Abstract
Background and Objective:
The human cytomegalovirus (HCMV) can persist lifelong as a latent infection and may result in a series of disorders. Associations with both Hodgkin’s disease and non-Hodgkin´s lymphomas have been reported. Expression of the unique long (UL)138 gene of HCMV is linked with the viral latency phase while that of the immediate-early (IE)1 gene is typical of the viral replication phase in patients. This study conducted to determine the prevalence of CMV latent infection in histological tissue samples from patients with Hodgkin’s and Non-Hodgkin´s lymphomas.
Material and Methods:
A cross sectional study was carried out with a total of 50 paraffin embedded tissues blocks, including 25 samples for Hodgkin’s disease and 25 samples for non-Hodgkin´s lymphomas. After RNA extraction and cDNA preparation, detection of IE1 mRNA was conducted by RT-PCR and identification of mRNA UL138 was achieved by nested PCR.
Results:
Among 25 cases of Non-Hodgkin´s lymphoma, 5 (20%) were positive for UL 138 and 1 (4%) for both IE1 and UL 138. Among 25 cases of Hodgkin only 1 (4%) was positive for UL 138 and all were negative for IE1
Conclusion:
A relatively high 20% rate of expression of UL 138 was detected in patients with non-Hodgkin´s lymphoma, so that latent CMV infection may play a role in development of the disease.
Keywords: Human Cytomegalovirus, nested PCR, Hodgkin disease, Non-Hodgkin lymphoma
Introduction
HCMV usually results in asymptomatic primary infection, and may persist lifelong in humans, and reactivates sporadically in phases of immunosuppression. In immunosuppressed patients, e.g., transplant recipients and AIDS patients, cytomegalovirus frequently causes severe diseases (do Carmo et al., 2014). It is estimated about 30-90 % of the Western countries and more than 90 % of the developing countries’ population are infected with cytomegalovirus (Crough et at., 2009: Lanzieri., 2014). The virus is transmitted via saliva, urine and breast milk or organ transplantation (Swanson et al., 2013). The incubation period is about 4–8 weeks (Schottstedt et al., 2010). Despite the infection of HCMV is commonly asymptomatic, symptoms similar to those of mononucleosis, namely fever, lymph node swelling, gastritis, esophagitis, and flu-like symptoms, are only rarely observed (Crough et at., 2009; Eskandar et al., 2015). HCMV belongs to genus Betaherpesvirinae of the family herpesviridae, HCMV genome is a linear, double-stranded DNA with 230 kb which encodes 200 gene products (Fasanya et al., 2016; Nakase et al., 2016; Petrucelli et al., 2012). HCMV is not known as an oncogenic virus, but observations demonstrate that HCMV in the latency phase was corelated with certain malignancy such as gastric canser and T-cell lymphoma (Ponticelli et al., 2011; Jin et al., 2014; Ballanger et al., 2009). Some observations have exhibited that HCMV may be associated with Hodgkin and Non-Hodgkin’s lymphomas (Irsai et al., 2012; Tafvizi et al., 2014; Kadry et al., 2014).
Hodgkin and Non-Hodgkin’s lymphomas are two malignant lymph nodes (Habibian et al., 2013). Hodgkin Disease is characterized by a small number of putative malignant mononuclear Hodgkin and multinucleated Reed-Sternberg cells among various cells, such as plasma cells, lymphocytes, eosinophils and neotrophils (Randa et al., 2009). Non-Hodgkin’s lymphoma includes some malignancies such as lymphoproliferative disorder, Burkitt’s and NK-T lymphoma (Bagirath et al., 1014).
After primary infection, HCMV will be entered in the latency phase, which in this phase its genome will become in the form of episome (Goodrum., 2007). There are some factors that implicate in the latency of HCMV. One of these factors is UL138. It is one of the few mRNA genes which express during the latency phase. Protein UL138 encoded by a polycistronic locus spanning UL133-UL138 within the ULb´ region, which contains genes involved in regulating latency, viral immune escape and cell tropism (Petrucelli et al., 2012 ;Goodrum., 2007; Montag et al., 2011). HCMV replication is regulated by subsets of genes, including immediate early genes (IEI), which has an important role in reactivation of latent state (Harwardt., 2016). The detection of UL138 indicates the latent HCMV infection and the detection of IE1 reveals the reactivation of latent CMV infection. The prevalence of Hodgkin and non-Hodgkin´s lymphoma have been reported in Iran (Hashemi-Bahremani., 2007). At present diagnosis of HCMV rely on HCMV IgG and IgM which does not describe the exact status of HCMVfection in in patients. On the other hand the perfomance of real-time PCR and PP65 tests show only the activation or reactivation of HCMV which are costly and not offerable by all patients. Thus this study was conducted to determine IE1mRNA by RT-PCR as a marker for replication or reactivation of HCMV and UL138 mRNA as a marker for latency of HCMV status. To prevent the outcome of HCMV infection in immunosuppression patients and improve treatment, the application of both tets should be implemented in patients with Hodgkin and non- Hodgkin´s lymphoma before treatment and during chemotherapy.
The prevalence of HCMV in patients with Hodgkin and non- Hodgkin´s lymphoma have not been evaluated in Iran, thus this study was conducted to determine the prevalence of HCMV in latency and reactivation phases in the patients with Hodgkin and non–Hodgkin´s lymphoma.
Materials and Methods
Patients and Specimens
The cross sectional study was carried out for the total of 50 paraffin embedded tissues blocks, including 25 (50%) samples for Hodgkin disease and 25 (50%) samples for non-Hodgkin´s lymphoma. All blocks of lymph node were collected from the department of pathology of Imam Khomeini and Shafa hospitals of the Ahvaz city during 2001-2011. The diagnosis of Hodgkin and Non-Hodgkin´s lymphoma were confirmed by the pathologist.
The proposal of this project was approved by the Ethics Committee of Ahvaz Jundishapur University of Medical Sciences.
Deparaffinization and RNA extraction
Initially sections of 5μm were prepared from each paraffin embedded block, after deparaffinazation of the tissues, RNA extraction were performed using High pure RNA paraffin kit (Roshe, Germany, cat. no. 3270289) according to the manufacturer´s instruction. Then complementary DNA (cDNA) was carried out for each extract.
Nested PCR for UL 138
The Nested PCR was preformed for the each sample in two rounds, in the first round, 25 µl of PCR mixing reaction containing 7μl of extracting mRNA, 2.5μl PCR buffer 10X (Roche), 0.5 μl dNTP 10mM (Roche), 1U Taq polymerase (Roche), 20 pmol of each primer sequence(table 1), distilled water up to 25µl, was subjected to Thermal Cycler (Peglab, Germany) and programmed in one cycle 94°C for 5 min, then 40 cycles; 94°C for 1 min, 52°C for one min, 72°C for 1 min and final extension: 72°C for 10 min. For the second round, 5 µl of PCR product of the first run was used in the PCR mixing reaction with the same component described in the first round, was subjected to Thermal Cycler and programmed; One cycle 94C for 5 min, then 35 cycle ; 94°C for 45s, 60° C for 45s, 72°C for 45s and final extension 72°C for 10 min. The primers and amplification conditions shows in Table 1. The final PCR products were subjected to electrophoresis in 2% agarose gel, stained with safe stain and visualized under ultraviolet transilluminator.
Table 1.
Primers and Amplification Conditions Used for Detection of UL138 and IE1
| Name | Primers (5 to 3) | PCR conditions | ||||
|---|---|---|---|---|---|---|
| Forward | Reverse | Annealing Temprature C | Annealing | Cycles n | Size bp | |
| UI138 | ATGGACGATCTGCCGCTGAA1 | TCACGTGTATTCTTGATGAT1 | 52 | 60 sec | 35 | 510 |
| IE1 | GCTTACCACTGGCACGACACCT | TACTCCCCGTACAGCTCGCAAC | 69 | 60 sec | 40 | 89 |
| AGCCTTCCCTAAGACCACCAAT | CATAGCAGCACAGCACCCGACA | 57 | 60 sec | 40 | 260 | |
PCR for IE1
The one step PCR was carried out for detection of IEI. The PCR reaction mixture of 25 µl containing 7μl of cDNA mRNA, 2.5μl PCR buffer 10X(Roche), 0.5 μl dNTP 10mM(Roche), 1 U Taq polymerase (Roche), 20 pmol of each primer sequence (table 1), distilled water up to 25 µl, was subjected to Thermal Cycler (Peglab, Germany). The thermal program was followed: 1 cycle, 94°C for 5 min, then 40 cycles :94°C for 1 min, 57°C for 1 min, 72°C for 1 min and final extension: 72°C for 10 min. The primers and amplification conditions are shown in Table 1. The final PCR products were subjected to electrophoresis in 2% agarose gel, stained with safe stain and visualized under ultraviolet transilluminator.
Results
Table 2 shows the profile of patients with Hodgkin and Non-Hodgkin lymphoma.
Table 2.
Profile of Patients with Hodgkin and Non-Hodgkin
| Category | Male | Female | Total |
|---|---|---|---|
| Hodgkin Lymphoma n = 25 | |||
| Mixed Cellularity (MC) | 4 (16%) | 8 (32%) | 12 (48%) |
| Lymphocyte Predominant (LP) | 3 (12%) | 2 (8%) | 5 (20%) |
| Nodular Sclerosis (NS) | 4 (16%) | 4 (16%) | 8 (32%) |
| Non-Hodgkin Lymphoma n= 25 | |||
| Diffuse Large Cell Type | 4 (16%) | 10 (40%) | 14 (56%) |
| Small Lymphocytic Lymphoma | 1 (4%) | 1 (4%) | |
| High Grade Immunoblastic Lymphoma | 1 (4%) | 1 (4%) | |
| Mixed Small and Large | 2 (8%) | 2 (8%) | |
| Malignant T Cell | 2 (8%) | 2 (8%) | 4 (16%) |
| Malignant B Cell | 1 (4%) | 1 (4%) | 2 (8%) |
| Diffuse Mixed Large Cell | 1 (4%) | 1 (4%) |
Table 3 shows the results of PCR for UL 138 and IE1 in Hodgkin and Non-Hodgkin groups.
Table 3.
Detection of UL138, I E1 in Subsets of Hodgkin and Non-Hodgkins Lymphoma
| Characteristic | Hodgkin (n=25) | Non-Hodgkin (n=25) | |||||
|---|---|---|---|---|---|---|---|
| UL138 | IE1 | Total | UL138 | IE1 | Total | ||
| Gender | Female | - | - | 14 | 3(12%),2DLBL*1MTC* | 1(4%)DLBL* | 11 |
| Male | 1(4%) | - | 11 | 2(8%), 1DLBL*1 MTC* | - | 14 | |
| Age group | <16 | - | - | 4 | - | - | 7 |
| >16 | 1(4%) | - | 21 | 5(20%) | 1(4%) | 18 | |
NS*, Nodular Sclerosis; DLBL*, Diffuse Large B-cell Lymphoma; MTC*
Figure 1 shows the results of nested PCR amplification of UL138 Gene.
Figure 1.
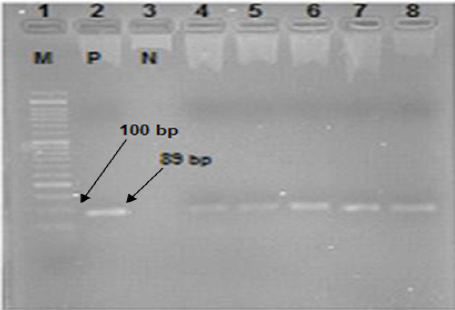
Results of Nested PCR Amplification of UL138 (Positive Band 89 bp) Lane 1:50-bp Size Marker, Lane 2 :Positive Control, Lane 3:Negative Control. Lane 4-8:Non-Hodgkin Samples Positive for UL138.
Discussion
Although the association of HCMV and cancer is controversial, but there are evidences revealed that progression of some tumors could be intensified by the regulatory protein named US28 protein encoded by HCMV during the phase of latent infection. HCMV protein, US28 in endogenously infected colorectal cancer (CRC) may constitute a novel antitumor approach (Cai et al.,2016). The another protein, which is encoded by the HCMV named UL138 found to have an important role in the establishment of the latency of the virus in some certain cells (Montag et al., 2011).
In our study the UL138 gene was detected in the five patients with Non-Hodgkin´s lymphoma, including two of 25 males and three of 25 females. Only one (4%) female patient with Non-Hodgkin´s lymphoma was found to be positive for IE1, while, in Hodgkin group only one (4%) male showed positive for UL 138 but no IE1 was detected.
The proteins pp65, IE1, US 28 are produced by HCMV and have been involved in numerous significant events in tumor progression (Ponticelli et al., 2011; Cai., 2016; Lucas., 2011). Pp65 is the most abundant virion protein and non-infectious viral particles that are assembled during active infection (Libard et al., 2014). Besides, the detection of IE1mRNA indicates the presence of active or reactivation of HCMV infection (Harwardt., 2016). High rate of 76% HCMV pp65 have been detected in biopsies of 29 patients with diffuse large B-cell lymphoma of the central nervous system (Libard., 2014) (CNS DLBCL). Subsequently high rate of 57%, 49% and 44% of HCMV pp65 Antigenemia have been detected in patients with Follicular and aggressive Non-Hodgkin lymphoma and with Hodgkin lymphoma respectively (Jacopo., 2014). While in our survey only one patient with subtype, diffuse large B-cell lymphoma was shown to be positive for IE1. It was exhibited the detection of HCMV UL138 gene is highly conserved in clinical strains and latency of HCMV infection (Grainger et al., 2010). Jinji et al., have detected UL138 in 11 of 58 (18.97%) of the patients with gastric cancer (Jinji et al., 2014). In our study, 5/25 (20%) of patients with Non-Hodgkin lymphoma showed positive for UL138, which is in accordance with Jinji et al finding (Jinji et al., 2014). In addittion, the other latent human cytomegalovirus proteins, US28, UL133, UL135, UL136 have been investigated in various types of cancer tissues (Cai., 2016; Jinji et al., 2014).
The achivement of both UL138 and IE1 detection are cost effective and easily can be performed at any common laboratory. Diagnosis of HCMV based on HCMV IgG does not clarify the exact status of HCMV infection. Although the presence of HCMV IgG seropositivity could be described as independent predictor of clinically relevant CMV infection (Drew et al., 2007). In our study the diagnosis of the HCMV activation replication was based on detection of IE1 mRNA by RT-PCR which is a qualitive test but to achive highly sensitivity performance it requires quantification realtime – PCR assay. While the diagnosis of the latent HCMV was based on detection of UL138 mRNA by Nested-PCR which is also a qualitive test but to achive higly sensitivity test it requires to fullfil quantification realtime – PCR assay. With the aforementioned data to improve treatment and management of HCMV infection, patients with Non-Hodgkin lymphoma should be screened for both UL138 rnRNA and IE1 mRNA before and during chemotherapy.
The association of other viruses, including the Epstein Barr virus (EBV), Hepatitis C virus (HCV), Human immunodeficiency virus (HIV) and hepatitis B (HBV) among the patients with non-Hodgkin´s lymphoma have been reported (Economides et al., 2016; Hoffmann et al., 2016; Engels et al., 2010). The evaluation of the mentioned viruses have not been studied in the present study but it requires to be evaluated in the further studies.
Conclusion, UL138 mRNA as a predictor of HCMV latency and IE1mRNA as a predictor of activation or reactivation of HCMV have been detected in patients with Hodgkin and Non-Hodgkin lymphoma. Both detection of UL138 mRNA and IE1 mRNA were carried out by PCR which are qualitive tets but to achive highly sensitivity performance it requirs to fullfil quantification realtime –PCR assay. High rate of 20% expression of UL 138 mRNA have been detected in the patients with non-Hodgkin´s lymphoma subtype T cell lymphoma, which indicates UL138 mRNA may play a possible role in development of Non-Hodgkin´s lymphoma, however it requires further investigation.
However the evaluation of other CMV latent genes or proteins, including US28, UL133, UL135, UL136 should be investigated in cancerous patients. To improve treatment and to prevent concequences of HCMV infection, the screening of the both UL138 mRNA and IE1mRNA of HCMV should be implemented for patients with Hodgkin disease and Non-Hodgkin lymphoma before treatment and during chemotherapy.
Acknowledgements
This study was done as a research project with 92,146 registration number in Health Research Institute, Infectious and Tropical Disease Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
Authors’ Contributions
Study concept and design: Manoochehr Makvandi, Alireza Samarbafzadeh; acquisitionof data: Hamide Mehravaran; analysis and interpretation of data: Manoochehr Makvandi and Hamide Mehravaran; drafting of the manuscript: Hamide Mehravaran, HadisKiani; critical revision of the manuscript for portant intellectual content: Manoochehr Makvandi; statisticalanalysis: Manoochehr Makvandiand, Hamide Mehravaran; Administrative, technical, and material support: Hamide Mehravaran, Niloofar Neisi, Seyedeh Zeinab Hoseini, Hashem Radmehr, Toran Shahani; Study supervision: Manoochehr Makvandi and Alireza Samarbafzadeh.
Funding/Support
This study was supported by Health Research Institute, infectious and Tropical disease Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
Financial Disclosures
The authors have no financial interest related to the material in the manuscript.
References
- Bagirath PV, Kumar JV, Arvind UD, et al. Aggressive extranodal peripheral T-cell non-Hodgkin’s lymphoma: A rare case report and review. J Oral Maxillofac Surg Med Pathol. 2014;18:80. doi: 10.4103/0973-029X.131918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballanger F, Bressollette C, Volteau C, et al. Cytomegalovirus: its potential role in the development of cutaneous T-cell lymphoma. Exp Dermatol. 2009;18:574–6. doi: 10.1111/j.1600-0625.2008.00817.x. [DOI] [PubMed] [Google Scholar]
- Cai ZZ, Xu JG, Zhou YH, et al. Human cytomegalovirus-encoded US28 may act as a tumor promoter in colorectal cancer. World J Gastroenterol. 2016;22:2789–98. doi: 10.3748/wjg.v22.i9.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crough T, Khanna R. Immunobiology of human cytomegalovirus: from bench to bedside. Clin Microbiol Rev. 2009;22:76–98. doi: 10.1128/CMR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Carmo AM, Santos FM, Ortiz-Agostinho CL, et al. Cytomegalovirus infection in inflammatory bowel disease is not associated with worsening of intestinal inflammatory activity. PLoS One. 2014;9:e111574. doi: 10.1371/journal.pone.0111574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew WL. Laboratory diagnosis of cytomegalovirus infection and disease in immunocompromised patients. Curr Opin Infect Dis. 2007;20:408–11. doi: 10.1097/QCO.0b013e32821f6010. [DOI] [PubMed] [Google Scholar]
- Economides MP, Mahale P, Turturro F, et al. Development of non-Hodgkin lymphoma as a second primary cancer in hepatitis C virus-infected patients with a different primary malignancy. Leuk Lymphoma. 2016;27:1–4. doi: 10.1080/10428194.2016.1196817. [DOI] [PubMed] [Google Scholar]
- Eskandar O, Smith G. Primary cytomegalovirus infection presenting with itching and obstetric cholestasis like picture in mid-trimester. Case Rep Obstet Gynecol. 2015;1:5–7. [Google Scholar]
- Engels EA, Cho ER, Jee SH. Hepatitis B virus infection and risk of non-Hodgkin lymphoma in South Korea: a cohort study. Lancet Oncol. 2010;11:827–34. doi: 10.1016/S1470-2045(10)70167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanya AA, Pedersen FT, Alhassan S, et al. Cytomegalovirus cutaneous infection in an immunocompromised patient. Cureus. 2016;8:e598. doi: 10.7759/cureus.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrum F, Reeves M, Sinclair J, et al. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood. 2007;110:937–45. doi: 10.1182/blood-2007-01-070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger L, Cicchini L, Rak M, et al. Stress-inducible alternative translation initiation of human cytomegalovirus latency protein pUL138. J Virol. 2010;84:9472–86. doi: 10.1128/JVI.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibian A, Makvandi M, Samarbafzadeh A, et al. Epstein-Barr Virus DNA frequency in paraffin embedded tissues of Non-Hodgkin lymphoma patients from Ahvaz, Iran. Jundishapur J Health Res. 2013;4:315–20. [Google Scholar]
- Harwardt T, Lukas S, Zenger M, et al. Human Cytomegalovirus Immediate-Early 1 Protein Rewires Upstream STAT3 to Downstream STAT1 Signaling Switching an IL6-Type to an IFNγ-Like Response. PLoS Pathog. 2016;12:e1005748. doi: 10.1371/journal.ppat.1005748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi-Bahremani M, Parwaresch MR, Tabrizchi H, et al. Lymphomas in Iran. Arch Iran Med. 2007;10:343–8. [PubMed] [Google Scholar]
- Hoffmann C, Schommers P, Wolf E, et al. CD4+and CD8+T-cell kinetics in aviremic HIV-infected patients developing Hodgkin or non-Hodgkin lymphoma. AIDS. 2016;13:753–60. doi: 10.1097/QAD.0000000000000980. [DOI] [PubMed] [Google Scholar]
- Irsai G, Tampu-Kiss T, Dezső B, et al. Complications of systemic cytomegalovirus infection in therapy-resistant Hodgkin’s lymphoma. Orv Hetil. 2012;13:751–5. doi: 10.1556/OH.2012.29364. [DOI] [PubMed] [Google Scholar]
- Jacopo M, Francesco M, Francesco S, et al. Impact of cytomegalovirus replication and cytomegalovirus serostatus on the outcome of patients with B Cell lymphoma after allogeneic stem cell transplantation. Biol Blood Marrow Transplant. 2014;20:885–90. doi: 10.1016/j.bbmt.2014.02.015. [DOI] [PubMed] [Google Scholar]
- Jin J, Hu C, Wang P, et al. Latent infection of human cytomegalovirus is associated with the development of gastric cancer. Oncol lett. 2014;4:898–904. doi: 10.3892/ol.2014.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Hu C, Wang P, et al. Latent infection of human cytomegalovirus is associated with the development of gastric cancer. Oncol lett. 2014;8:898–904. doi: 10.3892/ol.2014.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadry DY, Khorshed AM, Rashed RA, et al. Association of viral infections with risk of human lymphomas, Egypt. Asian Pac J Cancer Prev. 2016;17:1705–12. doi: 10.7314/apjcp.2016.17.4.1705. [DOI] [PubMed] [Google Scholar]
- Lanzieri TM, Dollard SC, Bialek SR, et al. Systematic review of the birth prevalence of congenital cytomegalovirus infection in developing countries. Int J Infect Dis. 2014;22:44–8. doi: 10.1016/j.ijid.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas KG, Bao L, Bruggeman R, et al. The detection of CMV pp65 and IE1 in glioblastoma multiforme. J Neurooncol. 2011;103:231–8. doi: 10.1007/s11060-010-0383-6. [DOI] [PubMed] [Google Scholar]
- Libard S, Popova SN, Amini RM, et al. Human cytomegalovirus tegument protein pp65 is detected in all intra- and extra- axial brain tumours independent of the tumour type or grade. PLoS One. 2014;9:e108861. doi: 10.1371/journal.pone.0108861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag C, Wagner JA, Gruska I, et al. The Latency-Associated UL138 Gene Product of Human Cytomegalovirus Sensitizes Cells to Tumor Necrosis Factor Alpha (TNF-α) Signaling by Upregulating TNF-αReceptor 1 Cell Surface Expression. J Virol. 2011;85:11409–21. doi: 10.1128/JVI.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase H, Herfarth H. Cytomegalovirus colitis, cytomegalovirus hepatitis and systemic cytomegalovirus infection: common features and differences. Inflamm Intest Dis. 2016;1:15–23. doi: 10.1159/000443198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli C. Herpes viruses and tumours in kidney transplant recipients. The role of immunosuppression. Nephrol Dial Transplant. 2011;26:1769–75. doi: 10.1093/ndt/gfr157. [DOI] [PubMed] [Google Scholar]
- Petrucelli A, Umashankar M, Zagallo P, et al. Interactions between proteins encoded within the human cytomegalovirus UL133-UL138 locus. J Virol. 2012;86:8653–62. doi: 10.1128/JVI.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randa El-Zein, Claudia M. Monroy, Carol J. Etzel, et al. Genetic Polymorphisms in DNA Repair Genes as Modulators of Hodgkin Disease Risk. Cancer. 2009;115:1651–59. doi: 10.1002/cncr.24205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottstedt V, Blümel J, Burger R, et al. Human cytomegalovirus (HCMV)–revised. Transfus Med Hemother. 2010;37:365–75. doi: 10.1159/000322141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson EC, Schleiss MR. Congenital cytomegalovirus infection: new prospects for prevention and therapy. Pediatr Clin North Am. 2013;60:335–49. doi: 10.1016/j.pcl.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafvizi F, Fard ZT. Detection of human cytomegalovirus in patients with colorectal cancer by nested-PCR. Asian Pac J Cancer Prev. 2014;15:1453–7. doi: 10.7314/apjcp.2014.15.3.1453. [DOI] [PubMed] [Google Scholar]


