Abstract
MicroRNAs (miRNAs), are a type of small non-coding RNAs, that induce mRNA degradation or repress translation by binding to the 3′-untranslated region (UTR) of its target mRNA. Some specific miRNAs, e.g. miR-93, have been discovered to be involved in pathological procedures by targeting some oncogenes or tumor suppressors in glioma. In the present study, real-time RT-PCR data was indicated the expression pattern and prognostic value of miR-93 in patients with types of Glioma. MiR-93 expression was significantly decreased in tumor tissue compared with normal group brain tissues (P<0.001). Low miR-93 expression was significantly correlated with progressive tumor grade (P=0.02). Moreover, multivariate analysis showed that miR-93 decreased expression (HR, 4.3; 95% CI, 0.8–17.2, P=0.02), advanced tumor grade (HR, 3.1; 95% CI, 0.2–13.9, P=0.04), for integrinβ8, level expression was inverse. Our data was shown that the down regulation of miR-93 was significantly correlated with unfavorable pathological features in patients with Glioma. Suggesting that decreased expression of miR-93can be used as a novel prognostic factor for this disease.
Keywords: miRNA93, integrinβ8, Glioblastoma multiform, Real time PCR
Introduction
Glioma is about 80% of malignant tumors in brain and the highest type of cancer in central nervous system. Studying on glioma shows that rapid growth and strong invasiveness, with high mortality and recurrence rates after surgical resection. Therapies including surgical resection, chemotherapy and radiotherapy, the prognosis of Glioblastoma patients still remain poor, with a median survival time of only 12 months. Therefore, it is urgent to explore the molecular mechanism involved in glioma for the development of effective therapeutic strategies (Chen et al., 2016). MicroRNAs (miRNAs) are non-coding small RNAs that contain approximately 19–25 nucleotides. They are transcribed from genomic DNAs to generate long primary transcripts, followed by modification by the RNase III-type enzymes Drosha and Dicer to produce pre-miRNAs and mature miRNAs (Mollaie et al., 2013). Mature miRNAs post-transcriptionally regulate the expression of target genes by directly binding to their 3′untranslated regions (3′-UTR) (Nesler et al., 2013). As a new class of regulatory molecules, miRNAs essential functions in many physiological and pathological processes, particularly carcinogenesis, and of these miRNAs, several function as oncogenic miRNAs, whereas others are tumor suppressors(Pogue et al., 2014; Galka-Marciniak et al., 2016; Jain and Das, 2016).
The advent of global genomic profiling techniques has enabled high throughput assessment of microRNA expression patterns in brain tumors (Li et al., 2014; Galka-Marciniak et al., 2016). Utilizing the advantages of this high throughput process, Ciafre et al. and Chan et al. were the first to document global microRNA expression profiles in glioblastoma human tissue samples(Chen et al., 2013a; Li et al., 2014; Chen et al., 2016; Galka-Marciniak et al., 2016). Ciafre et al. utilized microarray-based technology to profile the expression of 245 microRNAs in glioblastoma tissue samples(Mannino et al., 2008). Moreover, differentially expressed miRNAs in Glioblastoma multiform (GBM) have been screened, and several miRNAs were identified as specific biomarkers to evaluate lymph node metastasis and prognosis in patients with GBM (Shi et al., 2015; Lin et al., 2016). MiRNA-93-5p (miR-93) is from the miR-106b-25 cluster, a paralogue cluster of the miR-17-92 cluster. Located in intron 13 of the host gene MCM7 at chromosome 7q22, the miR106b-25 cluster consists of the highly conserved miR-106b, miR-93 and miR-25 (Chaulk et al., 2011; Sathyapalan et al., 2015). MiR-93 is differentially expressed in a variety of cancers, including cancers in the lung, Braine, stomach, colon, liver and breast. Differential expression of miR-93 and its correlation with metastasis have also been confirmed in primary osteosarcoma cells (Chen et al., 2013b). Another study found that over expression of miR-93 affects the growth and angiogenesis of human Glioblastoma cells. Currently, an increasing number of miR-93 target genes have been identified, such as P53 suggesting miR-93 may differentially affect the behaviors of tumors (Barry et al., 2015). Gliomas represent a heterogeneous group of malignancies of the central nervous system arising from glial cells. Glioblastoma multiform (GBM) is the most aggressive primary brain tumor with a median survival of approximately one year, and over 95% of patients surviving less than two years(Zadran et al., 2014; Abdel-Rahman and Fouad, 2015). GBM is the most aggressive of the Gliomas which are divided into four grades; unluckily, the most aggressive of these, grade 4 or Glioblastoma multiform (GBM), is also the most common in humans (Sturm et al., 2014). The clinical entity is classified based upon cell type, grade and location within the central nervous system. Glioma are named as a result of the suspected glial cell of origin and further characterized by the World Health Organization (WHO) grading system ranging from low grade (WHO grade I, II) to high grade (WHO grade III–IV)(Lynch et al., 2013). Glioma account for 80% of human malignant primary brain tumors. The most frequently diagnosed adult Glioma is GBM (WHO grade IV) (Binder et al., 2016). GBM remains the most common malignant adult primary brain tumor, with more than 17,000 cases diagnosed each year in the United States alone (Celis et al., 2015; Binder et al., 2016). GBMs affects all ages with a peak incidence between 50 and 60 years of age and a slight male predominance. Biologically, GBMs are highly aggressive, often with diffuse infiltration of the brain parenchyma making complete surgical resection difficult (Badhiwala et al., 2016). Despite advances in radiation therapy, chemotherapeutics and surgical interventions, the median survival for patients with newly diagnosed GBM is only 14.6 months. The majority of patients will experience disease recurrence with estimated 72% recurrence by 17 months per one study. The currently accepted treatment for GBM is chemo radiotherapy with temozolomide after maximal safe surgical resection (Bandak et al., 2014; Balca-Silva et al., 2015). GBM-induced angiogenesis and tumor cell invasion are influenced by a growth factors and extracellular matrix (ECM). Most mammalian cells communicate with protein components in the ECM via a family of cell surface receptors known as integrins (Liu et al., 2016). The five members of the αv integrin subfamily—αvβ1, αvβ3, αvβ5, αvβ6, and αvβ8—are expressed in neural and vascular cells of the brain and bind to arginine-glycine-aspartic acid (RGD) peptide motifs present in many shared ECM ligands. In particular, genetic studies in mice have revealed that αvβ8 integrin is a central regulator of angiogenesis in the developing brain (Saut et al., 2014). Targeted ablation of αv or β8 Integrin genes in embryonic neuro-epithelial cells causes brain-specific vascular phenotypes, including endothelial cell hyper proliferation, the formation of vessels with glomeruloid-like tufts, and intracerebral hemorrhage (Akimoto, 2000). These pathologies are due to defective activation of latent TGFβs, which are ECM-bound protein ligands for αvβ8 integrin. In neurogenic regions of the adult mouse brain, αvβ8 integrin is expressed in neural stem and progenitor cells where it governs neurogenesis and neuroblast migration (Rao et al., 2016). αvβ8 Integrin expression and functions are deregulated in various brain pathologies. For example, diminished expression of αvβ8 integrin and reduced TGFβ signaling are detected in brain arteriovenous malformations. Discovered that αvβ8 integrin–activated TGFβs suppress pathologic angiogenesis in mosaic mouse models of astrocytomas (Tchaicha et al., 2011). However, there are few reports of the role of miR-93 in GBM, particularly its relationship with GBM prognosis. Accordingly, to further clarify the role of miR-93 in GBM, we evaluated miR-93 expression in a large number of different types of GBM tissue samples. Here, we have analyzed expression level miRNA93 and its target αvβ8 Integrin in angiogenesis and tumor cell invasiveness in human GBM, using miRNA Taqman probe Real Time PCR assay.
Materials and Methods
Patients
Paraffin embedded blocks samples from patients with different types of brain tumor were collected during Jun 2001 -May2014, from Neurosurgery referral center in Shahid Bahonar hospital Kerman province, Iran. In total of 199 specimens were found, but 178 samples were entered to our study and others patients were lost due to inadequate, absence of histological material. Also 100 histopathologic samples were selected from peritumoral normal brain tissue as control group. The present study is based on a retrospective examination of GBM diagnostic biopsy or surgery samples from clinical cases, all original hematoxylin and eosin (H&E) slides and/or H and E recut from tissue blocks were reviewed.
In total, samples were screened for level expression of miR93 and its target Integrinβ8. This project was approved by the Neuroscience research center ethics committee of the Kerman University of Medical Sciences, Iran.
Deparaffination samples
Paraffinated blocks from the 178 tumor samples were cut in 5-μm sections and 8 sections/patient were collected in the same micro-centrifuge tube. Samples were de-waxed in 500 μl xylene, All micro-centrifuge tube located for 10 min in a 60 °C heated block and centrifuged at 8,000 rpm, supernatant was removed. This step was then repeated 3 times. Add 500 μl absolute ethanol, centrifuge at 10,000 rpm for 1 min, the samples were then dried in a 60°C heated block with open lids for 10-20 min for remove residual ethanol.
Tissue digestion
According to samples (Paraffinated blocks), 200-400 μl of Tissue Lysis Buffer was added to each tube [4 M Urea, 200 mMTris, 20 mMNaCl, 200 mM EDTA; PH=7.4 (25°C)]. To all tubes added 20-40 μl proteinase K, Samples were gently vortexes and located for 10 min in a 60°C heated block, and all samples were subsequently incubated at 37°C overnight.
RNA and miRNA Extraction
The next day, 200 μl of Binding Buffer [6 M Guanidine- Hcl, 10mM Urea, 10mM Tris-Hcl, 20% Tritonx-100(v/v); PH=4.4(25°C)] was added to each tube with gently vortex. Total RNA and specific miRNAs was extracted using RNAeasy mini kit and miRNA easy kit (Qiagen, Germany) according to the manufacturer’s instructions. Extracted RNA and miRNA pellets were resuspended in 100μl of pre-warmed Elution buffer and stored at -70°C until use.
Real time PCR
For determination expression level of miRNA93 and integrin, reverse transcription real time PCR (rReal Time PCR) was carried out by using the first strand cDNA synthesis kit by Revert AidcDNA synthesis kit (Thermoscientific, USA). Briefly, RNA samples were heated to 65°C for 10 minutes and then chilled on ice. First-strand cDNA reaction mix was added according to the manufacturer’s protocol. One μl of DTT solution, and 1 μl of random hexamer (N)6primer (0.2ug) were then added to the heat-denatured RNA. Samples were mixed properly by pipetting up and down several times and then incubated for 1 hour at 42°C. TaqMan microRNA assays used Human Panel-Early Access Kit (ABI, Forest City, CA) which includes 157 human miRNAs as well as 3 negative controls. To detect suppression of miR-93 expression by anti-miR-93, the following primers were used. (Table 1) Each forward primers used correspond to mature miRNA sequence according to miRBase database (http://microrna.sanger.ac.uk). Primers were modified with LNA (Locked Nucleic Acid) substitutions for increasing specificity and discriminating between miRNAs with a single base different nucleotide sequences. A Real time PCR was done for determination level expression of Integrinβ8 RNA, the QuantiTect Probe PCR Kit (Qiagen, Germany) used base on instruction kit.
Table 1.
Sequence Primer and Probes for Real Time Pcr in this Study
| Name | Sequence |
|---|---|
| hsa-miR-93-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTACCT |
| hsa-miR-93-F | ACACTCCAGCTGGGCAAAGTGCTGTTCGTGC |
| miR93 probe | CTA CCT GCA CGA ACA GCA CTT TG |
| URP | TGGTGTCGTGGAGTCG |
| U6 | F-CTCGCTTCGGCAGCACA |
| R-AACGCTTCACGAATTTGCGT | |
| Integrinβ8 | F- TCAGTTGATTCAATAGAATACC |
| R- CTGTGTATATGAATTTTAGCG | |
| Human β-actin_184 | F- AGAGCTACGAGCTGCCTGAC |
| R- AGCACTGTGTTGGCGTACAG |
URP, Universal reporter primer; RT, Reverse transcription; F, Forward; R: Reverse
Real-time PCR was carried out on an Rotor Gene 6,000 thermo cycler (Corbett research, Inc.) using the following conditions: 95 ºC for 10 min, followed by 40 cycles of 95 ºC for 15 s and 60 ºC for 1 min, followed by a hold at 4 ºC. Raw data can then be analyzed with REST Relative Quantification Software version 2.2.3 (Qiagen, Inc.), generally using the automatic cycle threshold (Ct) setting for assigning baseline and threshold for Ct determination. Relative expression (RE) of the sample gene was calculated using the ΔΔCT method. Real time PCR primers and probes were design for Integrinβ8 mRNA after alignment of these regions between all of them in EBML-EBI and as an internal control, U6 miRNA and β-actin were used for internal control that purchased from Metabion company (Germany).
Statistical analyses
Chi-square test or Fisher’s exact test was conducted using SPSS version 17 for the association between the level of expression mir93 and Integrin β8 in types of glioma tumors (values P=0.05 were considered statistically significant).
Results
One hundred seventy-eight patients with different grade of astrocytomas were selected during Jun 2001 -May 2014. Paraffinated blocks samples from these patients were selected for evaluation level gene expression of integrinβ8, miR-93 using Real Time PCR. From total 178 samples, 58 (32.5%) samples were female and 120 (67.5%) were male. Minimum and maximum age in all groups were 21 and 89 years old (Table 2). Of One hundred seventy-eight samples 21 (10.5%) grade II, 15 (7.5%) grade III, and 142 (71.3%) were grade IV or GBM. miR-93 was measured in all of 178 specimens using Real Time PCR, also in these samples miR-U6 was used for normalizing and calculation relative expression of miR-93. Different rate of miR-93 expression is shown in Figure 1 and it is clear that in other groups (low grade), the expression of miR-93 is normal and natural but when the tumor progress to III and IV grade, the rate of miR-93 dramatically drops which suggests that the reduction in amount of miR-93 may involve in vascular proliferation. In Figure 2, integrinβ8 mRNA expression in the different groups studied and seen the expression rate of integrinβ8 in the GBM patients is in the highest level (blue column) but as it is shown in the minimum levels of expression in low grade (grade II) were observed. Statistical analysis of results showed that there is an inverse significant correlation between miR-93 and tumor grade. In our study, an inverse relation between the expression levels of mir-93 and expression levels of integrinβ8 was found and it can be said that integrinβ8 that is a target for miR-93, was involved in tumors genesis and angiogenesis in tumors. Statistical analysis of results showed that a direct relation and significant correlation between sex and mean of expression of integrinβ8 in GBM tumors. As shown in Figure 3, mean expression integrinβ8 in men with GBM is more than woman, however no difference between expression of miR-93 in men and women. That is can be remarkable for another studies.
Table 2.
Basic Characteristic Samples in this Study
| Tumor grade | Type | N | Sex | ||
|---|---|---|---|---|---|
| M | F | Age (Mean±SD) | |||
| Grade II | Fibrilary astrocytoma Oligodendrogliomas | 21 | 13 | 8 | 21-55 (48.2±5.6) |
| Grade III | Anaplastic astrocytoma | 15 | 9 | 6 | 30-75(56±6.9) |
| Grade IV | Glioblastoma | 142 | 98 | 44 | 46-89(69.5±9.4) |
N, Number of cases
Figure 1.
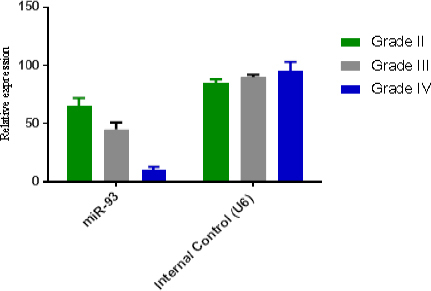
Relative Mirna Expression in Different Types of Tumors
Figure 2.
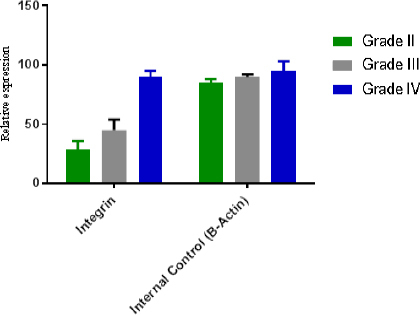
Relative RNA Expression in Different Types of Tumors
Figure 3.
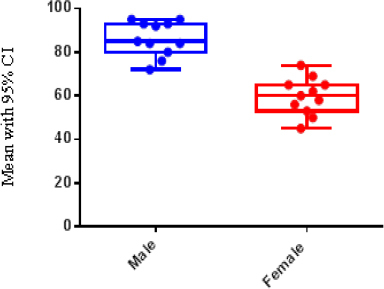
Mean Expression of Integrinβ8 in Male and Female
Discussion
The miR-17-92 is the well-characterized cluster. This cluster and its paralogs, miR-106, play important roles in cancer ; because they can repress expression of many tumor-associated proteins. The detailed role and underlying mechanism of miR-93 in the regulation of glioma growth and chemoresistance remain largely unclear. In the present study, we observed that miR-93 was significantly upregulated in glioma, and increased miR-93 levels were significantly associated with the advanced malignancy (Chaulk et al., 2011). Recently, miRNAs have been found to play key roles in the development and progression of glioma, such as miR-23b, miR-27b, miR-124, and miR-93(Stiefel et al., 2016). In the present study, we used real-time RT-PCR to examined the expression of miR-93 in glioma, and found that it was significantly upregulated in glioma tissues compared to normal brain tissues. Moreover, we showed that its up regulation was significantly associated with the malignant progression as well as poor prognosis of glioma patients, suggesting that miR-93 may play an oncogenic role in glioma. Jiang et al. also reported that the expression of miR-93 was markedly up regulated in glioma tissues, and that the miR-93 levels were significantly correlated with clinicopathological grade and overall survival in glioma, consistent with our findings (Jiang et al., 2015). Mature functional miRNAs of approximately 22 nucleotides that are generated from long primary miRNA transcripts control gene expression at the posttranscriptional level by degrading or repressing target mRNAs (Stiefel et al., 2016). These miRNAs regulate the expression of signaling molecules such as cytokines, growth factors, transcription factors, and pro apoptotic and anti apoptotic molecules (Sorensen et al., 2016). Recently, miRNAs were found to play a role in the differentiation of stem cells. Proper regulation of differentiation of stem cells is crucial to normal development and the avoidance of cancer (Sidiropoulos et al., 2016). GBM is the most aggressive adult brain tumor and, nevertheless the progresses in molecular therapy, its prognosis remains very poor. Identifying a miRNAs profile for GBM could be very useful for better clarify prognosis and researching new targeted drugs (Sun et al., 2013). For this reason, and for “opening” the anatomic pathology archives even to analysis of miRNAs expression in GBM, it is crucial determining if FFPE specimens are suitable for this type of analysis. The data in other studies were reported that αvβ8 integrin, via activation of its latent TGFβ protein ligands, plays central roles in regulating GBM-induced angiogenesis and perivascular tumor cell invasiveness (Tchaicha et al., 2011). Although various reports have shown elevated levels of TGFβ receptor signaling in GBM cells, the factors that activate latent TGFβs within the tumor microenvironment remain obscure(Sperber et al., 2014; Stiefel et al., 2016; Tang et al., 2016). miR-93 may promote the growth and metastasis of glioma. Indeed, Fang et al. found that miR-93 could enhance the tumor growth of glioma cells in vivo. They showed that miR-93-oveexpressing glioma cells induced the formation of blood vessels in the tumor tissue, which in return facilitated cell survival, resulting in enhanced tumor growth (Fang et al., 2011). In addition, they identified integrin-β8 as a target of miR-93, and found that higher levels of integrin-β8 were associated with cell death in tumor mass and in human glioblastoma (Fang et al., 2011). Our data also show that αvβ8 integrin is a positive regulator in invasive GBM cells. We propose that distinct populations of GBM cells selectively express high versus low levels of αvβ8 integrin, with directly determining tumor cell behaviors (Tezcan et al., 2014; Tezcan et al., 2015). Although the exact mechanisms that regulate β8 integrin expression in GBM cells remain unknown, in our report has shown that β8 integrin is a direct target for miR-93, with microRNA (miR)-mediated inhibition of β8 integrin expression correlating with enhanced angiogenesis and tumorigenicity. Therefore, it is enticing to speculate that during tumor growth and progression, subpopulations of GBM cells selectively express high levels of miR-93, thus leading to diminished levels of β8 integrin and the development of angiogenesis pathologies. We also postulate that invasive GBM cells express low levels of miR-93 and high levels of β8 integrin, leading to proinvasive behaviors likely via autocrine activation of TGFβ signaling pathways. Suggesting that genetic and epigenetic events may cooperatively control β8 integrin expression and functions in GBM-induced angiogenesis and tumor cell invasiveness (Figure 3). In conclusion, our study reveals an oncogenic role of miR-93 in the regulation of proliferation, cell cycle progression, colony formation, migration, invasion, and chemoresistance in glioma cells, possibly via directly targeting of β8 integrin.
Acknowledgments
The authors of this project are grateful to Kerman Virology Laboratory in faculty of medicine and their cooperation in collecting samples, and so thanks for Neuroscience research center ethics committee of the Kerman University of Medical Sciences in approve this project.
References
- Abdel-Rahman O, Fouad M. Irinotecan-based regimens for recurrent glioblastoma multiform:a systematic review. Expert Rev Neurother. 2015;15:1255–70. doi: 10.1586/14737175.2015.1101346. [DOI] [PubMed] [Google Scholar]
- Akimoto H. The possible role of adhesion molecule, alpha 3 integrin, in the synthesis of intracrescentic extracellular matrix in accelerated anti-GBM nephritis. Nihon Jinzo Gakkai Shi. 2000;42:1–10. [PubMed] [Google Scholar]
- Badhiwala JH, Nassiri F, Almenawer SA. GBM surgery in the elderly-time to be more aggressive? Clin Neurol Neurosurg. 2016;141:131–2. doi: 10.1016/j.clineuro.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Balca-Silva J, Matias D, do Carmo A, et al. Tamoxifen in combination with temozolomide induce a synergistic inhibition of PKC-pan in GBM cell lines. Biochim Biophys Acta. 2015;1850:722–32. doi: 10.1016/j.bbagen.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Bandak G, Jones BA, Li J, et al. Rituximab for the treatment of refractory simultaneous anti-glomerular basement membrane (anti-GBM) and membranous nephropathy. Clin Kidney J. 2014;7:53–6. doi: 10.1093/ckj/sft152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry SE, Chan B, Ellis M, et al. Identification of miR-93 as a suitable miR for normalizing miRNA in plasma of tuberculosis patients. J Cell Mol Med. 2015;19:1606–13. doi: 10.1111/jcmm.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder ZA, Wilson KM, Salmasi V, et al. Establishment and biological characterization of a panel of glioblastoma multiforme (GBM) and GBM variant oncosphere cell lines. PLoS One. 2016;11:e0150271. doi: 10.1371/journal.pone.0150271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis MA, Alegria-Loyola MA, Gonzalez-Aguilar A, et al. First Mexican consensus on recommendations of the multidisciplinary care of patients with glioblastoma multiforme (GBM):Mexican interdisciplinary group on neuro-oncology research (GIMINO) Gac Med Mex. 2015;151:403–15. [PubMed] [Google Scholar]
- Chaulk SG, Thede GL, Kent OA, et al. Role of pri-miRNA tertiary structure in miR-17~92 miRNA biogenesis. RNA Biol. 2011;8:1105–14. doi: 10.4161/rna.8.6.17410. [DOI] [PubMed] [Google Scholar]
- Chen Q, Lan W, Wang J. Mining featured patterns of MiRNA interaction based on sequence and structure similarity. IEEE/ACM Trans Comput Biol Bioinform. 2013a;10:415–22. doi: 10.1109/TCBB.2013.5. [DOI] [PubMed] [Google Scholar]
- Chen Q, Zhou Y, Richards AM, et al. Up-regulation of miRNA-221 inhibits hypoxia/reoxygenation-induced autophagy through the DDIT4/mTORC1 and Tp53inp1/p62 pathways. Biochem Biophys Res Commun. 2016;8:658–69. doi: 10.1016/j.bbrc.2016.04.090. [DOI] [PubMed] [Google Scholar]
- Chen R, Liu H, Cheng Q, et al. MicroRNA-93 promotes the malignant phenotypes of human glioma cells and induces their chemoresistance to temozolomide. Biol Open. 2016;5:669–77. doi: 10.1242/bio.015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Heneidi S, Lee JM, et al. miRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes. 2013b;62:2278–86. doi: 10.2337/db12-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Deng Z, Shatseva T, et al. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene. 2011;30:806–21. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- Galka-Marciniak P, Olejniczak M, Starega-Roslan J, et al. siRNA release from pri-miRNA scaffolds is controlled by the sequence and structure of RNA. Biochim Biophys Acta. 2016;1859:639–49. doi: 10.1016/j.bbagrm.2016.02.014. [DOI] [PubMed] [Google Scholar]
- Jain A, Das S. Synteny and comparative analysis of miRNA retention, conservation, and structure across Brassicaceae reveals lineage- and sub-genome-specific changes. Funct Integr Genomics. 2016;5:668–75. doi: 10.1007/s10142-016-0484-1. [DOI] [PubMed] [Google Scholar]
- Jiang L, Wang C, Lei F, et al. miR-93 promotes cell proliferation in gliomas through activation of PI3K/Akt signaling pathway. Oncotarget. 2015;6:8286–99. doi: 10.18632/oncotarget.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li C, Han J, et al. The detection of risk pathways, regulated by miRNAs, via the integration of sample-matched miRNA-mRNA profiles and pathway structure. J Biomed Inform. 2014;49:187–97. doi: 10.1016/j.jbi.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Lin X, Yang B, Liu W, et al. Interplay between PCBP2 and miRNA modulates ARHGDIA expression and function in glioma migration and invasion. Oncotarget. 2016;6:358–66. doi: 10.18632/oncotarget.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Han L, Dong Y, et al. EGFRvIII/integrin beta3 interaction in hypoxic and vitronectinenriching microenvironment promote GBM progression and metastasis. Oncotarget. 2016;7:4680–94. doi: 10.18632/oncotarget.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JC, Welling L, Escosteguy C, et al. Socioeconomic and educational factors interference in the prognosis for glioblastoma multiform. Br J Neurosurg. 2013;27:80–83. doi: 10.3109/02688697.2012.709551. [DOI] [PubMed] [Google Scholar]
- Mannino S, Molinari A, Sabatino G, et al. Intratumoral vs systemic administration of meta-tetrahydroxyphenylchlorin for photodynamic therapy of malignant gliomas:assessment of uptake and spatial distribution in C6 rat glioma model. Int J Immunopathol Pharmacol. 2008;21:227–31. doi: 10.1177/039463200802100126. [DOI] [PubMed] [Google Scholar]
- Mollaie HR, Monavari SH, Arabzadeh SA, et al. RNAi and miRNA in viral infections and cancers. Asian Pac J Cancer Prev. 2013;14:7045–56. doi: 10.7314/apjcp.2013.14.12.7045. [DOI] [PubMed] [Google Scholar]
- Nesler KR, Sand RI, Symmes BA, et al. The miRNA pathway controls rapid changes in activity-dependent synaptic structure at the Drosophila melanogaster neuromuscular junction. PLoS One. 2013;8:e68385. doi: 10.1371/journal.pone.0068385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue AI, Clement C, Hill JM, et al. Evolution of microRNA (miRNA) structure and function in plants and animals:Relevance to aging and disease. J Aging Sci. 2014;2:45–8. doi: 10.4172/2329-8847.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Manyam G, Rao G, et al. Integrative analysis of mRNA, microRNA, and protein correlates of relative cerebral blood volume values in GBM reveals the role for modulators of angiogenesis and tumor proliferation. Cancer Inform. 2016;15:29–33. doi: 10.4137/CIN.S33014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyapalan T, David R, Gooderham NJ, et al. Increased expression of circulating miRNA-93 in women with polycystic ovary syndrome may represent a novel, non-invasive biomarker for diagnosis. Sci Rep. 2015;5:16890–95. doi: 10.1038/srep16890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saut O, Lagaert JB, Colin T, et al. A multilayer grow-or-go model for GBM:effects of invasive cells and anti-angiogenesis on growth. Bull Math Biol. 2014;76:2306–33. doi: 10.1007/s11538-014-0007-y. [DOI] [PubMed] [Google Scholar]
- Shi R, Wang PY, Li XY, et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget. 2015;6:26971–81. doi: 10.18632/oncotarget.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidiropoulos KG, Ding Q, Pampalakis G, et al. KLK6-regulated miRNA networks activate oncogenic pathways in breast cancer subtypes. Mol Oncol. 2016;8:231–36. doi: 10.1016/j.molonc.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen SS, Nygaard AB, Christensen T. miRNA expression profiles in cerebrospinal fluid and blood of patients with Alzheimer’s disease and other types of dementia- an exploratory study. Transl Neurodegener. 2016;5:6–11. doi: 10.1186/s40035-016-0053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H, Beem A, Shannon S, et al. miRNA sensitivity to Drosha levels correlates with pre-miRNA secondary structure. RNA. 2014;20:621–31. doi: 10.1261/rna.043943.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiefel F, Fischer S, Sczyrba A, et al. miRNA profiling of high, low and non-producing CHO cells during biphasic fed-batch cultivation reveals process relevant targets for host cell engineering. J Biotechnol. 2016;225:31–43. doi: 10.1016/j.jbiotec.2016.03.028. [DOI] [PubMed] [Google Scholar]
- Sturm D, Bender S, Jones DT, et al. Paediatric and adult glioblastoma:multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14:92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Cao Y, Shi L, et al. Overexpressed miRNA-137 inhibits human glioma cells growth by targeting Rac1. Cancer Biother Radiopharm. 2013;28:327–34. doi: 10.1089/cbr.2012.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang NY, Chueh FS, Yu CC, et al. Benzyl isothiocyanate alters the gene expression with cell cycle regulation and cell death in human brain glioblastoma GBM 8401 cells. Oncol Rep. 2016;35:2089–96. doi: 10.3892/or.2016.4577. [DOI] [PubMed] [Google Scholar]
- Tchaicha JH, Reyes SB, Shin J, et al. Glioblastoma angiogenesis and tumor cell invasiveness are differentially regulated by beta8 integrin. Cancer Res. 2011;71:6371–81. doi: 10.1158/0008-5472.CAN-11-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezcan G, Tunca B, Bekar A, et al. microRNA expression pattern modulates temozolomide response in GBM tumors with cancer stem cells. Cell Mol Neurobiol. 2014;34:679–92. doi: 10.1007/s10571-014-0050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezcan G, Tunca B, Bekar A, et al. Ficus carica latex prevents invasion through induction of let-7d expression in GBM cell lines. Cell Mol Neurobiol. 2015;35:175–87. doi: 10.1007/s10571-014-0109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadran S, Remacle F, Levine R. Surprisal analysis of Glioblastoma Multiform (GBM) microRNA dynamics unveils tumor specific phenotype. PLoS One. 2014;9:e108171. doi: 10.1371/journal.pone.0108171. [DOI] [PMC free article] [PubMed] [Google Scholar]


