Abstract
Glycyl-tRNA synthetase (GARS; OMIM 600287) is one of thirty-seven tRNA-synthetase genes that catalyses the synthesis of glycyl-tRNA, which is required to insert glycine into proteins within the cytosol and mitochondria. To date, eighteen mutations in GARS have been reported in patients with autosomal-dominant Charcot-Marie-Tooth disease type 2D (CMT2D; OMIM 601472), and/or distal spinal muscular atrophy type V (dSMA-V; OMIM 600794). In this study, we report a patient with clinical and biochemical features suggestive of a mitochondrial respiratory chain (MRC) disorder including mild left ventricular posterior wall hypertrophy, exercise intolerance, and lactic acidosis. Using whole exome sequencing we identified compound heterozygous novel variants, c.803C>T; p.(Thr268Ile) and c.1234C>T; p.(Arg412Cys), in GARS in the proband. Spectrophotometric evaluation of the MRC complexes showed reduced activity of Complex I, III and IV in patient skeletal muscle and reduced Complex I and IV activity in the patient liver, with Complex IV being the most severely affected in both tissues. Immunoblot analysis of GARS protein and subunits of the MRC enzyme complexes in patient fibroblast extracts showed significant reduction in GARS protein levels and Complex IV. Together these studies provide evidence that the identified compound heterozygous GARS variants may be the cause of the mitochondrial dysfunction in our patient.
Introduction
Aminoacyl-tRNA synthetases (ARS) are ubiquitously expressed essential enzymes responsible for attaching amino acid residues to their cognate tRNA molecules, which is the first step of protein translation in the cytoplasm and mitochondria [1]. Human ARS proteins consist of three groups: cytoplasmic, mitochondrial, and bifunctional. For most ARS, the cytoplasmic and mitochondrial ARS for each amino acid are encoded by distinctly different nuclear genes. However, GARS (OMIM: 601472) and KARS (OMIM: 613641) encode both the cytoplasmic and mitochondrial ARS [2]. It is noteworthy that nuclear genes encode the mitochondrial ARSs that are then imported into mitochondria [2].
The cytoplasmic and mitochondrial isoforms of glycyl-tRNA synthetase, encoded by GARS, differ by a 54 amino acid N-terminal mitochondrial targeting sequence [3,4]. Mitochondrial GARS is required for the synthesis of 13 subunits of the MRC complexes, including subunits of Complex I, Complex III, Complex IV and Complex V [2]. The human GARS belongs to the class IIA aminoacyl- tRNA synthetases, with the cytosolic isoform having 685 amino acids and the mitochondrial isoform having 739 amino acids [4]. Both isoforms include the N-terminal WHEP-TRS domain composed of the amino acid residues 62–122, a catalytic domain (124–608), and a C terminal anticodon-binding domain (602–726) [4].
Missense mutations in GARS were first described in 2003 in association with Charcot-Marie-Tooth disease type 2D (CMT2D: OMIM 601472), and distal Spinal Muscular Atrophy type V (dSMA-V; OMIM 600794) in five families with atrophy and weakness of the hand muscles [5]. Several reports have since been published detailing pathogenic GARS missense mutations, mostly autosomal dominant, but some de novo [1,5–12] (Table 1).
Table 1. Clinical phenotypes associated with GARS variants in human.
| Kawakami et al. (2014) |
Del Bo et al. (2006) |
Antonellis et al. (2003) |
Rohkamm et al. (2007) |
Sivakumar et al. (2005) Antonellis et al. (2003) |
Sivakumar et al. (2005) Antonellis et al. (2003) |
Lee et al. (2012) |
Lee et al. (2012) |
Liao et al. (2015) |
Liao et al. (2015) |
Sivakumar et al. (2005) Antonellis et al. (2003) |
Hamaguchi et al. (2010) |
James et al. (2006) |
James et al. (2006) |
James et al. (2006) |
Klein et al. (2014) |
Sivakumar et al. (2005) |
Dubourg et al. (2006) |
McMillan at al. (2014) |
This Study | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
GARS variants |
c.815T>A p.Leu218Gln |
c.2016G>A p. Asp500Asn |
2094G>C p. Gly526Arg |
c.688 C > T p.Ala57Val |
c.730A>G p.Glu71Gly |
c.904C>T p.Leu129Pro |
c.598G>A p. Asp200Asn |
c.794C>T p. Ser265Phe |
c.598G>T p. Asp146Tyr |
c.875T>G p.Met238Arg |
c.1236G>C p.Gly240Arg |
c.893C>T p. Pro244Leu |
c.1358A>T p.Ile280Phe |
c.2313G>C p.Gly598Ala |
c.2260C>T p. Ser581Leu |
p.Ile334Phe |
p.His418Arg |
c.2094 A>G p.Gly526Arg |
c.1904C > T p.Ser635Leu c.1787G > A p.Arg596Gln |
c.803C>T p.Thr268Ile c.1234C>T p.Arg412Cys |
| Inheritance | AD | AD | AD | AD | AD | AD | AD | AD | De novo | De novo | AD | ND | AD | De novo | AD | AD | AD | AD | AR | AR |
| Protein domain potentially affected |
CD | Ins III | CD | WD | CD | CD | Ins I | Ins III | Ins I | CD | CD | CD | CD | ACBD | ACBD | Ins II | CD | CD | ACBD | CD |
| Phenotype | CMT2 | CMT2D/ dSMA-V |
dSMA-V | dHMN-V | CMT2D/ dSMA-V |
dSMA-V | dHMN- V | dHMN- V | CMT2 | CMT2 | CMT2D | CMT2 | dHMN- V | Infantile SMA | CMT2 | CMT2 | dSMA-V | dHMN-V | MRCD | MRCD |
| Cohort | 1 affected | 4 affected | 2 affected | 1 affected | 17 affected | 5 affected | 1 affected | 1 affected | 1 affected | 1 affected | 14 affected | 1 affected | 1 affected | 1 affected | 1 affected | 1 affected | 1 affected | 16 affected | 1 affected | 1 affected |
| Age at onset |
<2 Years | 10–35 Years (average) |
13–26 Years (average) |
12 Years | 18 Years (average) | 16.9 Years (average) |
15Years | 13Years | <6 months | 2 Years | 23 Years (average) |
10 Years | 11Years | <6 m | 27 Years | 24 Years | 26Years (average) |
23.3Years (average) |
7 Years | 6 Years |
| Consanguinity | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | No | NS | NS | NS | NS | NS | No | No | No |
| Ethnicity | Japan | Italy | Sephardic Jewish |
Ghana | Mongolia | Bulgaria | Korea | Korea | Taiwan | Taiwan | North America | Japan | UK | UK | UK | USA | UK/Australia | France | ND | UK/Australia |
| Clinical features | delayed onset of walking, slow running | atrophy and weakness of the hand muscles | atrophy and weakness of the hand muscles | atrophy and weakness of the hand muscles | atrophy and weakness of the hand muscles | atrophy and weakness of the hand muscles | atrophy and weakness of the hand muscles | atrophy and weakness of the hand muscles | delayed milestones, severe generalised weakness | delayed onset of walking, unsteady gait | atrophy and weakness of the hand muscles | slow running | distal limb muscle atrophy and weakness | delayed milestones, severe generalised weakness | distal limb muscle weakness | progressive unsteadiness walking | atrophy and weakness of the hand muscles | atrophy and weakness of the hand muscles or distal four limbs | leukoencephalopathy, lactic acidosis and myalgia | fatigue, exercise-lactic acidosis, mild cardiomyopathy |
| Functional studies | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | MRC enzymology, immunoblotting for GARS protein and subunits for MRC enzyme complexes |
ND = not done; NS–not stated; CD: Catalytic domain, DI: dimer interface, WD: WHEP domain, Ins I: Insertion I domain, Ins II: Insertion II domain, Ins III: Insertion III domain, ACBD: anticodon binding domain; AD: autosomal dominant, AR: autosomal recessive; CMT2: Charcot-Marie-Tooth hereditary neuropathy type 2; CMT2D: upper limb predominant CMT2; SMA: Spinal muscular atrophy; dSMA: distal SMA, dHMN: distal hereditary motor neuropathy type V, MRC: Mitochondrial respiratory chain, MRCD: Mitochondrial respiratory chain disorder.
However, McMillan and colleagues reported compound heterozygous variants in GARS for the first time, in a 12-year old girl with clinical manifestations suggestive of mitochondrial disease, including exercise-induced myalgia, non-compaction cardiomyopathy, lactic acidaemia, and abnormal T2 and T2 FLAIR hyperintensities in the periventricular and trigonal white matter bilaterally on magnetic resonance imaging (MRI)[13]. Herewith, we report the identification of compound heterozygous variants in the GARS gene in a 16 year-old female who presented with exercise-intolerance, mild cardiomyopathy and lactic acidosis. Also, our patient had clear evidence of a MRC enzyme deficiency, supporting an expansion of the phenotypic spectrum associated with GARS mutations to include mitochondrial disease in cases of autosomal recessive inheritance.
Material and methods
Patient clinical summary
The proband is a 16 year-old female of non-consanguineous Caucasian origin of British descent. She is the elder of two siblings with each parent being of elite athletic calibre (one represented their country in the Olympics). Difficulty with exercise was first noted at the age of 6 years when the proband started vomiting and became pale and lethargic on a cross-country walk. An echocardiogram and a 24 Holter assessment at the age of 9 years were normal. At this time, she completed isotonic exercise tests using the standard Bruce protocol, completing 8 minutes and 35 seconds, stopping due to breathlessness and a “sore chest.” She reached her anaerobic threshold early at 2.03 minutes with VO2 max being 57% of predicted. At 11 years of age, the Bruce exercise test was repeated by an exercise physiologist. She fatigued at 6.12 minutes having a VO2 max of 16.8 ml/kg/min (37% of predicted), with a very high respiratory exchange ratio (VCO2 /VO2) of 1.6.
The respiratory exchange ratio increases with increasing exercise intensity and is an indirect reflection of proportion of energy derived from carbohydrate metabolism vs. lipid oxidation. The high respiratory exchange ratio was consistent with CHO metabolism as the predominant energy source [14]. A respiratory exchange ratio > 1.1 is generally accepted as indicative of maximal exercise in adults and >1.0 in children [15]. Also, she had a low O2 pulse (defined as VO2/heart rate) of 3 mL/beat (41% of predicted) and a high ventilatory equivalent for O2 (as defined by VE/VO2) of 78 (169% of predicted), both of which are hallmarks of mitochondrial myopathy. Pre-exercise serum lactate was 2.7 mmol/L while after exercise this was 7.0 mmol/L (ref 0–2.0 mmol/L), with a ratio to pyruvate of 70. MRC enzyme assays were consequently performed demonstrating low Complex I, III and IV activity in skeletal muscle (18%, 17%, and 1% relative to citrate synthase respectively) and low Complex I and IV in liver (53% and 6% relative to citrate synthase respectively). There was no evidence of mitochondrial DNA depletion, deletion or duplications in these samples.
She performs academically well in mainstream education and participates recreationally in sailing. Systemic examination has been normal, in particular with normal tone, power and deep tendon jerk reflexes. She is post pubertal with weight on 40th percentile and height on 75th percentile. Electrolytes, liver function tests, full blood count and clotting, have been normal. At the age of 14 years, mild left ventricular posterior wall hypertrophy was identified measuring 13 mm with normal function.
Exome sequencing
Genomic DNA was isolated from whole blood of the proband and both parents. Exome capture and library preparation was performed using the Nextera Rapid Capture kit (Illumina, San Diego, CA). Captured libraries were sequenced on an Illumina HiSeq 2000 (2 ×100 nucleotides) to a depth such that a minimum 80% of targeted bases were sequenced to a read depth of 0020× or greater. Reads were aligned to the reference human genome (GRCh37) using BWA-MEM [16], and pedigree informed variant calling was performed using the Real Time Genomics (Hamilton, New Zealand) integrated analysis tool rtgFamily v3.6.2 [17]. All variants were annotated using SnpEff v4.2 [18], SnpEff GRCh37.72 database, dbSNP138, and dbNSFP v2.9. Rare variants (MAF <0.01) were identified and assessed as previously described for their potential to disrupt protein function under different inheritance models using a custom-built interpretation interface incorporating evidence including minor allele frequency, conservation, predicted pathogenicity, and disease association [19].
Variant validation
The GARS variants identified by WES analysis were confirmed by Sanger sequencing of DNA in the proband and her parents, using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit, and ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
In silico analyses
In silico analyses of the GARS variants were performed using PolyPhen2 (http://www.genetics.bwh.harvard.edu/pph2/), SIFT (http://www.sift.jcvi.org), MutationTaster (www.mutationtaster.org/), GVGD (www.agvgd.iarc.fr/), PhyloP (www.ccg.vital-it.ch/mga/hg19/phylop), PhastCons (www.compgen.bscb.cornell.edu), ExAC (http://exac.broadinstitute.org/) and HOPE (http://www.cmbi.ru.nl/hope).
Immunoblotting
Immunoblotting was performed as previously described [20], with the following modifications. Membranes were probed with 1:500 anti-OXPHOS (ab110411, Abcam) for 2 h at room temperature or with 1:1000 anti-GARS (ab89522, Abcam) overnight at 4°C. Protein loading was normalised to porin (1:5,000 anti-porin, ab14734, Abcam) for 2 h at room temperature. Densitometry was performed as previously described [20].
Spectrophotometric MRC enzyme activity
MRC enzyme activities in liver and skeletal muscle biopsies from the patient were determined as previously described [21].
Complex I and IV dipstick enzyme activity assays in fibroblasts
Complex I and IV dipstick enzyme activities were determined as previously described [20].
Statistical analyses
Graphpad Prism 5.03 was used for all statistical analyses. Statistical analyses were performed using the non-parametric Mann-Whitney U test. A P value less than 0.05 was considered to be statistically significant.
Results
The presence of exercise intolerance and lactic acidosis in our patient led to an initial evaluation of MRC enzymology in skeletal muscle and liver extracts. Complex IV activity was almost undetectable in skeletal muscle, with a marked deficiency of Complex I and Complex III, suggestive of a defect in mitochondrial DNA maintenance or expression (Table 2). In the liver, Complex IV activity was markedly deficient and Complex I activity borderline low. Complex II and citrate synthase were elevated in both muscle and liver (Table 2). Additionally, we evaluated the activity of the MRC complexes I and IV in patient fibroblast extracts where we observed that the % residual activity relative to protein (% protein) were 75% (P = 0.0004) in Complex I and 45% (P < 0.0001) in Complex IV, compared to controls (Table 3). These results were consistent with the MRC enzymology results in patient skeletal muscle and liver homogenates, although the MRC deficiency was not as severe in fibroblasts.
Table 2. Spectrophotometric MRC enzyme diagnostic data in skeletal muscle, liver and fibroblasts.
Patient liver and muscle samples. Data are expressed as activity relative to protein and as % CS ratio, which represents % of the normal control mean value when expressed relative to Citrate Synthase. Bold characters indicate clinically significant abnormal values (H–high, L–low). Complex I (CI), NADH-coenzyme Q1 oxidoreductase; Complex II (CII), succinate-coenzyme Q1 oxidoreductase; Complex III (CIII), decylbenzylquinol-cytochrome c oxidoreductase; Complex IV, cytochrome c oxidase (CIV).
| Enzyme Activity | % CS Ratio | |||
|---|---|---|---|---|
| Muscle | Patient | (Ref Range) | Patient | (Ref Range) |
| CI (nmol/min/mg) | 18 | 19–72 | 18 L | 36–269 |
| CII (nmol/min/mg) | 142 H | 26–63 | 126 | 52–156 |
| CIII (/min/mg) | 13.1 | 12.8–50.9 | 17 L | 62–185 |
| CIV (/min/mg) | 0.21 L | 3.3–9.1 | 1 L | 36–192 |
| CS (nmol/min/mg) | 320 H | 85–179 | - | - |
| Liver | ||||
| CI (nmol/min/mg) | 6 | 8–11 | 53 | 65–137 |
| CII (nmol/min/mg) | 116 H | 54–73 | 158 | 59–127 |
| CIII (/min/mg) | 11.7 | 5.2–10.3 | 127 | 77–127 |
| CIV (/min/mg) | 0.05 L | 0.5–0.9 | 6 L | 75–134 |
| CS (nmol/min/mg) | 33 | 26–31 | - | - |
Table 3. Dipstick MRC enzyme data from cultured fibroblasts.
Enzyme activity data are expressed as % residual activity relative to protein (% protein).
| Enzyme Activity % | ||
|---|---|---|
| Skin fibroblasts | Patient | Control |
| Complex I | 75% (p = 0.0004) | 100% |
| Complex IV | 45% (p < 0.0001) | 100% |
Mitochondrial deletion/duplication screening was undertaken, but no structural abnormalities of the mitochondrial genome were identified. Thus, we performed whole exome sequencing and identified compound heterozygous variants, c.803C>T [p.(Thr268Ile), rs2230310] and c.1234C>T [p.(Arg412Cys), rs770924455], in GARS (NM_002047.3). Sanger sequencing, with each parent carrying one of the mutations (Fig 1A and 1B) confirmed both variants. Similarly, to the previous study by McMillan et al. (2014), neither parent showed symptoms or signs of CMT2D or dSMA-V, which are associated with autosomal dominant GARS mutations, but this has not been formally excluded by nerve conduction studies.
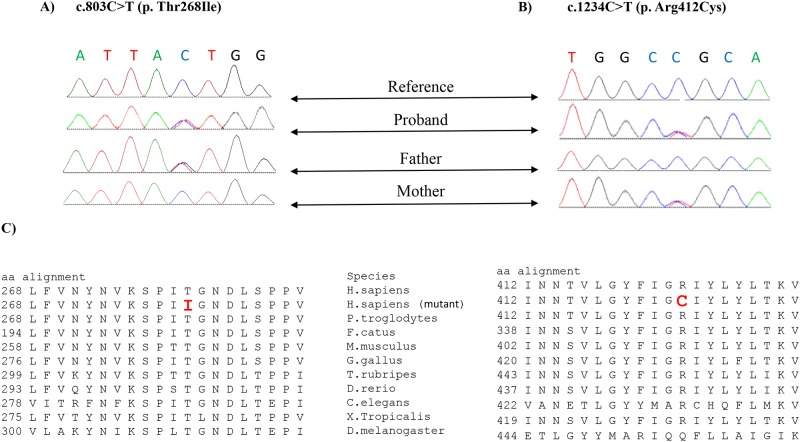
Fig 1.
A) Sanger sequencing profile of GARS from the proband and parents showing c.803C>T; p.(Thr268Ile) variant is heterozygous in the proband and the father. B) Sanger sequencing profile of GARS from the proband and parents showing c.1234C>T; p.(Arg412Cys) variant is heterozygous in the proband and the mother. C) Evolutionary sequence conservations of the altered amino acid residues p.Thr268 and p. Arg412 are denoted in bold red in boxes.
Both variants were considered rare with the minor allele frequency of the c.803C>T variant being 0.0031 and for the c.1234C>T variant being 0.000016 in the ExAC database (Table 4). In silico review of both variants (c.803C>T and c.1234C>T) predicted them to affect protein function with the most damaging score by SIFT, PolyPhen-2, Mutation-Taster and GVGD (Table 4).
Table 4. In silico analyses of the GARS variants identified in this study.
| Position | Pathogenicity prediction | Grade of conservation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Gene | Exon | cDNA | Protein | Sift | GVGD | Polyphen-2 | Mutationtster | PhyloP | PhastCons | MAF (ExAC) | Genotype | Inheritance |
| Proband | GARS | 7 | c.803C>T | p.(Thr268Ile) | Damaging | Class C65 (most likely) | Probably damaging | disease causing | 4.754 | 1.00 | 0.003174 | Heterozygous | Paternal |
| 10 | c.1234C>T | p.(Arg412Cys) | Damaging | Class C65 (most likely) | Probably damaging | disease causing | 1.78 | 1.00 | 0.000016 | Heterozygous | Maternal | ||
N/A: not applicable; MAF: minor allele frequency; PhyloP: values vary between -14 and +6 (Sites predicted to be conserved are assigned positive scores); PhastCons: values vary between 0 and 1. The closer the value is to 1, the more probable the nucleotide is conserve.
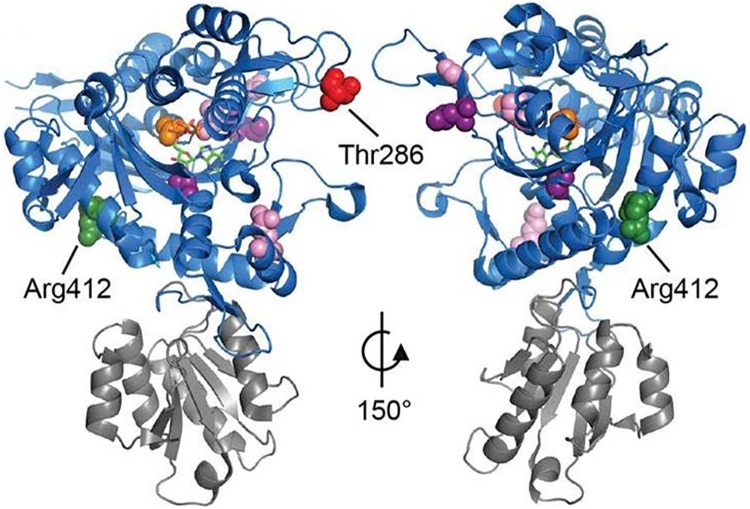
Both amino acid positions (p.Thr268 and p.Arg412) are highly conserved across many species (Fig 1C). In silico modelling predicted the amino acid changes (p.Thr268Ile and p.Arg412Cys) to affect the overall chemical and physical properties of the GARS protein (http://www.cmbi.ru.nl/hope/). Both Thr268 and Arg 412 lie in the catalytic domain that synthesizes glycyl adenylate and transfers glycine to its cognate tRNA [22]. Thus, mutations in these positions are likely to disturb the protein synthesis process (http://www.cmbi.ru.nl/hope/). The wild-type residue Thr268 is smaller and less hydrophobic compared to the mutant amino acid (Ile), while the wild-type residue Arg 412 is larger, positively charged, and less hydrophobic compared to the neutral mutant amino acid, Cys (http://www.cmbi.ru.nl/hope/) (Fig 2).
Fig 2. Relative position and conservation of GARS mutations.
Model of GARS protein structure showing catalytic domain (blue) and anticodon binding domain (grey). Residues mutated in the proband are displayed as red (Thr268) and green (Arg412) spheres. ATP (sticks) and glycine (orange spheres) are seen in the active site pocket. Pink and purple residues indicate previously reported pathogenic mutations in CMT2D and dSMA-V respectively [6,27,28]. Model based on PDB structure 2ZT7.
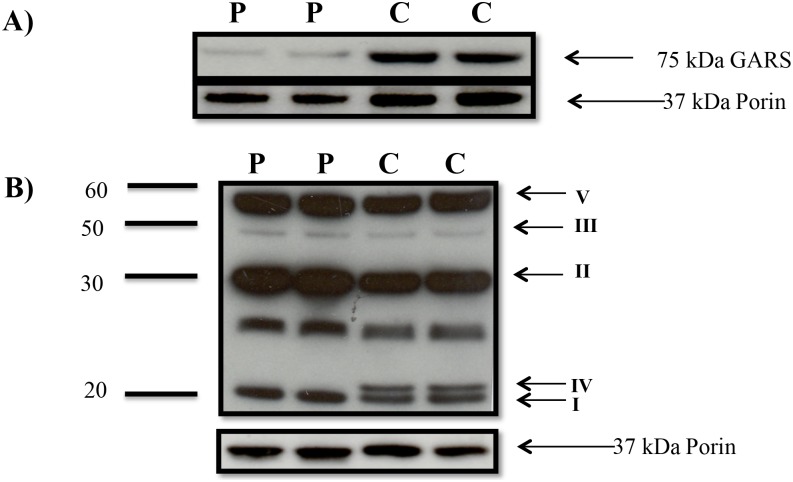
Since GARS is required for synthesis of mitochondrial DNA-encoded subunits of Complex I, III, IV and V, we examined GARS protein levels and its pathogenic effect on MRC enzyme complex levels in patient fibroblast extracts. Immunoblot analysis revealed a 300% reduction in GARS protein levels in patient fibroblasts compared to a healthy control using ImageJ version 1.49 [23] (Fig 3A). Immunoblot analysis of one subunit from each MRC enzyme complex revealed undetectable levels of the mitochondrial DNA-encoded Complex IV COXII subunit in patient fibroblasts, with no reduction in the levels of the other complexes, (Fig 3B).
Fig 3.
A) Immunoblot analysis of cultured fibroblasts lysates indicated reduction in expression of GARS protein in patient (P) compared to control (C). Samples containing 20 μg of total protein per lane were loaded in duplicate. Porin was used as a loading control. B) Each data point is immunoblot showing OXPHOS expression in cultured fibroblasts from the proband (P) compared to controls (C) in duplicate with a total of 30 μg of protein in each lane. Porin was used as a loading control.
Discussion
Here we report the identification of compound heterozygous GARS variants in a patient with exercise-intolerance, mild cardiomyopathy and lactic acidosis. GARS encodes both cytoplasmic and mitochondrial glycyl-tRNA synthetases which are required for protein synthesis in their respective subcellular locations [2]. Previous reports of GARS mutations have largely been autosomal dominant mutations associated with neuromuscular symptoms including atrophy and weakness of the hand muscles [1,5–12,24]. This phenotype closely resembles those caused by autosomal dominant mutations in cytoplasmic ARS, such as AARS (OMIM: 613287), HARS (OMIM: 616625), YARS (OMIM: 608323) and MARS (OMIM: 616280), which have been associated with distal motor neuropathy or polyneuropathies in children and adults [13]. It is thus likely that the neuropathic phenotype in autosomal dominant GARS is due to effects on the cytoplasmic GARS rather than mitochondrial GARS, although possible effects on mitochondrial GARS have not been investigated in these cases.
There is only one previously reported case of autosomal recessive inheritance in GARS where the patient displayed some clinical features similar to those seen in our patient. McMillan and colleagues reported a 12-year old girl with clinical and some biochemical features of a systemic mitochondrial disease including exercise-induced myalgia, non-compaction cardiomyopathy, persistent elevation of blood lactate, and white matter changes on brain MRI, who had compound heterozygous mutations (c.1904C>T; p.Ser635Leu and c.1787G>A; p.Arg596Gln) in GARS [13]. Neither patient with compound heterozygous variations in GARS displayed neuropathy but rather had clinical features which more closely resembled those caused by mutations in mitochondrial ARS. Mitochondrial ARS mutations are associated with a wide phenotypic spectrum, but clinical features may include leukoencephalopathy, cardiomyopathy and lactic acidosis [2].
Patients with compound heterozygous GARS variants also share some phenotypic overlap with patients reported to have variants in MT-TG, which encodes mt-tRNAGly, the cognate tRNA that mitochondrial GARS attaches to glycine. MT-TG variants have been associated with hypertrophic cardiomyopathy and exercise intolerance [25,26].
Mitochondrial ARS mutations mainly affect mitochondrial protein synthesis [2]. Our patient displayed reduced activity of Complex I, III and IV in skeletal muscle consistent with a mitochondrial protein synthesis defect, as a number of subunits of these complexes are mitochondrially encoded. There appeared to be some tissue specific effects, with liver and fibroblasts less affected. The reduced activity of MRC complexes most likely resulted from the reduced levels and/or activity of GARS. Immunoblotting revealed a reduction in the level of the GARS protein in the patient fibroblasts, consistent with the in silico predictions, and suggests that the mutant GARS is less stable. Given that both variants are in the catalytic domain, it is also likely that the residual GARS have reduced activity.
Our results suggest the mitochondrial function of GARS is affected by the compound heterozygous variants we identified in the patient. While the clinical features of the McMillan case were consistent with a mitochondrial disorder, no evidence of a mitochondrial protein synthesis defect was presented and the patient had normal MRC enzyme activities in muscle [13].
The difference in GARS phenotypes observed in cases of autosomal dominant versus autosomal recessive inheritance may be a consequence of gain of function versus loss of function effects of the mutations [24]. In a mouse study, dominant mutations in Gars caused gain of function, with a neuropathic phenotype that could not be corrected by overexpression of wild-type Gars. Mice with homozygous Gars mutations or a missense mutation in combination with a null allele, displayed a more severe neurological phenotype resulting from loss of function [24]. In our study, we have demonstrated that the compound heterozygous GARS variants are also associated with loss of function. In summary, the compound heterozygous GARS variants identified in our patient resulted in reduced GARS protein levels and MRC enzyme deficiency. We recommend GARS should be added to the list of genes that should be considered in cases of exercise-intolerance and lactic acidosis.
Acknowledgments
We thank the Queensland Center for Medical Genomics and the IMB Sequencing Core for their assistance. This research was supported by Australian NHMRC grant 1026891 to J.C., an NHMRC Principal Research Fellowship to D.R.T., and an Australian Mitochondrial Disease Foundation (AMDF) PhD Scholarship to M.N. Finally, we also gratefully acknowledge donations to J.C. by the Crane and Perkins families.
Data Availability
The Sanger sequencing results for the mutations are described in the paper and the chromatograms are included in Figure 1. The whole exome sequence data (fastq files) cannot be made publicly available as data sharing was not included in the consent at the time the family was recruited to the project. Requests for these data can be sent to the corresponding author. This research project was administered by the Sydney Children's Hospitals Network Human Research Ethics Committee. Alternatively requests could be made to the Governance Officer (SCHN-Governance@health.nsw.gov.au) for release of data.
Funding Statement
We thank the Queensland Center for Medical Genomics and the IMB Sequencing Core for their assistance. This research was supported by Australian NHMRC grant 1026891 to J.C., an NHMRC Principal Research Fellowship to D.R.T., and an Australian Mitochondrial Disease Foundation (AMDF) PhD Scholarship to M.N. Finally, we also gratefully acknowledge donations to J.C. by the Crane and Perkins families.
References
- 1.Kawakami N, Komatsu K, Yamashita H, Uemura K, Oka N, Takashima H, et al. (2014) [A novel mutation in glycyl-tRNA synthetase caused Charcot-Marie-Tooth disease type 2D with facial and respiratory muscle involvement]. Rinsho Shinkeigaku 54: 911–915. [DOI] [PubMed] [Google Scholar]
- 2.Diodato D, Ghezzi D, Tiranti V (2014) The Mitochondrial Aminoacyl tRNA Synthetases: Genes and Syndromes. Int J Cell Biol 2014: 787956 10.1155/2014/787956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grice SJ, Sleigh JN, Motley WW, Liu JL, Burgess RW, Talbot K, et al. (2015) Dominant, toxic gain-of-function mutations in gars lead to non-cell autonomous neuropathology. Hum Mol Genet 24: 4397–4406. 10.1093/hmg/ddv176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motley WW, Talbot K, Fischbeck KH (2010) GARS axonopathy: not every neuron's cup of tRNA. Trends Neurosci 33: 59–66. 10.1016/j.tins.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao YC, Liu YT, Tsai PC, Chang CC, Huang YH, Soong BW, et al. (2015) Two Novel De Novo GARS Mutations Cause Early-Onset Axonal Charcot-Marie-Tooth Disease. PLoS One 10: e0133423 10.1371/journal.pone.0133423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, et al. (2003) Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet 72: 1293–1299. 10.1086/375039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sivakumar K, Kyriakides T, Puls I, Nicholson GA, Funalot B, Antonellis A, et al. (2005) Phenotypic spectrum of disorders associated with glycyl-tRNA synthetase mutations. Brain 128: 2304–2314. 10.1093/brain/awh590 [DOI] [PubMed] [Google Scholar]
- 8.James PA, Cader MZ, Muntoni F, Childs AM, Crow YJ, Talbot K (2006) Severe childhood SMA and axonal CMT due to anticodon binding domain mutations in the GARS gene. Neurology 67: 1710–1712. 10.1212/01.wnl.0000242619.52335.bc [DOI] [PubMed] [Google Scholar]
- 9.Rohkamm B, Reilly MM, Lochmuller H, Schlotter-Weigel B, Barisic N, Schols L, et al. (2007) Further evidence for genetic heterogeneity of distal HMN type V, CMT2 with predominant hand involvement and Silver syndrome. J Neurol Sci 263: 100–106. 10.1016/j.jns.2007.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamaguchi A, Ishida C, Iwasa K, Abe A, Yamada M (2010) Charcot-Marie-Tooth disease type 2D with a novel glycyl-tRNA synthetase gene (GARS) mutation. J Neurol 257: 1202–1204. 10.1007/s00415-010-5491-x [DOI] [PubMed] [Google Scholar]
- 11.Lee HJ, Park J, Nakhro K, Park JM, Hur YM, Choi BO, et al. (2012) Two novel mutations of GARS in Korean families with distal hereditary motor neuropathy type V. J Peripher Nerv Syst 17: 418–421. 10.1111/j.1529-8027.2012.00442.x [DOI] [PubMed] [Google Scholar]
- 12.Klein CJ, Middha S, Duan X, Wu Y, Litchy WJ, Gu W, et al. (2014) Application of whole exome sequencing in undiagnosed inherited polyneuropathies. J Neurol Neurosurg Psychiatry 85: 1265–1272. 10.1136/jnnp-2013-306740 [DOI] [PubMed] [Google Scholar]
- 13.McMillan HJ, Schwartzentruber J, Smith A, Lee S, Chakraborty P, Bulman DE, et al. (2014) Compound heterozygous mutations in glycyl-tRNA synthetase are a proposed cause of systemic mitochondrial disease. BMC Med Genet 15: 36 10.1186/1471-2350-15-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ten Harkel AD, Takken T, Van Osch-Gevers M, Helbing WA (2011) Normal values for cardiopulmonary exercise testing in children. Eur J Cardiovasc Prev Rehabil 18: 48–54. 10.1097/HJR.0b013e32833cca4d [DOI] [PubMed] [Google Scholar]
- 15.Arena R, Myers J, Williams MA, Gulati M, Kligfield P, Balady GJ, et al. (2007) Assessment of functional capacity in clinical and research settings: a scientific statement from the American Heart Association Committee on Exercise, Rehabilitation, and Prevention of the Council on Clinical Cardiology and the Council on Cardiovascular Nursing. Circulation 116: 329–343. 10.1161/CIRCULATIONAHA.106.184461 [DOI] [PubMed] [Google Scholar]
- 16.Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cleary JG, Braithwaite R, Gaastra K, Hilbush BS, Inglis S, Irvine SA, et al. (2014) Joint variant and de novo mutation identification on pedigrees from high-throughput sequencing data. J Comput Biol 21: 405–419. 10.1089/cmb.2014.0029 [DOI] [PubMed] [Google Scholar]
- 18.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanderver A, Simons C, Helman G, Crawford J, Wolf NI, Bernard G, et al. (2016) Whole exome sequencing in patients with white matter abnormalities. Ann Neurol 79: 1031–1037. 10.1002/ana.24650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nafisinia M, Guo Y, Dang X, Li J, Chen Y, Zhang J, et al. (2016) Whole Exome Sequencing Identifies the Genetic Basis of Late-Onset Leigh Syndrome in a Patient with MRI but Little Biochemical Evidence of a Mitochondrial Disorder. JIMD Rep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frazier AE, Thorburn DR (2012) Biochemical analyses of the electron transport chain complexes by spectrophotometry. Methods Mol Biol 837: 49–62. 10.1007/978-1-61779-504-6_4 [DOI] [PubMed] [Google Scholar]
- 22.Li R, Macnamara LM, Leuchter JD, Alexander RW, Cho SS (2015) MD Simulations of tRNA and Aminoacyl-tRNA Synthetases: Dynamics, Folding, Binding, and Allostery. Int J Mol Sci 16: 15872–15902. 10.3390/ijms160715872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasband WS (1997–2016) ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA.https://imagej.nih.gov/ij/.
- 24.Motley WW, Seburn KL, Nawaz MH, Miers KE, Cheng J, Antonellis A, et al. (2011) Charcot-Marie-Tooth-linked mutant GARS is toxic to peripheral neurons independent of wild-type GARS levels. PLoS Genet 7: e1002399 10.1371/journal.pgen.1002399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merante F, Tein I, Benson L, Robinson BH (1994) Maternally inherited hypertrophic cardiomyopathy due to a novel T-to-C transition at nucleotide 9997 in the mitochondrial tRNA(glycine) gene. Am J Hum Genet 55: 437–446. [PMC free article] [PubMed] [Google Scholar]
- 26.Nishigaki Y, Bonilla E, Shanske S, Gaskin DA, DiMauro S, Hirano M (2002) Exercise-induced muscle "burning," fatigue, and hyper-CKemia: mtDNA T10010C mutation in tRNA(Gly). Neurology 58: 1282–1285. [DOI] [PubMed] [Google Scholar]
- 27.Del Bo R, Locatelli F, Corti S, Scarlato M, Ghezzi S, Prelle A, et al. (2006) Coexistence of CMT-2D and distal SMA-V phenotypes in an Italian family with a GARS gene mutation. Neurology 66: 752–754. 10.1212/01.wnl.0000201275.18875.ac [DOI] [PubMed] [Google Scholar]
- 28.Abe A, Hayasaka K (2009) The GARS gene is rarely mutated in Japanese patients with Charcot-Marie-Tooth neuropathy. J Hum Genet 54: 310–312. 10.1038/jhg.2009.25 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Sanger sequencing results for the mutations are described in the paper and the chromatograms are included in Figure 1. The whole exome sequence data (fastq files) cannot be made publicly available as data sharing was not included in the consent at the time the family was recruited to the project. Requests for these data can be sent to the corresponding author. This research project was administered by the Sydney Children's Hospitals Network Human Research Ethics Committee. Alternatively requests could be made to the Governance Officer (SCHN-Governance@health.nsw.gov.au) for release of data.