Abstract
HPV 16 intratypic sequence variations has been recognized in association with oncogenic potential diverge and geographic distribution. This study aimed to investigate nucleotide modifications and optimization of HPV 16 E7 regions from Iranian infected women. Cervical biopsies from 79/163 HPV 16 positive cancer patients detected in our study were analyzed by PCR in a couple of cloning of a complete ORF of the E7 gene, and sequencing. The most frequently observed variant was C196T in E7 which led to an amino acid change of R66W. In addition, only one common variant T234G was identified from all specimens, but it did not lead to any amino acid change. We also detected nucleotide variations A86G, and C188T in samples. Among 99 codons in E7 gene, 56 codons were improved for Lactococcus lactis subsp. cremoris MG1363 resulting in a reduced G+C content from 43.1% to 34.0%. Also, the AT%, ENC, and CAI values were 66, 20±1.1, and 1.000 instead of 56.90, 60 ±1.1, and 0.406 respectively. Finally we constructed expression vector pNZ8148 encoding optimized E7 oncoprotein of HPV 16. This study declared for the first time, the genetic variations of HPV 16 E7 in IRAN. We conclude that plasmid pNZ8148-HPV 16-opti E7 can be potential vaccine candidates in the future.
Keywords: Human papillomaviruses, HPV 16, E7, lactococcus lactis
Introduction
Cervical cancer is the second important cause of cancer deaths between women worldwide, and it is famous that high-risk types of human papillomaviruses (HPVs) are associated with more than 90% of cervical cancers (Muñoz et al., 2003). According to previous publications, most of the cases of invasive cervical cancers harbor HPV 16 and HPV 18. In detail, E6 and E7 oncogenes have been revealed to contribute to tumor development by functionally deactivating two significant cellular tumor inhibitor proteins, p53 and retinoblastoma. So far, E6 and E7 appear to be the best promising target molecules for therapeutic vaccines because they are constitutively expressed in cervical cancer cells (Zur Hausen, 2002).
The HPV 16 genome has been expansively studied for nucleotide and a number of HPV 16 variants have been identified in separate geographical locations and racial groups. In particular, HPV 16 is distributed into four distinct phylogenetic branches. These four different clusters are designated as (i) European–Asian, (ii) African I, (iii) African II and (iv) North American/Asian American (Cornet et al., 2012).
Findings from several studies recommend that viral codon usage adapted to resemble host gene codon usage can lead to an improved expression of the viral protein. Synonymous codons encoding a specific amino acid are not used with equal frequency irrespective of the degeneracy of the genetic code due to a phenomenon known as codon bias (Ikemura, 1981). The frequencies of codon usage are found to be species specific and also specific across genomes and inside the same genome. It is an evolutionary effect. Codon usage pattern is influence by numerous factors such as compositional bias (GC percent), mutation pressure, natural selection, gene length, expression level, replication, RNA stability, hydrophobicity and hydrophilicity of the protein (Powell et al., 2003).
Food grade lactic acid bacteria (LAB) are progressively used as oral delivery vehicles for therapeutic and prophylactic proteins. In the previous two periods, genetic tools for the model LAB, L. lactis, were established (Ravn et al., 2000). Moreover L. lactis genome is completely sequenced. In order to optimize testing of candidate proteins in LAB, especially genetic protocol need to be optimized to permit the maximum level of protein expressed per cell (Oddone et al., 2007).
Hence, the current study focuses on E7 regions of HPV 16 in order to examine the nucleotide variability among different HPV 16 variants that circulate in the Iranian population. Also, we aimed to optimize a HPV 16 E7 nucleotide sequence using codon usage table from Lactococcus lactis subsp. cremoris MG1363. Regarding replacement of wild-type E7 genes with codon-optimized E7 genes, we created novel E7 plasmid DNA cassettes.
Materials and Methods
Study Design and Population
The study population includes Iranian samples of 166 female between 18 and 62 years of age who were enrolled in a study of HPV infection at the Keyvan specialty virology laboratory between May 2015 and April 2016. This group was selected from patients suffering from cervical carcinoma. Written informed consent was obtained from all participants. The samples of cervical cancer cells were obtained with a Cervex-Brush. The biopsy specimens were fixed in 20% buffered formalin and embedded in paraffin for analyses.
HPV DNA Detection
The tissue fragments were deparaffinized in xylene and rehydration in 95% ethanol, DNA was extracted from the tissues via QIAamp Tissue kit following the manufacturer’s instruction (Qiagen GmbH, Hilden, Germany). Genomic DNA of the samples is used in PCR using the MY09 and MY11 degenerate consensus primers. DNA quality and lack of PCR inhibitors in samples were verified using beta-globin PCR assay (Keyvani et al., 2016).
INNO-LiPA assay
Broad-spectrum HPV DNA amplification was achieved with the PCR fragment (SPF10) primer set supplied by the INNO-LiPA HPV Genotyping Extra kit (Fujirebio, Gent, Belgium). The SPF10 primers amplify a 65 bp segment from the L1 region of the HPV genome. The primer sequences and HPV PCR settings were described previously (Kleter et al., 1998). The results of hybridization were evaluated visually by comparing to the standard grid. This type of the assay includes probes for 32 HPV genotypes.
Sequence analysis of Iranian HPV16 E7 region
DNA fragment from the HPV16 genome spanning nucleotides 7,604 to 7,900 and covering the E7 open reading frames (ORFs) was PCR amplified with primers 5 ’- CCATGGCATGGAGATACACCTACATT -3 ’ and 5 ’- GAGCTCTGGTTTCTGAGAACAGATGG -3 ’. Underlines demonstrate the NcoI and SacI sites respectively. The mixture was incubated for 10 min at 94°C, 40 cycles of 50 s at 56°C, and 40 cycles of 50 s at 72°C, with a final extension of 15 min at 72°C. For PCR product subcloning, 303 bp fragments placed within the E7 HPV gene were cloned into the pTZ57R/T vector (InsTAclone PCR cloning kit, Thermo Scientific). Plasmid was extracted from colonies containing the insert of interest and was sequenced using the universal M13 primer (Bioneer, Korea). To detect the existence of HPV intratype variants within the similar E7 genes of HPV16 isolate, a multiple sequence alignment was accomplished among all the reads that had been clustered in the same HPV type, by using ClustalW, and containing also the reference sequence of the corresponding genotype (ACCESSION: NC_001526).
Codon usage optimization
Based on the integration of codons which are favorably utilized by L. lactis, we took a strategy of rewriting the native gene of HPV 16 pursuant to L. lactis subsp. cremoris MG1363 preferred codon usage to improve the expression of the HPV 16 E7 gene from the recombinant host. At the same time, to regulate G+C content percentage, the latter was considered more close to actual total G+C content percentage in L. lactis. Furthermore, the ENC and CAI values were calculated separately. The effective number of codons (ENC) is usually used parameter to calculate the usage bias of synonymous codons. The ENC value ranges from 20 to 61. Also the codon adaptation index (CAI) is a very widely used factor to measure the comparative adaptedness of the codon usage of a gene to the codon usage of the highly expressed genes. Its value ranges from 0 to 1 (Wright, 1990). Accordingly, the coding sequences of Iranian HPV 16 E7 gene (in FASTA format) having correct codons with an exact multiple of three bases were subjected to codon usage analysis. Finally, the codon usage in the HPV 16 E7 gene was evaluated for L. lactis subsp. cremoris MG1363. It was estimated by an online CAI calculator OPTIMIZER (http://genomes.urv.es/OPTIMIZER) with the codon usage table of L. lactis subsp. cremoris MG1363 tabulated at Kazusa database (http://www.kazusa.or.jp/codon/). The NcoI and SacI sites were added to the 5 ’ and 3 ’ ends of the optimized HPV 16 E7 gene. The resultant fragment was constructed in a PMD18 vector via Biomatik Company (Biomatik Corporation, Cambridge, Canada).
Construction of shuttle vector
The optimized E7 gene, encoding the E7 oncoprotein from HPV 16, was obtained as a 291 bp DNA fragment by digesting plasmid PMD18 with restriction enzymes NcoI and SacI (New England Biolabs). The resultant DNA fragment was ligated into the NcoI/SacI site of pNZ8148 shuttle vector (MoBiTec, Germany). The subsequent plasmid (named pNZ8148-HPV 16-opti E7) was transformed to chemically competent E. coli MC1061 cells (MoBiTec, Germany). Bacteria were plated onto Luria–Bertani plates having 10µg/mL chloramphenicol. Validity of the constructed plasmid was determined by sequencing of pNZ8148-HPV 16-opti E7 vector.
Results
Detection of HPV in clinical samples
Out of 166 formalin-fixed paraffin embedded cervical cancer samples, 163 were found to be HPV positive. MY09/11 PCR could not identify the existence of HPV DNA in 1.80% (3/166) of samples. All 166 cervical carcinomas were β-globin gene positive. All HPV positive samples (163/166, 98.19%) were subjected to typing assay via INNO-LiPA. The results showed that the most common HPV type was HPV 16 with a prevalence of 48.46% (79/163) followed by 9.81% HPV 31, 7.97% HPV 39, 6.74% HPV 51, 6.13% HPV 53, 5.52% HPV 18, and HPV 66, 3.06% HPV 58, 2.45% HPV 35, 1.84% HPV 52, 1.22% HPV 56, 0.61% HPV 45, and HPV 68. A single HPV type was discovered in 53.37% (87/163) of the genotyped samples, and multiple HPV types were identified in the remaining 46.62% (76/163). Among multiple infections, HPV 31, HPV 53, and HPV 66 followed by HPV 16, were the most commonly detected types.
Nucleotide analysis
Sequencing of clones from independent PCR reactions exposed the E7 gene consists of 303 bp in length. Sequence analysis of the HPV 16 E7 gene revealed that this ORF appears to be more weakly conserved among Iranian cervical samples. Hence, four nucleotide modifications were recognized in the current report. The results demonstrated that the most predominant variant was C196T which led to an amino acid change of R66W in 72.39% of the specimens. Also, the nucleotide variations A86G, and C188T were detected in cervical samples. These variations induce the amino acid changes N29S, and S63F, respectively. The common variant detected from all the cervical samples was T234G, but it did not lead to any amino acid (Threonine) change. The details of E7 mutations are summarized in Table 1.
Table 1.
Sequence Variation Analysis of HPV16 E7 Orfs in Iranian Cervical Samples
| Position | Wild Type Codon | Wild Type Amino Acid | Mutant Codon | Mutant Amino Acid | Rate |
|---|---|---|---|---|---|
| 85-87 | AAT | Asparagine (N) | AGT | Serine (S) | (17/163) 10.42% |
| 187-189 | TCT | Serine (S) | TTT | Phenylalanine (F) | (94/163) 57.66% |
| 196-198 | CGG | Arginine (R) | TGG | Tryptophan (W) | (118/163) 72.39% |
| 232-234 | ACT | Threonine (T) | ACG | Threonine (T) | (163/163) 100.0% |
Codon bias of E7 oncogene of HPV16 in L. lactis subsp. cremoris MG1363
The native HPV 16 E7 gene contains 297 bp with 99 codons and an overall G+C content of 43.10%. Of the 99 codons present, 39.39% of these had a G or a C at the third base location. In contrast, GC% content in the third position of codons in L. lactis subsp. cremoris MG1363 was 26.78%. From Figure 1, it was suggested that the distribution of A, T, G, and C% among the E7 oncogene codons was unequal in specie of L. lactis subsp. cremoris MG1363. The number of codon usage frequency presented in L. lactis subsp. cremoris MG1363 described in the Figure 2. We found 56 of the total 99 codons that are infrequently used in L. lactis subsp. cremoris MG1363 including TTA/TTG/CTC/CTA/CTG (L), ATC/ATA (I), GTA/GTG (V), TCT/AGT/AGC (S), CCT/CCC/CCG (P), ACC/ACA/ACG (T), GCC/GCA (A), TAC (Y), CAC (H), CAG (Q), AAG (K), GAC (D), GAG (E), TGC (C), AGA (R), GGC/GGA (G) (Table 2). Consider the following optimization, G+C content reduced from 43.1% for the native gene to 34.0% for the adapted form. Also the AT%, ENC, and CAI values were 66, 20±1.1, and 1.000 instead of 56.90, 60 ±1.1, and 0.406 respectively. Overall, alignment between wild type and optimized E7 genes exhibited in Figure 3.
Figure 1.
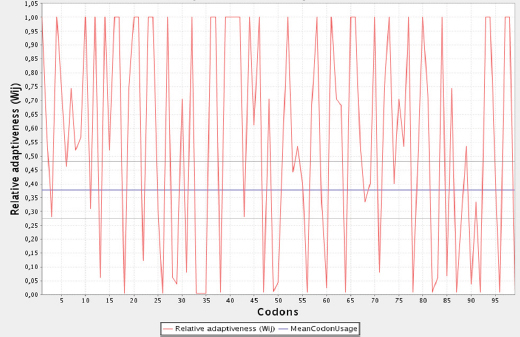
The Scatter Diagram of Relative Addictiveness against Codons of Wild Type E7 Gene of HPV16 in Lactococcus Lactis Subsp. Cremoris MG1363
Figure 2.
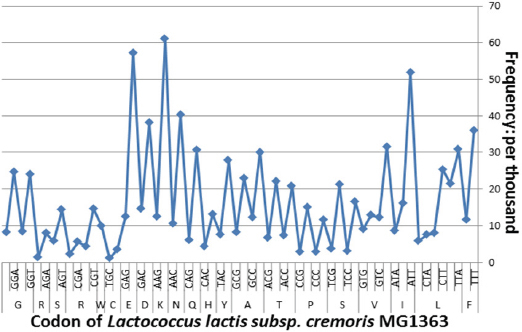
Overall Codon Usage Frequency of Lactococcus Lactis Subsp. Cremoris MG1363 Genes
Table 2.
Codon Usage Table for the Wild Type and Codon Optimized Iranian E7 Gene Of HPV16 Based on the Relative Codon Usage Frequencies of Lactococcus Lactis Subsp. Cremoris MG1363
| Amino Acid | Codon | Host fraction | Wild Type Number | Optimized Number |
|---|---|---|---|---|
| F | TTT | 0.76 | 2 | 2 |
| TTC | 0.24 | 0 | 0 | |
| L | TTA | 0.31 | 3 | 0 |
| TTG | 0.22 | 4 | 0 | |
| CTT | 0.26 | 1 | 11 | |
| CTC | 0.08 | 1 | 0 | |
| CTA | 0.08 | 1 | 0 | |
| CTG | 0.06 | 1 | 0 | |
| I | ATT | 0.68 | 3 | 5 |
| ATC | 0.21 | 1 | 0 | |
| ATA | 0.11 | 1 | 0 | |
| V | GTT | 0.48 | 0 | 4 |
| GTC | 0.19 | 0 | 0 | |
| GTA | 0.2 | 3 | 0 | |
| GTG | 0.14 | 1 | 0 | |
| S | TCT | 0.25 | 1 | 0 |
| TCC | 0.05 | 0 | 0 | |
| TCA | 0.33 | 1 | 5 | |
| TCG | 0.06 | 0 | 0 | |
| P | CCT | 0.36 | 1 | 0 |
| CCC | 0.09 | 1 | 0 | |
| CCA | 0.46 | 3 | 6 | |
| CCG | 0.09 | 1 | 0 | |
| T | ACT | 0.36 | 1 | 9 |
| ACC | 0.13 | 1 | 0 | |
| ACA | 0.39 | 5 | 0 | |
| ACG | 0.12 | 2 | 0 | |
| A | GCT | 0.41 | 1 | 3 |
| GCC | 0.17 | 1 | 0 | |
| GCA | 0.31 | 1 | 0 | |
| GCG | 0.11 | 0 | 0 | |
| Y | TAT | 0.78 | 2 | 4 |
| TAC | 0.22 | 2 | 0 | |
| H | CAT | 0.75 | 3 | 4 |
| CAC | 0.25 | 1 | 0 | |
| Q | CAA | 0.84 | 4 | 5 |
| CAG | 0.16 | 1 | 0 | |
| N | AAT | 0.79 | 1 | 1 |
| AAC | 0.21 | 0 | 0 | |
| K | AAA | 0.83 | 1 | 2 |
| AAG | 0.17 | 1 | 0 | |
| D | GAT | 0.72 | 5 | 10 |
| GAC | 0.28 | 5 | 0 | |
| E | GAA | 0.82 | 4 | 9 |
| GAG | 0.18 | 5 | 0 | |
| C | TGT | 0.76 | 4 | 7 |
| TGC | 0.24 | 3 | 0 | |
| W | TGG | 1 | 1 | 1 |
| R | CGT | 0.4 | 1 | 2 |
| CGC | 0.12 | 0 | 0 | |
| CGA | 0.15 | 0 | 0 | |
| CGG | 0.06 | 0 | 0 | |
| S | AGT | 0.22 | 1 | 0 |
| AGC | 0.09 | 2 | 0 | |
| R | AGA | 0.22 | 1 | 0 |
| AGG | 0.04 | 0 | 0 | |
| G | GGT | 0.37 | 1 | 5 |
| GGC | 0.13 | 1 | 0 | |
| GGA | 0.38 | 3 | 0 | |
| GGG | 0.13 | 0 | 0 |
Figure 3.

Nucleotide Alignment between the Adapted and Wild-Type E7 Gene Sequence along with Amino Acid Composition.
Construction of shuttle plasmid
E. coli-L. lactis shuttle vector was created using the fragments of pNZ8148 shuttle vector and ligated as outlined in materials and methods section. To check the sequence around the NcoI and SacI locations, digestion using enzymes were carried out. Bands of 3135 and 303 bp are shown in Figure 4, revealing that pNZ8148 containing the expression cassettes pNZ8148-HPV 16-opti E7 was successfully constructed. Sequence analysis confirmed the presence of optimized E7 gene in the pNZ8148.
Figure 4.
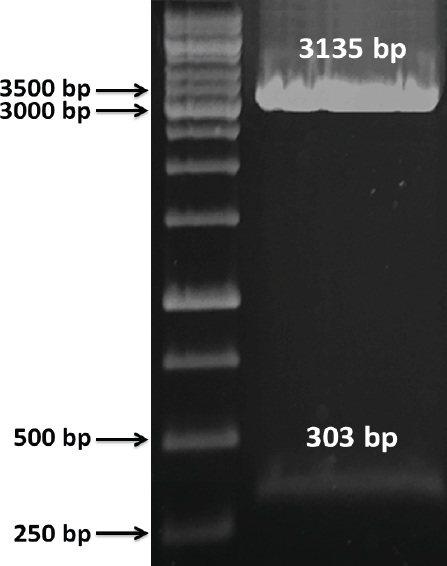
The Double Digestion Patterns of the pNZ8148-HPV16-optiE7 Shuttle Plasmid via NcoI and SacI Restriction Endonuclease
Discussion
In the current study, we characterized and optimized the nucleotide modifications of the HPV 16 isolates amongst infected Iranian women with cervical cancer based on the E7 viral oncogenes. To date, there are no molecular epidemiological reports on HPV 16 E7 gene variants accomplished in IRAN. A recent meta-analysis reported frequency of HPV DNA in 89.7% of invasive cervical cancer case from Eastern Asia; HPV 16, 18, 58, 52, 33, 31, 45, and 59 were the greatest usual HPV types identified (Singh et al., 2016). Former examinations in diverse regions of Iran proven HPV type 16 as the highest oncogenic type of HPV associated with cervical cancer (Salehi-Vaziri et al., 2016). Our results in accordance with earlier results of IRAN show that HPV 16 was the most common High-risk subtypes, occurring in 79/163 cases (48.46%). Also, HPV 31 was the second most common subtype detected (16/163, 9.98%).
Amino acid mutations in HPV 16 E7 variants should be considered in the vaccine design and evasion of the natural humoral and cellular immune response (Wu et al., 2006). Furthermore, any change in the transforming gene E7 may lead to different biological function and oncogenicity of the proteins encoded, which can affect the natural history of HPV infection. Several studies, have reported a high incidence of E7 gene mutation (65 to 75%) in cervical cancer patients (de Boer et al., 2004). Remarkably, a recent report from China showed that 100% of cervical cancer cases had nucleotide variations in the HPV 16 E7 region (Wu et al., 2006). Boumba et al., (2015) explained two common silent variations and described mutations N29S among isolates from Asia, and Africa (23%). The significance of this amino acid change was recommended by Zehbe et al., (1998) due to its location in an immunoreactive area. Moreover, they founded the D21N mutation (G622A). Tsakogiannis et al., recognized seven new amino acid substitutions H2Y, M12K, A42T, M84I, G85S, G85D, and T86I within the E7 gene in cervical samples (Tsakogiannis et al., 2013). Our data are in agreement with previous studies representing the amino acid replacements are presented in Iranian women with cervical cancer. We observed a weakly conserved E7 region; as high as 72.39 % of patients displayed sequence variation. Our results explain the amino acid substitutions N29S (10.42 %) was located at the N-terminal domain of the E7 oncoprotein because of the nucleotide changes A86G. Also, the amino acid substitutions S63F (57.66 %), and R66W (72.39 %) were positioned at the C-terminal domain of the E7 oncoprotein due to the nucleotide changes C188T, and R196W respectively. All of Iranian cervical cancer showed variation T234G in the form of silent mutation, thus having no effect on protein.
Codon optimization is another important aspect of this study. It has been widely and successfully used to improve the expression level of foreign protein in different expression system such as P. pastoris, E. coli BL21 (DE3), Aspergillus niger, and mammalian cells. The basic genetics features of L. lactis look a lot like those of E. coli, so yields are also likely to be influenced by Shine–Dalgarno sequences, plasmid copy numbers, promoters, and mRNA secondary structures (Oddone et al., 2007). Separately from the prospect of an increased yield, elimination of rare codons has the potential of improving the quality of the expressed protein and creating it less prone to immature turnover. It is known that rare codons can disturb mRNA stability and its translation rate, and gene containing A+ T-rich sequence of terminator consensus-like may cause fortuitous polyadenylation, and cryptic splice site may result in truncated products. Therefore, some elements should be considered before designing an optimized synthetic gene for expression (Wright, 1990). In detail, optimized G+C content, irrespective of corresponding codon optimization, can contribute to increase translational efficiency in the heterologous expression. Usually, the high G+C content can result in incorrect pair and unsuccessful expression (Sinclair and Choy, 2002). There are exist several literature about codon optimization in L. lactis such as interleukin-2 (CAI 0.22), and interleukin-6 (CAI 0.3) from Mus musculus, egg white lysozyme (CAI 0.23), and Tetanus toxin fragment C (CAI 0.33) from Gallus domesticus, and Clostridium tetani, respectively, peptidase pep N (CAI 0.55), and peptidase pep X (CAI 0.49) from lactobacillus helveticus, and Urease ureB (CAI 0.51) from Helicobacter pylori (Fuglsang, 2003). The data are important in light of the CAI values of the naturally occurring CAI-values of genes in L. lactis. Therefore, it becomes clear that CAI-values of 0.3 or lower are very unusual. Thus, the presented cases such as interleukin-2, interleukin-6, and hen egg white lysozyme provide some very worthy definition of genes that subject to codon optimization in order to get higher yields (Fuglsang, 2003). Talarico et al., (2005) also detected the expression of a pyruvate decarboxylase from the Gram positive bacterium S. ventriculi in B. megaterium was successful with a CAI value of 0.60. But genes isolated from other organisms like A. pasteurianus, Zymononas mobilis, and Saccharomyces cerevisiae were not expressed. The CAI values were merely 0.19, 0.32, and 0.34, respectively. Sinclair and Choy reported a consistent result that reduction of GC content would lead to 7.5% increase in the expression of human glucocerebrosidase in Pichia pastoris cells (Sinclair and Choy, 2002).
In the present study, we designed synthetic construct coding human papillomavirus 16 E7 gene. Most of codons of the synthetic gene are copied from the most frequently used codons in L. lactis subsp. cremoris MG1363 to prevent the possible depletion of tRNAs encoding the same amino acid. Regarding the fact, the ENC, and CAI value of E7 antigen estimated 20±1.1, and 1.000, respectively, which suggest that the high expression level of E7 antigen in bacterium L. lactis and thereby enhance the immunogenicity of DNA vaccines against human papillomavirus 16 (HPV 16). In our study, the GC content of E7 gene was reduced from 43.1% to 34% because L. lactis subsp. cremoris MG1363 has low GC content (36.72%).
In conclusions, findings confirmed the predominance of the amino acid modifications among Iranian cervical samples, and may offer fundamental information for evolving concepts in the diagnosis and as well as designing vaccines for a specific population. To the best of our knowledge, this is the first study describing nucleotide variations and codon usage within the E7 region of HPV 16 genomes from the Iranian population.
Conflict of interest
The authors have no conflicts of interest to declare.
Acknowledgments
The authors would like to thank Sedigheh Taghinezhad Saroukalaei for technical support and performing the INNO-LiPA assay in the study. This work was financially supported by the grants of the Keyvan virology specialty laboratory (KVSL) in cooperation with Islamic Azad University, Science and Research Branch (SRBIAU) (Tehran, Iran).
References
- Boumba LMA, Assoumou SZ, Hilali L, et al. Genetic variability in E6 and E7 oncogenes of human papillomavirus Type 16 from Congolese cervical cancer isolates. Infect Agent Cancer. 2015;10:15–21. doi: 10.1186/s13027-015-0010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet I, Gheit T, Franceschi S, et al. Human papillomavirus type 16 genetic variants: phylogeny and classification based on E6 and LCR. J Virol. 2012;86:6855–61. doi: 10.1128/JVI.00483-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer MA, Peters LA, Aziz MF, et al. Human papillomavirus type 16 E6, E7, and L1 variants in cervical cancer in Indonesia, Suriname, and The Netherlands. Gynecol Oncol. 2004;94:488–94. doi: 10.1016/j.ygyno.2004.05.037. [DOI] [PubMed] [Google Scholar]
- Fuglsang A. Lactic acid bacteria as prime candidates for codon optimization. Biochem Biophys Res Commun. 2003;312:285–91. doi: 10.1016/j.bbrc.2003.10.120. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. italic>J Mol Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Keyvani H, Saroukalaei ST, Mohseni AH. Assessment of the human cytomegalovirus UL97 gene for identification of resistance to ganciclovir in Iranian immunosuppressed patients. Jundishapur J Microbiol. 2016;9:31733–38. doi: 10.5812/jjm.31733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleter B, van Doorn L-J, ter Schegget J, et al. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am J Pathol. 1998;153:1731–9. doi: 10.1016/S0002-9440(10)65688-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz N, Bosch FX, de Sanjosé S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–27. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- Oddone GM, Lan CQ, Rawsthorne H, et al. Optimization of fed-batch production of the model recombinant protein GFP in Lactococcus lactis. Biotechnol Bioeng. 2007;96:1127–38. doi: 10.1002/bit.21192. [DOI] [PubMed] [Google Scholar]
- Powell JR, Sezzi E, Moriyama EN, et al. Analysis of a shift in codon usage in Drosophila. J Mol Evol. 2003;57:214–25. doi: 10.1007/s00239-003-0030-3. [DOI] [PubMed] [Google Scholar]
- Ravn P, Arnau J, Madsen SM, et al. The development of TnNuc and its use for the isolation of novel secretion signals in Lactococcus lactis. Gene. 2000;242:347–56. doi: 10.1016/s0378-1119(99)00530-2. [DOI] [PubMed] [Google Scholar]
- Salehi-Vaziri M, Sadeghi F, Hashemi FS, et al. Distribution of human papillomavirus genotypes in Iranian women according to the severity of the cervical lesion. Iran Red Crescent Med J. 2016;18:24458–63. doi: 10.5812/ircmj.24458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair G, Choy FY. Synonymous codon usage bias and the expression of human glucocerebrosidase in the methylotrophic yeast, Pichia pastoris. Protein Expr Purif. 2002;26:96–105. doi: 10.1016/s1046-5928(02)00526-0. [DOI] [PubMed] [Google Scholar]
- Singh G, Sharma D, Gupta N. Seroepidemiology of human papilloma virus and HPV vaccination in cervical cancer–Era of new hope: A brief review of article. Int J Med Paediatr Oncol. 2016;2:88–91. [Google Scholar]
- Talarico LA, Gil MA, Yomano LP, et al. Construction and expression of an ethanol production operon in Gram-positive bacteria. Microbiology. 2005;151:4023–31. doi: 10.1099/mic.0.28375-0. [DOI] [PubMed] [Google Scholar]
- Tsakogiannis D, Papadopoulou A, Kontostathi G, et al. Molecular and evolutionary analysis of HPV16 E6 and E7 genes in Greek women. J Med Microbiol. 2013;62:1688–96. doi: 10.1099/jmm.0.055491-0. [DOI] [PubMed] [Google Scholar]
- Wright F. The effective number of codons used in a gene. Gene. 1990;87:23–9. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Wu Y, Chen Y, Li L, et al. Analysis of mutations in the E6/E7 oncogenes and L1 gene of human papillomavirus 16 cervical cancer isolates from China. J Gen Virol. 2006;87:1181–8. doi: 10.1099/vir.0.81649-0. [DOI] [PubMed] [Google Scholar]
- Zehbe I, Wilander E, Delius H, et al. Human papillomavirus 16 E6 variants are more prevalent in invasive cervical carcinoma than the prototype. Cancer Res. 1998;58:829–33. [PubMed] [Google Scholar]
- Zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–50. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]


