Abstract
Background:
Over-expression of p16INK4a protein is a biomarker for human papillomavirus (HPV)-associated cervical cancer. However, absence of p16INK4a protein expression in HPV-associated cancer of the oral cavity and oropharynx has been reported. Among a number of possible reasons for this is methylation, which is frequently noted in the promoter region of p16INK4a and is associated with silencing of the gene and disease severity.
Methods:
We investigated the relationships between p16INK4a protein expression, HPV infection and methylation status of the p16INK4a promoter in cancers of the oral cavity and oropharynx. Fifty-three formalin-fixed paraffin-embedded (FFPE) cancer tissue samples from the oral cavity (49 cases) and oropharynx (4 cases) were studied. P16INK4a protein expression was determined using immunohistochemical staining (IHC). Additional oral tissues lacking squamous intraepithelial lesions (SILs), and cervical tissues with high-level SILs, were used as negative and positive controls, respectively. High-risk HPV infection was detected using HPV E6/E7 mRNA in situ hybridization. Methylation status of the p16INK4a promoter was investigated using sodium bisulfite treatment and methylation-specific PCR (MS-PCR).
Results:
HPV infection was found in 40.8% (20/49) and 50.0% (2/4) of oral cavity and oropharynx cancers, respectively. Promoter methylation of p16INK4a occurred in 73.6 % of all cases and differed significantly in frequency between HPV-positive (90.9%, 20/22) and HPV-negative (61.3%, 19/31) samples. Expression of p16INK4a was found in 35.8% (19/53) and commonly detected in samples with p16INK4a unmethylation (79.5%). Interestingly, the silencing of p16INK4a (64.2%, 34/53) was significantly associated with methylation status (91.2%, 31/34), especially in HPV-infected samples in which the p16INK4a promoter was methylated (52.9%, 18/34).
Conclusions:
This result demonstrated high frequency of p16INK4a promoter methylation status in HPV-associated HNSCC subsets that could influence the silent p16INK4a expression and might promote disease severity.
Keywords: Human papillomavirus, promoter methylation, oral cavity, oropharynx, p16INK4a
Introduction
Cancers of the oral cavity, nasal cavity, sinonasal tract, pharynx and larynx are subsets of head and neck cancers. Approximately 90% of these cancers are squamous cell carcinomas. More than 550,000 cases of head and neck squamous cell carcinoma (HNSCC) occur globally each year, causing around 300,000 deaths (Jemal et al., 2011). It is possible that environmental risk factors or cofactors play an important role in onset of HNSCC. Such factors include genetic susceptibility, nutritional factors, tobacco and alcohol interaction as well as chewing of betel quid and areca nut which is common in Asian countries (Krishna Rao et al., 2013). Infections with human papillomavirus (HPV) are also found increasingly in HNSCC, especially tonsillar or oropharyngeal subsites (Dahlstrand and Dalianis, 2005; Mirghani et al., 2015) The prevalence of HPV in HNSCC differs regionally. It is higher in western countries than in Asian countries (Kreimer et al., 2005). Prevalence of HPV also differs among HNSCC subsites: 16%, 55% and 26% prevalence in cancers of the oral cavity, oropharynx and larynx, respectively (Gillison et al., 2000). Biological distinctions among HPV-positive and -negative HNSCC cases have been reported.
Infection with high-risk HPV (HR-HPV) is well-known as a cause of cervical cancer. HPV-positive oral squamous cell carcinoma (OSCC) is characterized by inactivation of p53 and retinoblastoma protein (pRb) via HPV oncoproteins E6 and E7, respectively. E7 overexpression induces the expression of a protein that slows cell-cycle, cyclin-dependent kinase inhibitor protein 2A (CDKN2A or p16INK4a) by disruption of interaction between pRb and E2F and release of E2F. In contrast, tobacco-related oropharyngeal cancer is characterized by p53 mutation and down-regulation of CDKN2A (encoding p16INK4a). HPV-positive OSCC seems to be more responsive to chemotherapy and radiation than is HPV-negative disease (Jung et al., 2010).
Elevated expression of p16INK4a, a surrogate marker of HPV-related cervical neoplasia, occurs in nearly 100% of such cancers to an extent proportional to cervical lesion severity (Kalof and Cooper, 2006). Overexpression of p16INK4a, reported in 24-61% of HNSCC cases, is associated with good prognosis and response to treatment with chemotherapy and radiation (Rautava et al., 2012). Overexpression of p16INK4a varies across subsets of HNSCC. It has been reported in 30% of cases involving the oral cavity, but in up to 86.3% of cases of oropharynx cancer (Bixofis et al., 2014; Zafereo et al., 2016). Previous studies have reported that not all HPV-positive HNSCCs showed p16INK4a overexpression (Nemes et al., 2006; Bishop et al., 2012). Some studies showed a strong association between HPV infection and p16INK4a overexpression, but 37-52% of HPV-positive HNSCC were p16INK6a -negative and associated with poor prognosis (Ukpo et al., 2011; Lingen et al., 2013; Melkane et al., 2014). In such cases, epigenetic events may influence p16INK4a expression. Promoter methylation of p16INK4a might explain the lack of p16INK4a overexpression in many cancers (Sanchez-Cespedes et al., 2000).
Expression of the p16INK4a gene, located at the INK4A/ARF locus on chromosome 9p21, can be controlled by many molecular pathways. Increase of transcriptional factors or host oncoproteins can up-regulate expression of this gene (Sharpless and DePinho, 1999). Some reports have shown that the INK4A/ARF locus is controlled by epigenetic mechanisms such as histone modification and promoter methylation pathways. Promoter methylation of p16INK4a is found commonly in prostate, colorectal and gastric cancers and the degree of methylation is proportional to disease severity (Goto et al., 2009).
HR-HPV oncoproteins are involved in up-regulation processes of the DNA methyltransferase (DNMTs) enzymes DNMT1 and DNMT3b. HPV16 E6 is identified as a factor for induction of DNMT1 expression in SiHa and CaSki cells (Au Yeung et al., 2010). HPV16 E7 was also found to up-regulate DNMT1 expression in a human keratinocyte cell line (Laurson et al., 2010). HPV18-transfected organotypic raft culture showed the expression of DNMT1 and DNMT3b in basal and suprabasal layers (Leonard et al., 2012). Moreover, promoter methylation of various genes accumulated in HR-HPV-infected keratinocytes with HPV integration form before cell immortalization (Henken et al., 2007).
Promoter methylation of p16INK4a gene is a frequent event in primary HNSCCs and plays a major role in the silencing of this gene during tumor development (Demokan et al., 2012). Although promoter methylation leads to down-regulation of p16INK4a expression, in oral cancer cell lines treated with a DNMT inhibitor (5-aza-deoxycytidine), re-activation of p16INK4a expression was shown (Yakushiji et al., 2003).
To explore the INK4A/ARF promoter methylation affecting p16INK4a protein expression in HPV-associated HNSCC subsets (cancers of oral cavity and oropharynx), we first investigated HPV infection in formalin-fixed paraffin-embedded (FFPE) tissues of HNSCC subsets from northeastern Thai patients. Presence of HPV infection was determined by HR-HPV E6/E7 mRNA in situ hybridization (HR-HPV E6/E7 mRNA ISH). The methylation status of the p16INK4a promoter was evaluated by sodium bisulfite treatment and methylation-specific polymerase chain reaction (MS-PCR). Finally, the expression of p16INK4a protein was observed with immunohistochemistry (IHC) staining.
Materials and Methods
Patients and Samples
Studied samples were 53 FFPE tissue samples collected from various HNSCC subsets including cancer of the oral cavity (49 cases) and oropharynx (4 cases). Additional 6 FFPE tissue samples from the oral cavity with no squamous intraepithelial lesions (No-SIL) and FFPE cervical tissues with high-SILs were included as negative and positive controls for p16INK4a IHC, respectively. All samples were collected from participating patients at Srinagarind Hospital, Faculty of Medicine, Khon Kaen University and Khon Kaen Central Hospital. This study was approved by Khon Kaen University Ethics Committee in Human Research (No.HE521344).
High risk (HR)-HPV E6/E7 mRNA detection by in situ hybridization (ISH)
RNAscope® Reagent Kit (Advanced Cell Diagnostics, Hayward, CA) was used to detect 18 types of HR- HPV E6/E7 RNA as described in the manufacturer’s instructions. This detection by ISH was performed on a tissue microarray (TMA) platform. FFPE blocks were cut into 5 µm sections that were put on glass slides and stained with hematoxylin and eosin (H&E) dyes. The tumor areas were identified using a light microscope. To prepare the TMA blocks, FFPE tissues were punched with a biopsy-gauge needle in at least two areas of the tumor. Punched tumor tissues were re-embedded in paraffin. Five µm sections of re-embedded TMAs were cut and put on SuperFrost Plus glass slides. TMA slides were de-paraffinized in xylene and hybridized with a cocktail of 18 HR-HPV E6/E7 mRNA probes (HPV type 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59,66, 68, 73 and 82) and hybridization signal was developed with the RNAscopeTM 2.0 HD (26) system (Advanced Cell Diagnostic, Hayward, CA, USA). The human cyclophilin B probe (PPIB), which is the internal control targeting a constitutively expressing gene and bacterial dihydrodipicolinate reductase (DapB) negative control probe were also used. HPV E6/E7 mRNA hybridization signals were observed under a light microscope. Specific signals were observed as brown dots in the cytoplasm or in both cytoplasm and nucleus of tumor cells.
p16INK4a Immunohistochemical staining (IHC)
Sections 2 μm thick were cut from each FFPE block and adhered to SuperFrost Plus glass slides (Menzel-Gläser, Braunschweig, Germany). The sections were de-paraffinized in xylene (5 min x 3 washes) followed by re-hydration in absolute ethanol (5 min x 2), 95% ethanol (5 min), 70 % ethanol (5 min) and distilled water (at least 10 min). IHC was performed using a CINtec histology kit (mtm labs, Heidelberg, Germany) following the manufacturer’s instructions. The stained tissues were observed under a light microscope. In cells positive for p16INK4a, stain was present in nuclei or nuclei/cytoplasm. Over 500 tumor cells were evaluated in each sample and scored according to the intensity of p16INK4a staining: 0=no expression; 1=1-20% of cells were stained; 2=21-40% of cells were stained and 3=41-100% of cells were stained (Pande et al., 1998). In cervical tissues used as controls, cells were scored in three categories; no expression, focal staining and diffuse staining for p16INK4a.
Sodium bisulfite modification and p16INK4a-methylation-specific polymerase chain reaction (MS-PCR)
Before performing the experiments, control DNA for the p16INK4a MS-PCR was prepared and tested. DNA was extracted from SiHa cells, which are known to be unmethylated. The methylated DNA control was prepared by treatment of 1 µg of SiHa DNA with 100 units of M. SssI DNA CpG methyltransferase (New England Biolabs, Ipswich, MA, USA) together with S-adenosylmethionine (Monzon et al., 1998) substrate and incubated at 37°C for 16 hours for complete methylation. Fifty-three selected DNA samples from HNSCC subsets (HPV-positive or HPV-negative) were treated using the sodium bisulfite modification method. A quantity (200-500 ng) of DNA was treated with Epitect Bisulfite kit (QIAGEN, Hilden, Germany) following the instruction manual. The treated DNA samples were checked by PCR using p16INK4a wild-type primers (p16INK4a WT) to monitor the gene modification after bisulfite treatment (no amplification should occur). The primers for p16INK4a MS-PCR were designed to distinguish methylated from unmethylated DNA and produced amplicons of 150 and 151 bp, respectively. The details of each primer pair are shown in Table SI (Herman et al., 1996) All MS-PCR reactions were performed with Platinum Taq polymerase (Life Technologies). PCR products were separated on a 2% agarose gel and visualized under UV light.
Statistical analysis
Statistical analysis was done using SPSS version 16.0 (IBM, Armonk, NY, USA). Relationships of HPV infection, p16INK4a expression and p16INK4a promoter methylation were analyzed using the Chi-square test. Any P-value < 0.05 was considered as statistically significant. The impact of potential factors on p16INK4a methylation was evaluated by univariate analysis.
Results
Characteristics of studied samples
Table 1 describes the characteristics of samples used in this study. Median age of patients was 70 years (ranging from 41 to 90 years). Female patients were in the majority (69.8%).
Table 1.
Characteristics of Samples (n=53)
| Characteristics | Number (%) |
|---|---|
| Sex | |
| Female | 37 (69.8) |
| Male | 16 (30.2) |
| Anatomical site | |
| Oral cavity | 49 (92.5) |
| Buccal mucosa | 33 (62.3) |
| Tongue | 3 (5.7) |
| Lip | 3 (5.7) |
| Floor of mouth | 5 (9.4) |
| Gum | 2 (3.8) |
| Hard palate | 3 (5.7) |
| Oropharynx | 4 (7.5) |
| Soft palate | 2 (3.8) |
| Uvular | 2 (3.8) |
| Histological classification | |
| Well differentiated | 36 (67.9) |
| Moderately differentiated | 9 (17.0) |
| Poorly differentiated | 8 (15.1) |
| HPV status | |
| HPV positive | 22 (41.5) |
| HPV negative | 31 (58.5) |
| Molecular features of p16INK4a in patients | |
| p16INK4a methylation | 39 (73.6) |
| p16INK4a expression | 19 (35.8) |
HPV infection in HNSCC subsets
A total of 53 HNSCC subset samples and 6 oral samples with No-SIL were tested for HPV infection by detection of HR-HPV E6/E7 mRNA expression using the ISH method. The mRNA hybridization was performed with different three probes; DapB (Figure 1A.), PPIB (Figure 1B.) and HR-HPV E6/E7 mRNA (Figure 1C.). HPV infection was found in 41.5% (22/53) of HNSCC subsets (Table 1). All oral samples with No-SIL were negative for HPV infection.
Figure 1.
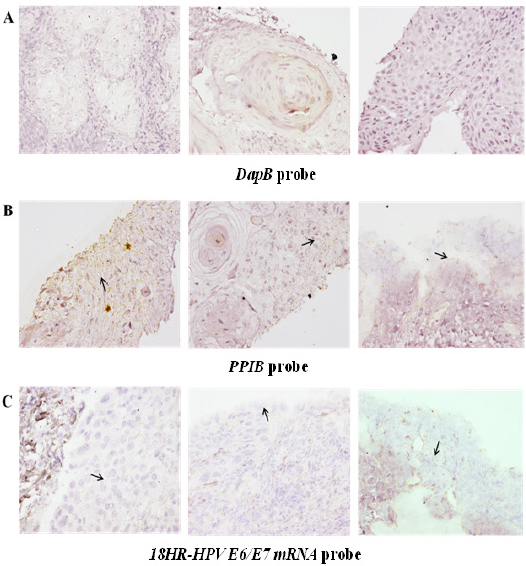
HPV Detection in Various HNSCC Subsets. RNA in situ hybridization methods (RISH) were used to identify the genotypes of HR-HPV E6/E7 present. Probes for 17 different genotypes were used. (A) DapB and (B) PPIB probes were used as internal negative and positive controls respectively for the in situ hybridization. (C) Hybridization signals of HR-HPV E6/E7 mRNA are shown. The signals appear as brown dots and are indicated with black arrows. DapB: bacterial dihydrodipicolinate reductase, PPIB: human peptidylpropyl isomerase B, HR-HPV: high-risk human papillomavirus.
Methylation of p16INK4a promoter in HNSCC subsets
Methylation was observed on the p16INK4a promoter located at the INK4A/ARF locus by MS-PCR. The PCR products were separated using agarose gel electrophoresis and visualized under ultraviolet light (Figure 2A.). The association between p16INK4a promoter methylation and HPV infection was investigated. Fifty-three HNSCC subset samples were evaluated according to their HPV infection status: 22 positive cases and 31 negative cases. Methylation of the p16INK4a promoter methylation was found in 73.6% (39/53) of all samples. The promoter of p16INK4a gene was more frequently methylated in HPV-positive (90.9%) than in HPV-negative (61.3%) HNSCC cases (P<0.001) (Figure 2B. and Table 2.). This result demonstrated that a high frequency of p16INK4a methylation in HNSCC subsets is associated with HPV infection.
Figure 2.
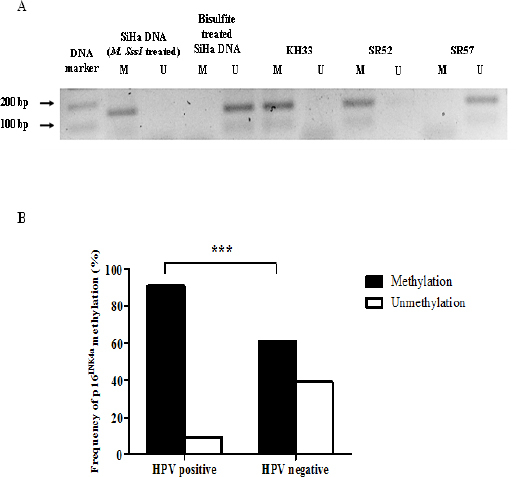
p16INK4a Promoter Methylation in HNSCC Samples. DNAs of OSCCs were treated with sodium bisulfite before evaluation of p16INK4a promoter methylation by MS-PCR. (A) Representative examples of MS-PCR for p16INK4a. PCR products (approx 150 bp) were separated in an agarose gel. SiHa DNA treated with DNA methylatransferase (M. SssI) was used as a positve control of p16INK4a methylated promoter. (B) Frequency of p16INK4a promoter methylation catagorized by HPV infection status. MS-PCR: methylation-specific polymerase chain reaction, M. SssI, DNA methyltranferase; U, unmethylation; M, methylation
Table 2.
HPV Infection and p16INK4a Promoter Methylation in HNSCC Subset Samples (n=53)
| HPV infection | p16INK4a (%) | P | OR | 95% CI | |
|---|---|---|---|---|---|
| M | U | ||||
| Positive | 20 (90.9) | 2 (9.1) | <0.0001 | 6.46 | 2.9-14.3 |
| Negative | 19 (61.3) | 12 (38.7) | |||
| Total | 39 (73.6) | 14 (26.4) | |||
M, methylated; U, unmethylated
p16INK4a protein expression in HNSCC subsets with HPV infection and p16INK4a methylation
To elucidate the relationship of HPV associated p16INK4a methylation to p16INK4a protein expression in HNSCC subsets, p16INK4a expression was investigated in oral cavity SCC (49 cases) and oropharynx SCC (4 cases). The correlation between p16INK4a expression and HPV-associated p16INK4a methylation is shown in Table 3. P16INK4a protein expression was demonstrated in controls (FFPE cervical tissues) and showed 3 patterns; no-expression, focal and diffuse expression (Figure 3A.). Expression of p16INK4a in FFPE HNSCC subsets showed different patterns and at least 500 cells per section with positive staining of nuclei/cytoplasm were scored (Figure 3B.). P16INK4a expression in HNSCC subsets was 35.8% (19/53, Table 3.). Interestingly, p16INK4a expression was significantly more frequent in HPV-negative (78.9%, 15/19) than in HPV-positive HNSCC subsets (21.1%, 4/19) (P<0.05). Lack of p16INK4a expression was significantly associated with a higher frequency of methylation (91.2%, 31/34), especially in HPV-associated cases 52.9% (18/34). This result indicates that silencing p16INK4a protein expression in HPV-infected HNSCC subsets is associated with p16INK4a promoter methylation.
Table 3.
HPV Status and Molecular Features of p16INK4a in HNSCC Subset Samples (n=53)
| HPV infection | Total | P | OR | 95% CI | ||
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| Positive p16INK4a expression (n=19) | ||||||
| p16INK4a (M) | 2 (5.1) | 6 (15.4) | 8 (20.5) | 0.53 | 1.52 | 0.47-4.89 |
| p16INK4a (U) | 2 (14.3) | 9 (64.3) | 11 (79.5) | |||
| Negative p16INK4a expression (n=34) | ||||||
| p16INK4a (M) | 18 (52.9) | 13 (38.2) | 31 (91.2) | <0.001 | 26.4 | 1.49-467.8 |
| p16INK4a (U) | 0 (0) | 3 (8.8) | 3 (8.8) | |||
| Total | 22 (41.5) | 31 (58.5) | 53 (100) | |||
M, methylated; U, unmethylated
Figure 3.
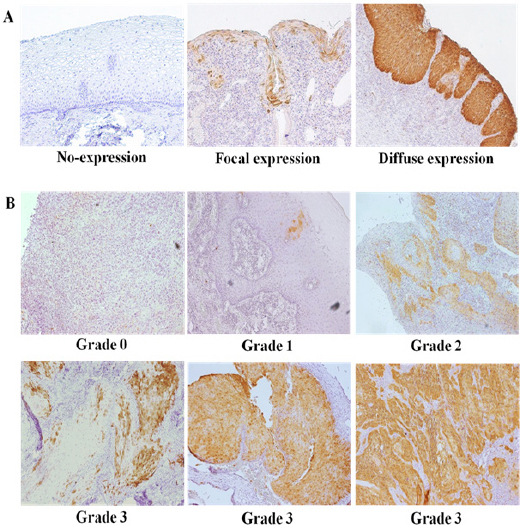
Immunohistochemical (IHC) Staining to Demonstrate Expression of p16INK4a in Representative Cervical and Oral Tissues. (A) IHC expression in cervical tissues exhibited 3 patterns: no-expression, focal expression and diffuse expression. (B) IHC staining patterns of p16INK4a fell into 4 categories: At least 500 tumor cells, in which nuclei and cytoplasm were stained, were counted in each section
Discussion
This study investigated relationships between HPV-infected HNSCC subsets in northeastern Thai patients and molecular features of p16INK4a. We found that 90.9% of HPV-infected HNSCC cases exhibited p16INK4a promoter methylation. A possible effect of epigenetic involvement via promoter methylation was further investigated using p16INK4 IHC. This study demonstrated that 91.2% of promoter methylation in most HNSCC subsets lacked an express p16INK4a. Interestingly, approximately 50% of HPV-associated HNSCC cases exhibiting p16INK4a methylation lacked p16INK4a protein expression.
Reported prevalences of HPV infection in HNSCC patients range from 0 to 75%, depending on sample types and methods used (Mirghani et al., 2015). We found an HPV prevalence in HNSCC subsets of 41.5% when a “gold standard” diagnostic method was used (Table 1). Although many studies have used a combination of methods to increase sensitivity and specificity of HPV detection in OSCC samples, detection of viral transcripts using ISH is suggested to be the most reliable approach. This method can indicate viral transcriptional activity and increased sensitivity of HPV detection since more copies of target mRNA are presented than of viral genomes (Weinberger et al., 2006). Evans et al., (2014) reported that the patterns of HPV E6/E7 staining in cervical intraepithelial neoplasia (CIN) related to grade of cervical lesion. In addition, a pattern of multiple dot-like staining in nucleus and cytoplasm suggests the transformative phase of HPV. Therefore, HPV E6/E7 RNA ISH might indicate an important role for the transformative phase of HPV infection in development of oral carcinogenesis.
Previous reports showed considerable variation of p16INK4a expression (approximately 18.75-86.3%) in OSCC. In part, this may be a function of different geographical regions and/or antibody clone used (Buajeeb et al., 2009; Fischer et al., 2010; Thomas and Primeaux, 2012; Bixofis et al., 2014). In this study, p16INK4a expression was detected in 35.8% of HNSCC cases (Table 1. and Table 3.). A previous report from Thailand noted expression of p16INK4a in 18.75% of OSCC (a subset of HNSCC) cases and in 26.7% of oral leukoplakia tissues, but no expression of p16INK4a was detected in any cases of dysplasia or in normal mucosa (Buajeeb et al., 2009). HPV infection is a significant factor influencing p16INK4a expression via oncogenes (Bussu et al., 2013). A positive relationship between HPV DNA, E6/E7 mRNA and p16INK4a expression in OSCC samples are 96.4% and 78.7%, respectively (Ukpo et al., 2011). However, as many as 41.8% of HPV-positive OSCC cases lacked expression of p16INK4a (Junor et al., 2012). This matches our study, in which half of all HPV-infected HNSCC cases lacked p16INK4a expression (Table 3): we demonstrated that p16INK4a expression was more frequent in HPV-negative HNSCC cases (15/19) than in HPV-infected HNSCC cases (4/19).
In this study, about half of HPV-positive HNSCC cases did not express p16INK4a. Promoter methylation has been reported as a key factor controlling p16INK4a expression in many cancers, such as colorectal and prostate cancers (Goessl et al., 2000; Goto et al., 2009). In addition to the p16INK4a promoter, methylation of many other promoters, such as those for retinoic acid receptor-beta (RARβ), p14ARF, p15INK4a and the E-cadherin family, has been reported in HNSCC cases (Viswanathan et al., 2003; Shaw et al., 2006). We found a high frequency of methylation of the p16INK4a promoter (73.6%) in HNSCC cases, most of which (91.2%) were found in the p16INK4a-negative group (Table 3.). Interestingly, we found expression of p16INK4a in eight cases exhibiting promoter methylation (20.5%). Similar findings have been reported for cervical carcinomas, in which promoter hypermethylation did not affect p16INK4a expression (Nehls et al., 2008). Moreover, hypermethylation of p14ARF, located at the same locus as p16INK4a, is usually high in oral cancer and not correlated with p16INK4a promoter methylation status (Esteller et al., 2000). We have also found a high frequency (98.21%) of methylation of the p14ARF promoter (data not shown). Promoter methylation was found in 52.9% of HNSCC cases that were HPV positive/p16INK4a-negative (Table 3). Previous evidence has demonstrated that HPV oncoproteins can up-regulate the DNA methyltransferase enzymes (DNMTs) including DNMT1 and DNMT3b (D’Costa et al., 2012; Leonard et al., 2012). Therefore, HPV infection involving expression of viral oncoproteins may influence methylation status in HPV-positive HNSCC subsets. In HPV-negative HNSCC subsets, other risk factors, such as chewing of betel quid and areca nut, may affect promoter hypermethylation (Lee-Chen et al., 1996; Lai and Lee, 2006). The study of arecoline in mouse models showed suppression of RARβ, which regulates DNMT expression in human HNSCC (Lai et al., 2014). Previous evidence has indicated good prognosis and better survival rate in HPV-positive and p16INK4a-expressing HNSCC cases (Deng et al., 2014). Moreover, methylation of p16INK4a promoter was associated with poor prognosis in anal and colorectal and cancers (Kim et al., 2005; Koerber et al., 2014). HNSCC cases that are HPV-positive and lacking p16INK4a expression might experience a worse prognosis due to DNMT expression and promoter methylation. However, our sample size was small. Larger sample sizes are required for further investigation.
In conclusion, aberrant p16INK4a expression was usually associated with promoter methylation in HNSCC cases. Methylation of the p16INK4a promoter was also commonly found in HPV-positive HNSCC cases lacking expression of p16INK4a, a situation that may be related to disease severity.
Funding Statement
This work was supported by research grant from Faculty of Medicine (grant number I54141), Graduated School (grant number 511H1201) and Khon Kaen University (grant number 573003).
Statement conflict of Interest
The authors have no conflicts of interest to declare.
Acknowledgments
The chemicals, antibodies for p16INK4a IHC including research site were kindly provided by Prof. Dr. Magnus von Knebel Doeberitz and Dr. Miriam Reuschenbach from the Department of Applied Tumor Biology, Institute of Pathology, University of Heidelberg, Germany. We would like to acknowledge Prof. David Blair for editing the MS via Publication Clinic KKU, Thailand.
References
- Au Yeung CL, Tsang WP, Tsang TY, et al. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol Rep. 2010;24:1599–604. doi: 10.3892/or_00001023. [DOI] [PubMed] [Google Scholar]
- Bishop JA, Ma XJ, Wang H, et al. Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol. 2012;36:1874–82. doi: 10.1097/PAS.0b013e318265fb2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bixofis RB, Sassi LM, Patussi C, et al. Significance of p16 positive expression in oropharyngeal cancers. Asian Pac J Cancer Prev. 2014;15:10289–92. doi: 10.7314/apjcp.2014.15.23.10289. [DOI] [PubMed] [Google Scholar]
- Buajeeb W, Poomsawat S, Punyasingh J, et al. Expression of p16 in oral cancer and premalignant lesions. J Oral Pathol Med. 2009;38:104–8. doi: 10.1111/j.1600-0714.2008.00710.x. [DOI] [PubMed] [Google Scholar]
- Bussu F, Sali M, Gallus R, et al. HPV infection in squamous cell carcinomas arising from different mucosal sites of the head and neck region. Is p16 immunohistochemistry a reliable surrogate marker? Br J Cancer. 2013;108:1157–62. doi: 10.1038/bjc.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’ Costa ZJ, Jolly C, Androphy EJ, et al. Transcriptional repression of E-cadherin by human papillomavirus type 16 E6. PLoS One. 2012;7:e48954. doi: 10.1371/journal.pone.0048954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrand HM, Dalianis T. Presence and influence of human papillomaviruses (HPV) in Tonsillar cancer. Adv Cancer Res. 2005;93:59–89. doi: 10.1016/S0065-230X(05)93002-9. [DOI] [PubMed] [Google Scholar]
- Demokan S, Chuang A, Suoglu Y, et al. Promoter methylation and loss of p16(INK4a) gene expression in head and neck cancer. Head Neck. 2012;34:1470–5. doi: 10.1002/hed.21949. [DOI] [PubMed] [Google Scholar]
- Deng Z, Hasegawa M, Aoki K, et al. A comprehensive evaluation of human papillomavirus positive status and p16INK4a overexpression as a prognostic biomarker in head and neck squamous cell carcinoma. Int J Oncol. 2014;45:67–76. doi: 10.3892/ijo.2014.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Tortola S, Toyota M, et al. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res. 2000;60:129–33. [PubMed] [Google Scholar]
- Evans MF, Peng Z, Clark KM, et al. HPV E6/E7 RNA in situ hybridization signal patterns as biomarkers of three-tier cervical intraepithelial neoplasia grade. PLoS One. 2014;9:e91142. doi: 10.1371/journal.pone.0091142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer CA, Kampmann M, Zlobec I, et al. p16 expression in oropharyngeal cancer: its impact on staging and prognosis compared with the conventional clinical staging parameters. Ann Oncol. 2010;21:1961–6. doi: 10.1093/annonc/mdq210. [DOI] [PubMed] [Google Scholar]
- Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–20. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- Goessl C, Krause H, Muller M, et al. Fluorescent methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res. 2000;60:5941–5. [PubMed] [Google Scholar]
- Goto T, Mizukami H, Shirahata A, et al. Aberrant methylation of the p16 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2009;29:275–7. [PubMed] [Google Scholar]
- Henken FE, Wilting SM, Overmeer RM, et al. Sequential gene promoter methylation during HPV-induced cervical carcinogenesis. Br J Cancer. 2007;97:1457–64. doi: 10.1038/sj.bjc.6604055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jung AC, Briolat J, Millon R, et al. Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int J Cancer. 2010;126:1882–94. doi: 10.1002/ijc.24911. [DOI] [PubMed] [Google Scholar]
- Junor E, Kerr G, Oniscu A, et al. Benefit of chemotherapy as part of treatment for HPV DNA-positive but p16-negative squamous cell carcinoma of the oropharynx. Br J Cancer. 2012;106:358–65. doi: 10.1038/bjc.2011.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalof AN, Cooper K. p16INK4a immunoexpression: surrogate marker of high-risk HPV and high-grade cervical intraepithelial neoplasia. Adv Anat Pathol. 2006;13:190–4. doi: 10.1097/00125480-200607000-00006. [DOI] [PubMed] [Google Scholar]
- Kim BN, Yamamoto H, Ikeda K, et al. Methylation and expression of p16INK4 tumor suppressor gene in primary colorectal cancer tissues. Int J Oncol. 2005;26:1217–26. [PubMed] [Google Scholar]
- Koerber SA, Schoneweg C, Slynko A, et al. Influence of human papillomavirus and p16(INK4a) on treatment outcome of patients with anal cancer. Radiother Oncol. 2014;113:331–6. doi: 10.1016/j.radonc.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Kreimer AR, Clifford GM, Boyle P, et al. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14:467–75. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- Krishna Rao SV, Mejia G, Roberts-Thomson K, et al. Epidemiology of oral cancer in Asia in the past decade--an update (2000-2012) Asian Pac J Cancer Prev. 2013;14:5567–77. doi: 10.7314/apjcp.2013.14.10.5567. [DOI] [PubMed] [Google Scholar]
- Lai KC, Lee TC. Genetic damage in cultured human keratinocytes stressed by long-term exposure to areca nut extracts. Mutat Res. 2006;599:66–75. doi: 10.1016/j.mrfmmm.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Lai ZL, Tsou YA, Fan SR, et al. Methylation-associated gene silencing of RARB in areca carcinogens induced mouse oral squamous cell carcinoma. Biomed Res Int. 2014;2014:378358. doi: 10.1155/2014/378358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurson J, Khan S, Chung R, et al. Epigenetic repression of E-cadherin by human papillomavirus 16 E7 protein. Carcinogenesis. 2010;31:918–26. doi: 10.1093/carcin/bgq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Chen SF, Chen CL, Ho LY, et al. Role of oxidative DNA damage in hydroxychavicol-induced genotoxicity. Mutagenesis. 1996;11:519–23. doi: 10.1093/mutage/11.5.519. [DOI] [PubMed] [Google Scholar]
- Leonard SM, Wei W, Collins SI, et al. Oncogenic human papillomavirus imposes an instructive pattern of DNA methylation changes which parallel the natural history of cervical HPV infection in young women. Carcinogenesis. 2012;33:1286–93. doi: 10.1093/carcin/bgs157. [DOI] [PubMed] [Google Scholar]
- Lingen MW, Xiao W, Schmitt A, et al. Low etiologic fraction for high-risk human papillomavirus in oral cavity squamous cell carcinomas. Oral Oncol. 2013;49:1–8. doi: 10.1016/j.oraloncology.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Melkane AE, Auperin A, Saulnier P, et al. Human papillomavirus prevalence and prognostic implication in oropharyngeal squamous cell carcinomas. Head Neck. 2014;36:257–65. doi: 10.1002/hed.23302. [DOI] [PubMed] [Google Scholar]
- Mirghani H, Amen F, Moreau F, et al. Do high-risk human papillomaviruses cause oral cavity squamous cell carcinoma? Oral Oncol. 2015;51:229–36. doi: 10.1016/j.oraloncology.2014.11.011. [DOI] [PubMed] [Google Scholar]
- Monzon J, Liu L, Brill H, et al. CDKN2A mutations in multiple primary melanomas. N Engl J Med. 1998;338:879–87. doi: 10.1056/NEJM199803263381305. [DOI] [PubMed] [Google Scholar]
- Nehls K, Vinokurova S, Schmidt D, et al. p16 methylation does not affect protein expression in cervical carcinogenesis. Eur J Cancer. 2008;44:2496–505. doi: 10.1016/j.ejca.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Nemes JA, Deli L, Nemes Z, et al. Expression of p16(INK4A), p53, and Rb proteins are independent from the presence of human papillomavirus genes in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102:344–52. doi: 10.1016/j.tripleo.2005.10.069. [DOI] [PubMed] [Google Scholar]
- Pande P, Mathur M, Shukla NK, et al. pRb and p16 protein alterations in human oral tumorigenesis. Oral Oncol. 1998;34:396–403. doi: 10.1016/s1368-8375(98)00024-4. [DOI] [PubMed] [Google Scholar]
- Rautava J, Kuuskoski J, Syrjanen K, et al. HPV genotypes and their prognostic significance in head and neck squamous cell carcinomas. J Clin Virol. 2012;53:116–20. doi: 10.1016/j.jcv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Esteller M, Wu L, et al. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000;60:892–5. [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Liloglou T, Rogers SN, et al. Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br J Cancer. 2006;94:561–8. doi: 10.1038/sj.bjc.6602972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Primeaux T. Is p16 immunohistochemistry a more cost-effective method for identification of human papilloma virus-associated head and neck squamous cell carcinoma? Ann Diagn Pathol. 2012;16:91–9. doi: 10.1016/j.anndiagpath.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Ukpo OC, Flanagan JJ, Ma XJ, et al. High-risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol. 2011;35:1343–50. doi: 10.1097/PAS.0b013e318220e59d. [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Tsuchida N, Shanmugam G. Promoter hypermethylation profile of tumor-associated genes p16, p15, hMLH1, MGMT and E-cadherin in oral squamous cell carcinoma. Int J Cancer. 2003;105:41–6. doi: 10.1002/ijc.11028. [DOI] [PubMed] [Google Scholar]
- Weinberger PM, Yu Z, Haffty BG, et al. Molecular classification identifies a subset of human papillomavirus-associated oropharyngeal cancers with favorable prognosis. J Clin Oncol. 2006;24:736–47. doi: 10.1200/JCO.2004.00.3335. [DOI] [PubMed] [Google Scholar]
- Yakushiji T, Uzawa K, Shibahara T, et al. Over-expression of DNA methyltransferases and CDKN2A gene methylation status in squamous cell carcinoma of the oral cavity. Int J Oncol. 2003;22:1201–7. [PubMed] [Google Scholar]
- Zafereo ME, Xu L, Dahlstrom KR, et al. Squamous cell carcinoma of the oral cavity often overexpresses p16 but is rarely driven by human papillomavirus. Oral Oncol. 2016;56:47–53. doi: 10.1016/j.oraloncology.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


