugs at 1 mM (filled symbols) or at 100 nM (open symbols). For the human samples, a total of 5 different tumors were tested for MLN2480 and 4 different tumors were tested for vemurafenib (Table S2). The data shown depict cumulative cell counts (day 7/day 1) for 7 biological replicate experiments (mouse). Each symbol represents the average cell counts (day 7/day 1) of a single tumor. MLN2480 suppressed proliferation of the KIAA1549:BRAF murine model cells (mean ± SEM;*P < .05. One way ANOVA with Tukey’s multiple comparison test, 7 slices for each group) and their human tumor counterparts (mean ± SEM; DMSO vs. MLN, ** P = .0006, paired t-test, n = 7 tumors;). Vemurafenib is without effect on the mouse cells (mean ± SEM; P > .05, one way ANOVA with Tukey’s multiple comparison test) and has no discernible effect on the human tumor cells (DMSO vs VMRF, P = .185, paired t-test, n = 4 tumors; *MLN vs VMRF, P = .0305, paired t-test, n = 3 tumors). Scale bar, 50 uM.
To monitor the impact of RAF inhibitors on PLGA cell proliferation, we developed an organotypic assay23 in which fresh surgical isolates of primary pediatric brain tumors can be expanded for 1 to 2 weeks when seeded upon sections of neonatal mouse brain. As shown in Fig. 4C, D, the growth responses of our KIAA1549 murine model cells to type I and type II RAF inhibitors on this slice overlay system are identical to those observed in neurosphere cell cultures. The type II inhibitor MLN2480 inhibits growth, whereas the type I inhibitor vemurafenib does not. In the same assay, MLN2480 suppressed the growth of WHO grade I juvenile pilocytic astrocytomas with the truncation/fusion BRAF mutation, whereas vemurafenib has no significant effect (Fig. 4D, Supplementary Table S2).
MLN2480 Does Not Trigger Paradoxical Activation or “Rebound” ERK Signaling
As indicated in Supplementary Figs S6, S7, the type II drugs studied here duplicate desirable “paradox-breaking” features when tested on BRAF-transformed neural progenitor cells. In extended time course experiments with our V600E-transformed neural stem cells, type I RAF inhibitors give rise to the stereotypical rebound ERK signal. However, the type II inhibitors MLN2480 and AZD628 do not trigger this response in neural progenitors transformed with either of the BRAF oncoproteins (Supplementary Figs S6C, D; S7C, D).
Paradoxical activation is triggered when type I inhibitors are delivered at therapeutic concentrations (~1 µM and ~0.1 µM for vemurafenib and dabrafenib, respectively; Fig. 1) to murine neural progenitor cells that have been stably transduced with the HRASG12V oncoprotein (Supplementary Fig. S6E, 7E). Under identical conditions, paradoxical activation is not observed in cells treated with type II inhibitors at therapeutic concentrations (~1 µM for both drugs; Supplementary Fig. S6). However, we note that both MLN2480 and AZD628 give some paradoxical activation at subtherapeutic doses (0.01–0.1 µM). This paradoxical response to low doses of the type II drugs was also observed in studies by Yao et al,19 and may actually underlie the observed therapeutic window of these agents (Fig. 1) on BRAF mutant versus BRAF wild-type cells.
MLN2480 Is an Equipotent Inhibitor of Catalysis in BRAF Monomers and Dimers
To resolve the molecular mechanism(s) whereby MLN2480 (and AZD628) inhibits catalysis of BRAF monomers and dimers, we examined the effect of the drugs on BRAF dimerization, allosteric activation, and catalysis. We first observed, by co- immunoprecipitation experiments (Supplementary Fig. S8), that MLN2480 does not inhibit dimerization. Indeed, as shown by co-immunoprecipitation experiments, MLN2480 (and AZD628) actually promotes dimerization of epitope-tagged KIAA1549:BRAF subunits in a fashion similar to that observed previously for type I inhibitors.10,15
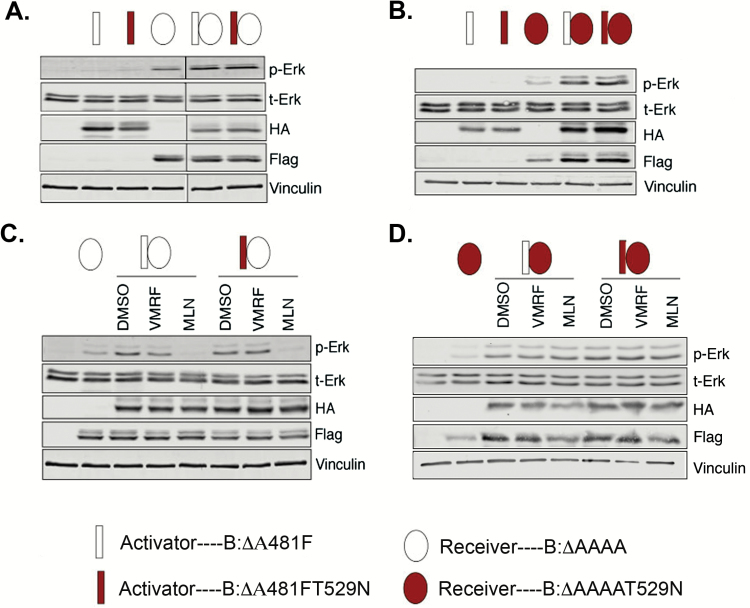
To address allosteric activation and catalysis, we transfected HEK293 cells with informative combinations of “activator” and “receiver” mutants of wild-type BRAF. As shown schematically in Fig. 5, the activator BRAF mutant is kinase-dead due to an A481F substitution in the catalytic domain but is still competent for transactivation of wild-type BRAF subunits in a dimeric configuration.35,36 The receiver BRAF mutant is catalytically active but incapable of activating a dimeric partner RAF due to a quadruple alanine substitution in the acidic domain (residues 446–449) N-terminal to the kinase domain.35 A variant of both the activator and inhibitor mutants was constructed with the gatekeeper mutation (T529N) so as to render the corresponding proteins incapable of interacting with RAF inhibitors (Fig. 2).
Fig. 5.
Type II RAF inhibitors block BRAF catalytic activity. BRAF cDNAs were genetically engineered to encode activators (rectangles) or receivers (ovals). The gatekeeper mutation (T529N) was introduced into either activator or receiver mutants to prevent drug binding (red rectangles or ovals). Both vemurafenib (VMRF) and MLN2480 (MLN) were used at 1 µM. (A) Optimum signaling requires receiver and activator present, as indicated by increased P-ERK. (B) Same as (A) but with gatekeeper mutation in the receiver subunit. (C) MLN2480 (MLN) but not vemurafenib (VMRF) can block signaling via BRAF dimers. The action of MLN2480 is unaffected by insertion of the gatekeeper mutation into the activator subunit. (D) Introduction of the gatekeeper mutation into the receiver subunit prevents the action of MLN2480.
Optimal RAF signaling requires coexpression of activator and receiver mutants (Fig. 5A, B). MLN2480 (and AZD628) blocks signaling if the receiver subunit is capable of binding the inhibitor, but does not block signaling if the receiver subunit has the gateway mutation. Conversely, both drugs inhibit signaling irrespective of whether or not the activator subunit carries the gatekeeper mutation (Fig. 6C, D). The type I RAF inhibitor vemurafenib cannot prevent signaling from any combination of these wild-type BRAF constructs. Collectively these results suggest that the antagonistic functions of MLN2480 and AZD628 on proliferation and signal transduction are exerted exclusively at the level of RAF catalysis.
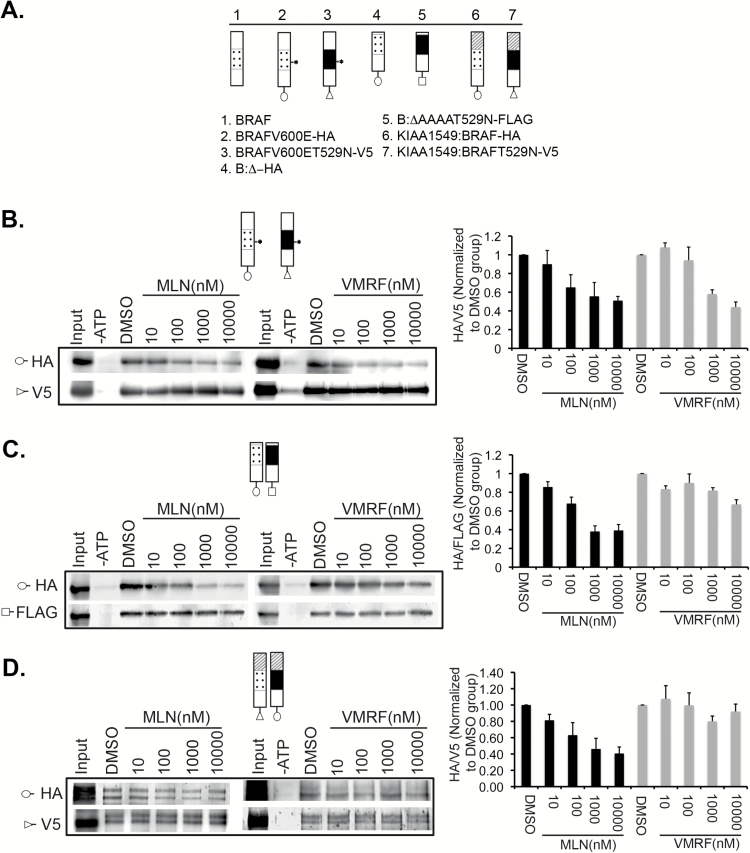
Fig. 6.
Differential interaction of type I and type II RAF inhibitors with BRAF monomers and dimers. (A) Indicated constructs were cotransfected into HEK293 cells. The gatekeeper mutation variants were used as internal controls, * denotes BRAFV600E. (B) Interaction of drugs with BRAF monomers (V600E). Representative immunoblots are shown to left and quantification of triplicate experiments is to right. (C, D) Data for the N-terminal truncation mutant and for the KIAA1549:BRAF, respectively, as in (B).
The physical interaction of MLN2480 with preformed BRAF monomers and dimers in vitro was assessed by a variant of the KiNativ assay wherein binding of RAF antagonists to a specific target is monitored by competition with biotinylated ATP and displayed by western blot.29 For this assay, in addition to the V600E (monomeric) and KIAA1549 (dimeric) BRAF oncoproteins, we also tested an amino terminal deletion mutant of BRAF wherein elimination of the autoinhibitory RAS binding domain leads to constitutive activity as a dimer35 (Fig. 6A). As indicated in Fig. 6B–D, MLN2480 binds with equal affinity to BRAF monomers and to preformed BRAF dimers, whereas vemurafenib binds only to BRAFV600E subunits. We conclude that the type II RAF inhibitor MLN2480 functions exclusively at the level of catalysis and that it circumvents paradoxical activation because it binds with equal affinity to monomeric and dimeric forms of BRAF.
Discussion
Overall, roughly 75% of all PLGAs express either the V600E or the KIAA1549 version of BRAF oncoprotein—generally on a mutually exclusive basis.6–9 Unfortunately, vemurafenib and dabrafenib, small molecule RAF inhibitors currently approved by the FDA for malignant melanoma, are ill-suited for treatment of PLGAs. Neither vemurafenib nor dabrafenib (both type I RAF inhibitors) is very active on BRAF oncoproteins, such as KIAA1549, that function as dimers. Moreover, both of these drugs can trigger paradoxical activation of signal transduction through wild-type RAF and/or BRAF oncoproteins that function as dimers.10,11,13–15,37 Finally, the blood–brain and blood–tumor barriers remain intact within many PLGAs and the brain penetrance of vemurafenib and dabrafenib is poor. Recent studies have highlighted a set of RAF inhibitors that resolve the first and second issues.17–20 In studies summarized here we identify another pair of RAF dimer-competent RAF inhibitors with comparable “paradox-breaking” properties. One of the drugs we tested, MLN2480, also addresses the third problem of brain and tumor penetrance and is active on patient-derived PLGA cells.
The proposed mechanism for paradox-breaking RAF antagonists described by Girotti et al17 was dual RAF and Src family inhibition. However, as shown in Fig. 2 and Supplementary Fig. S4, neither MLN2480 nor AZD628 would appear to target Src family kinases. Instead, our in vitro drug engagement assays (Fig. 6) support the paradox-breaking mechanism proposed by Yao et al for the RAF inhibitor BGB659.19 These investigators, using methodology quite different from our chemical proteomics experiments, found that BGB659, like MLN2480 and AZD628, circumvents paradoxical activation by binding with equal affinity to monomeric or dimeric BRAF subunits. The paradox-breaking, dimer-competent RAF inhibitors described here and by 3 of the 4 other groups17–19 are all type II RAF inhibitors, which target the DFG-out conformation of the kinase. However, the paradox-breaking drugs described by Zhang et al20 appear to be type I RAF inhibitors that circumvent paradoxical activation because, in marked contrast to vemurafenib, dabrafenib, and the type II drugs studied here, they do not induce formation of RAF dimers.
As potential therapeutic agents, one price that is paid for the broader action spectrum of dimer-competent RAF inhibitors is a reduction of the therapeutic window. The therapeutic window for type I RAF inhibitors that target the V600E BRAF oncoprotein appears to be a fringe benefit of negative cooperation at the ATP-binding site. Negative cooperation makes it difficult for type I RAF inhibitors to occupy both subunits of a wild-type RAF dimer. The V600E oncoprotein is active as a monomer that can be sufficiently occupied by type I drugs at concentrations below that which is needed to saturate both subunits of a RAF dimer.10,15 The type II inhibitors studied here and elsewhere are functional on dimeric BRAF oncoproteins and, in principle, should function equally well on wild-type RAF dimers. However, as shown in Fig. 1 and Supplementary Table S1, the half-maximal inhibitory concentration (IC50) for type II inhibitors on the V600E and KIAA1549 oncoproteins is actually ~10-fold lower than the IC50 for normal progenitor cells cultured in the presence of EGF and FGF.
The molecular mechanism underlying this therapeutic window is not completely clear to us at the present time. However, one plausible explanation is suggested by the studies by Yao et al on the type II inhibitor BGB659.19 These authors observed a similar ~10-fold differential in IC50 for BGB659 on BRAF oncoproteins relative to wild-type BRAF and speculated that the therapeutic window reflects opposing effects of BGB659 on catalysis (suppressed by drug) and dimerization (conditionally enhanced by drug). In their model, the sole effect of BGB659 in cells transformed by BRAF oncoproteins is suppression of catalysis because levels of active RAS (essential for dimerization) are suppressed by the RAF-mediated feedback inhibition of RAS. In contrast, in cells with moderate levels of active RAS (eg, wild-type cells exposed to growth factors or RAS-transformed cells), low doses of BGB659 initially stimulate signaling by promoting formation of RAF dimers wherein one dimer subunit is unoccupied by drug and available for catalysis. These partially dosed dimers account for low dose, drug-induced activation as well as the somewhat reduced efficacy in wild-type cells. Our own data on the dimerization (Supplementary Fig. S8) and catalytic responses to MLN2480 and AZD628 (Fig. 5) are in accord with this model. Note that neither of the type II inhibitors tested here seem to target ARAF, one of the 3 RAF family members that might be involved in physiologically relevant RAF signaling in healthy cells (Fig. 2D and Supplementary Fig. S4).
Irrespective of molecular mechanism, it is possible that a 10-fold therapeutic window will prove useful in clinical practice. Our data suggest that MLN2480 might have therapeutic benefit on children and young adults with BRAF mutant low-grade astrocytomas. Pediatric diffuse leptomeningeal glioneuronal tumors also often harbor BRAF mutations. Therefore, further testing should be done to determine if this compound also penetrates the cerebrospinal fluid and could therefore be used to target these rare but infiltrative tumors.38 Finally, the brain penetrance of MLN2480 suggests that it might have therapeutic benefit not only on pediatric astrocytomas, but also on common adult RAF mutant tumors that metastasize to brain.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This work was supported by A Kid’s Brain Tumor Cure Foundation (C.D.S), National Cancer Institute SPORE Grant P50CA165962 (C.D.S.), the US National Institutes of Health (NIH) (P01CA142536) (R.A.S.), the US NIH Director’s New Innovator Award (1DP2OD007383-01), and the Daniel E. Ponton Fund for the Neurosciences (N.Y.A). Additional support was provided by the Stop & Shop Pediatric Brain Tumor Program and the Andryziak Low-Grade Glioma Research Fund (M.W.K.).
Conflict of interest statement. In compliance with Harvard Medical School and Partners Healthcare guidelines on potential conflict of interest, we disclose that N.Y.A. is a scientific advisor to BayesianDx and inviCRO.
Supplementary Material
References
- 1. Holderfield M, Deuker MM, McCormick F, et al. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014; 14(7):455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cin H, Meyer C, Herr R, et al. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol. 2011; 121(6):763–774. [DOI] [PubMed] [Google Scholar]
- 3. Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008; 68(21):8673–8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sievert AJ, Jackson EM, Gai X, et al. Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single-nucleotide polymorphism-based genotype arrays results in a novel BRAF fusion gene. Brain Pathol. 2009; 19(3):449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lito P, Pratilas CA, Joseph EW, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell. 2012; 22(5):668–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009; 4(11):e7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011; 121(3):397–405. [DOI] [PubMed] [Google Scholar]
- 8. Zhang J, Wu G, Miller CP, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013; 45(6):602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schiffman JD, Hodgson JG, VandenBerg SR, et al. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010; 70(2):512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poulikakos PI, Zhang C, Bollag G, et al. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010; 464(7287):427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sievert AJ, Lang SS, Boucher KL, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci U S A. 2013; 110(15):5957–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pollack IF, Jakacki RI. Childhood brain tumors: epidemiology, current management and future directions. Nat Rev Neurol. 2011; 7(9):495–506. [DOI] [PubMed] [Google Scholar]
- 13. Holderfield M, Merritt H, Chan J, et al. RAF inhibitors activate the MAPK pathway by relieving inhibitory autophosphorylation. Cancer Cell. 2013; 23(5):594–602. [DOI] [PubMed] [Google Scholar]
- 14. Heidorn SJ, Milagre C, Whittaker S, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010; 140(2):209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hatzivassiliou G, Song K, Yen I, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010; 464(7287):431–435. [DOI] [PubMed] [Google Scholar]
- 16. Zhao Z, Wu H, Wang L, et al. Exploration of type II binding mode: a privileged approach for kinase inhibitor focused drug discovery? ACS Chem Biol. 2014; 9(6):1230–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Girotti MR, Lopes F, Preece N, et al. Paradox-breaking RAF inhibitors that also target SRC are effective in drug-resistant BRAF mutant melanoma. Cancer Cell. 2015; 27(1):85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng SB, Henry JR, Kaufman MD, et al. Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell. 2015; 28(3):384–398. [DOI] [PubMed] [Google Scholar]
- 19. Yao Z, Torres NM, Tao A, et al. BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell. 2015; 28(3):370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang C, Spevak W, Zhang Y, et al. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature. 2015; 526(7574):583–586. [DOI] [PubMed] [Google Scholar]
- 21. Mehta S, Huillard E, Kesari S, et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011; 19(3):359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun Y, Meijer DH, Alberta JA, et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011; 69(5):906–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chadwick EJ, Yang DP, Filbin MG, et al. A brain tumor/organotypic slice co-culture system for studying tumor microenvironment and targeted drug therapies. J Vis Exp. 2015; 105:e53304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ligon KL, Huillard E, Mehta S, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007; 53(4):503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jonas O, Calligaris D, Methuku KR, et al. First in vivo testing of compounds targeting group 3 medulloblastomas using an implantable microdevice as a new paradigm for drug development. J Biomed Nanotechnol. 2016; 12(6):1297–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Felix CA, Slavc I, Dunn M, et al. p53 gene mutations in pediatric brain tumors. Med Pediatr Oncol. 1995; 25(6):431–436. [DOI] [PubMed] [Google Scholar]
- 27. Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov. 2014; 13(12):928–942. [DOI] [PubMed] [Google Scholar]
- 28. Khazak V, Astsaturov I, Serebriiskii IG, et al. Selective Raf inhibition in cancer therapy. Expert Opin Ther Targets. 2007; 11(12):1587–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patricelli MP, Szardenings AK, Liyanage M, et al. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007; 46(2):350–358. [DOI] [PubMed] [Google Scholar]
- 30. Whittaker S, Kirk R, Hayward R, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci Transl Med. 2010; 2(35):35ra41. [DOI] [PubMed] [Google Scholar]
- 31. Liu X, Ide JL, Norton I, et al. Molecular imaging of drug transit through the blood–brain barrier with MALDI mass spectrometry imaging. Sci Rep. 2013; 3:2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ligon KL, Alberta JA, Kho AT, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004; 63(5):499–509. [DOI] [PubMed] [Google Scholar]
- 33. Otero JJ, Rowitch D, Vandenberg S. OLIG2 is differentially expressed in pediatric astrocytic and in ependymal neoplasms. J Neurooncol. 2011; 104(2):423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tanaka Y, Sasaki A, Ishiuchi S, et al. Diversity of glial cell components in pilocytic astrocytoma. Neuropathology. 2008; 28(4):399–407. [DOI] [PubMed] [Google Scholar]
- 35. Hu J, Stites EC, Yu H, et al. Allosteric activation of functionally asymmetric RAF kinase dimers. Cell. 2013; 154(5):1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu J, Yu H, Kornev AP, et al. Mutation that blocks ATP binding creates a pseudokinase stabilizing the scaffolding function of kinase suppressor of Ras, CRAF and BRAF. Proc Natl Acad Sci U S A. 2011; 108(15):6067–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013; 19(11):1401–1409. [DOI] [PubMed] [Google Scholar]
- 38. Dodgshun AJ, SantaCruz N, Hwang J, et al. Disseminated glioneuronal tumors occurring in childhood: treatment outcomes and BRAF alterations including V600E mutation. J Neurooncol. 2016; 128(2):293–302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.