Abstract
Preeclampsia is a leading cause of perinatal maternal–foetal mortality and morbidity. The aim of this study is to identify the key microRNAs and genes in preeclampsia and uncover their potential functions. We downloaded the miRNA expression profile of GSE84260 and the gene expression profile of GSE73374 from the Gene Expression Omnibus database. Differentially expressed miRNAs and genes were identified and compared to miRNA-target information from MiRWalk 2.0, and a total of 65 differentially expressed miRNAs (DEMIs), including 32 up-regulated miRNAs and 33 down-regulated miRNAs, and 91 differentially expressed genes (DEGs), including 83 up-regulated genes and 8 down-regulated genes, were identified. The pathway enrichment analyses of the DEMIs showed that the up-regulated DEMIs were enriched in the Hippo signalling pathway and MAPK signalling pathway, and the down-regulated DEMIs were enriched in HTLV-I infection and miRNAs in cancers. The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) enrichment analyses of the DEGs were performed using Multifaceted Analysis Tool for Human Transcriptome. The up-regulated DEGs were enriched in biological processes (BPs), including the response to cAMP, response to hydrogen peroxide and cell-cell adhesion mediated by integrin; no enrichment of down-regulated DEGs was identified. KEGG analysis showed that the up-regulated DEGs were enriched in the Hippo signalling pathway and pathways in cancer. A PPI network of the DEGs was constructed by using Cytoscape software, and FOS, STAT1, MMP14, ITGB1, VCAN, DUSP1, LDHA, MCL1, MET, and ZFP36 were identified as the hub genes. The current study illustrates a characteristic microRNA profile and gene profile in preeclampsia, which may contribute to the interpretation of the progression of preeclampsia and provide novel biomarkers and therapeutic targets for preeclampsia.
Introduction
Preeclampsia (PE) is a prevalent disease characterized by hypertension and proteinuria, and it affects approximately 5%-8% of pregnancies worldwide[1]. Accumulating evidence has demonstrated that multiple genes and cellular pathways contribute to the occurrence and development of PE [2].
MicroRNAs (miRNAs) are small non-coding RNAs of approximately 19–23 nucleotides that can bind to the 3’ untranslated region of target mRNAs resulting in the degradation and translation inhibition of the mRNA, thereby regulating gene expression at the post-transcriptional level. Reportedly, up-regulated miR-210 in the placenta has been associated with the pathogenesis of PE[3], and miR-1233 might be a potential biomarker of early PE[4].
High-throughput platforms such as microarrays are increasingly valued for the analysis of miRNA and gene expression in PE. Many miRNA expression profile and gene expression profile studies on PE have been performed using microarray technology; for example, Zhu et al[5] identified 11 overexpressed microRNAs and 23 under-expressed microRNAs in PE compared to that in normal controls. Zhang et al[6] found that miR-515 family members were related to PE through the inhibition of key genes in human trophoblast differentiation. The previous studies on miRNA expression profiles in PE all had their limitations. First, all of the reported studies focused one or several of the differentially expressed miRNAs; none of them focused on the relationship between all of the differentially expressed miRNAs with PE. Second, miRbase (http://microrna.sanger.ac.uk), PicTar (http://pictar.mdc-berlin.de), TargetScan (http://www.targetscan.org) and MiRTarget2 (http://mirdb.org) were usually used to identify the target genes of the miRNAs, but the calculation principles and methods of each database are quite different, leading to a high false-positive rate. Therefore, we combined the miRNA expression profile GSE84260 with the gene expression profile GSE73374 to uncover the key miRNAs and genes that contribute to the pathology of PE and, thus, provide novel insights into potential biomarkers for PE prognosis and therapeutic strategies.
Materials and methods
Microarray data
The miRNA expression profile GSE84260 and the gene expression profile GSE73374 were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The GSE84260 dataset based on GPL15018 (Agilent Human miRNA V16.0 Microarray) contained 32 samples, including 16 PE placenta samples and 16 normal placenta samples. The GSE73374 dataset based on GPL16686 (Affymetrix Human Gene 2.0 ST Array) contained 36 samples, including 19 PE placenta samples and 17 normal placenta samples.
Identification of differentially expressed miRNAs and genes and the DEMI-DEG regulatory network
Firstly, after the raw data from the miRNA profile and gene profile underwent background correction, quartile normalization and probe summarization with the limma R package [7–8], we used a classical t test to identify the miRNAs that were differentially expressed between the two groups with cutoff values |log2 FC| ≥ 1 and p values < 0.05 and to identify the genes that were differentially expressed with the cutoff values |log2FC| ≥ 0.5 and p values < 0.05. Secondly, the MiRWalk 2.0 database, which provides the largest available collection of miRNA-target interactions [9], was used to identify target genes of the differentially expressed miRNAs identified from the GSE84260 dataset. Thirdly, we downloaded the miRNA-mRNA information from the MiRWalk 2.0-validated miRNA-gene interaction information retrieval system, in which all of the genes had been identified as target genes of the miRNAs. The intersection of the target genes from the miRNA-mRNA information and the identified differentially expressed genes from the GSE73374 dataset was selected as the final set of differentially expressed genes (DEGs). Lastly, by comparing the DEGs with the miRNA-mRNA information, we were able to identify the miRNAs that target the DEGs, and those miRNAs were selected as the final differentially expressed miRNAs (DEMIs). By mapping the DEMIs and DEGs using Cytoscape (version: 3.2.0)[10], we obtained the DEMI-DEG regulatory network.
Functional enrichment analyses of the DEMIs and DEGs
Pathway enrichment analyses of the DEMIs were performed by utilizing the in-plug clusterProfiler from the limma R package. The Gene ontology (GO), a method for annotating genes [11], and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, which presenting the systematic analysis of gene functions[12] enrichment analyses were performed utilizing the MATHT (http://www.biocloudservice.com) to identify potential biological processes and pathways in which the DEGs are involved. P<0.05 was considered statistically significant.
Integration of the protein-protein interaction (PPI) network
DEGs were mapped to the Search Tool for the Retrieval of Interacting Genes (STRING version: 10.0)[13], an online tool utilized to evaluate the PPI information. Interactions with a combined score > 0.4 were selected as significant. The integrated regulatory networks were constructed using the Cytoscape software.
Results
Identification of DEMIs and DEGs and the DEMI-DEG regulatory network
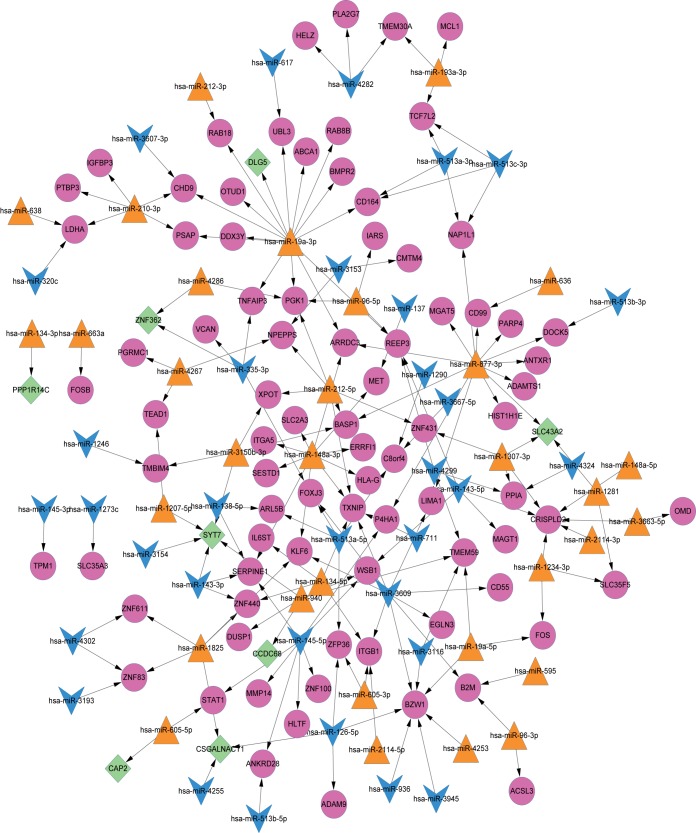
A total of 65 differently expressed microRNAs (DEMIs), 32 up-regulated miRNAs and 33 down-regulated miRNAs, and 91 differently expressed genes (DEGs), 83 up-regulated genes and 8 down-regulated genes, were finally identified. Data for the 65 DEMIs are provided in S1 Table. In the DEMI-DEG regulatory network, there were 156 nodes and 184 interactions (Fig 1). The interaction degrees for the DEMI-DEG regulatory network represent the number of the interactions between the DEMIs and DEGs. Those DEMIs and DEGs with high interaction degrees were identified as hub nodes in the DEMI-DEG regulatory network. The top 10 DEMIs and DEGs with high degrees from the DEMI-DEG regulatory network are shown in Table 1 and Table 2.
Fig 1. The DEMI-DEG regulatory network.
Orange triangles represent up-regulated DEMIs(32); blue arrows represent the down-regulated DEMIs(33); red cycles represent the up-regulated DEGs(83); green rhombus represent the down-regulated DEGs(8).
Table 1. Top 10 hub DEMIs from the DEMI-DEG regulatory network.
| miRNA | hsa-miR-19a-3p | hsa-miR-877-3p | hsa-miR-148a-3p | hsa-miR-3609 | hsa-miR-145-5p |
| Description | upmiRNA | upmiRNA | upmiRNA | downmiRNA | downmiRNA |
| degree | 15 | 13 | 10 | 9 | 8 |
| miRNA | hsa-miR-212-5p | hsa-miR-1825 | hsa-miR-210-3p | hsa-miR-940 | hsa-miR-134-5p |
| Description | upmiRNA | upmiRNA | upmiRNA | upmiRNA | upmiRNA |
| degree | 6 | 5 | 5 | 5 | 4 |
Table 2. Top 10 hub genes from the DEMI-DEG regulatory network.
| Gene | BZW1 | CRISPLD2 | TXNIP | SYT7 | TMEM59 | SERPINE1 | REEP3 | PGK1 | ITGB1 | ZNF83 |
|---|---|---|---|---|---|---|---|---|---|---|
| Description | upgene | upgene | upgene | downgene | upgene | upgene | upgene | upgene | upgene | upgene |
| Degree | 7 | 6 | 5 | 5 | 4 | 4 | 4 | 4 | 4 | 3 |
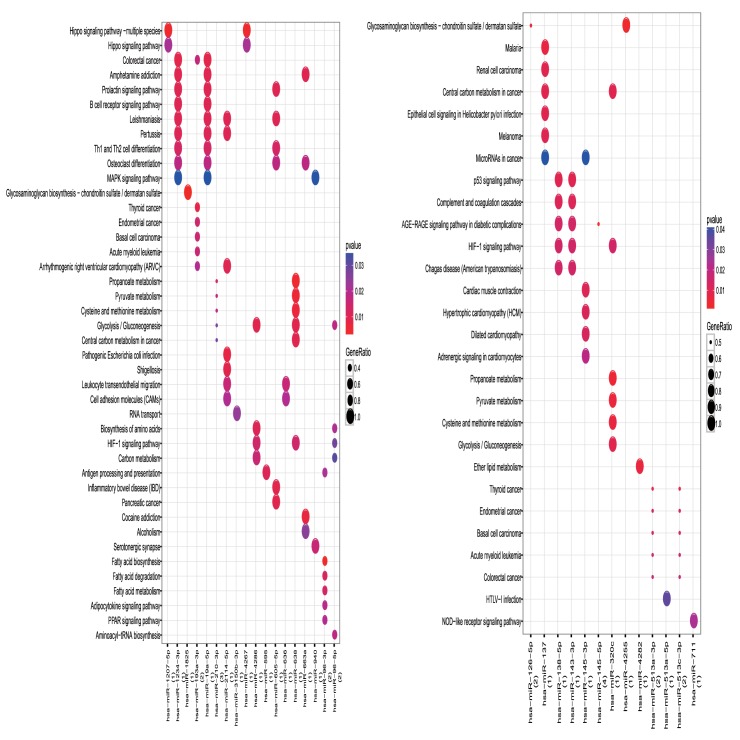
Pathway enrichment analyses of the DEMIs
KEGG pathway analyses indicated that the up-regulated DEMIs were enriched in 150 pathways such as the Hippo signalling pathway and MAPK signalling pathway. The down-regulated DEMIs were enriched in 73 pathways such as HTLV-I infection and miRNAs in cancers (Fig 2).
Fig 2. Pathway enrichment analyses of DEMIs.
The left is up-regulated DEMIs and the right is down-regulated DEMIs. Red: p value is small; Blue: p value is large; the size of the bubbles means the enrichment, larger bubbles means larger generatio.
Functional enrichment analyses of the DEGs
We uploaded all DEGs to MATHT to identify the GO categories and KEGG pathways of the DEGs. The functional enrichment analysis results showed that the down-regulated DEGs (8) were not enriched in any of the categories or pathways. The GO analysis results showed that the up-regulated DEGs were mainly involved in biological processes (BP) such as the response to cAMP (Table 3). GO molecular function (MF) analysis indicated that the up-regulated DEGs were mainly involved in protein binding and growth factor binding (Table 3). In addition, for the cell component (CC) analysis, the up-regulated DEGs were significantly enriched in the extracellular exosome and membrane (Table 3). KEGG pathways analyses showed that up-regulated DEGs (83) were significantly enriched in the Hippo signalling pathway and pathways in cancer.
Table 3. Pathway and Gene ontology analysis of the up-regulated DEGs associated with PE (TOP5).
| ID | Name | Count | PValue | |
|---|---|---|---|---|
| PATHWAY | hsa04514 | Cell adhesion molecules (CAMs) | 4 | 3.44E-02 |
| PATHWAY | hsa04390 | Hippo signaling pathway | 4 | 4.02E-02 |
| PATHWAY | hsa05200 | Pathways in cancer | 6 | 4.61E-02 |
| PATHWAY | hsa05140 | Leishmaniasis | 3 | 4.98E-02 |
| PATHWAY | hsa05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) |
3 | 4.98E-02 |
| GO_BP | GO:0051591 | response to cAMP | 5 | 6.06E-05 |
| GO_BP | GO:0042542 | response to hydrogen peroxide | 5 | 9.12E-05 |
| GO_BP | GO:0033631 | cell-cell adhesion mediated by integrin | 3 | 1.27E-04 |
| GO_BP | GO:0071222 | cellular response to lipopolysaccharide | 6 | 1.80E-04 |
| GO_BP | GO:0042493 | response to drug | 8 | 5.24E-04 |
| GO_CC | GO:0070062 | extracellular exosome | 27 | 1.33E-04 |
| GO_CC | GO:0016020 | membrane | 23 | 1.73E-04 |
| GO_CC | GO:0005925 | focal adhesion | 8 | 1.77E-03 |
| GO_CC | GO:0009897 | external side of plasma membrane | 6 | 2.60E-03 |
| GO_CC | GO:0000139 | Golgi membrane | 8 | 1.60E-02 |
| GO_MF | GO:0005515 | protein binding | 55 | 9.25E-04 |
| GO_MF | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 8 | 1.17E-03 |
| GO_MF | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding |
3 | 4.88E-03 |
| GO_MF | GO:0019838 | growth factor binding | 3 | 6.70E-03 |
| GO_MF | GO:0004386 | helicase activity | 4 | 6.89E-03 |
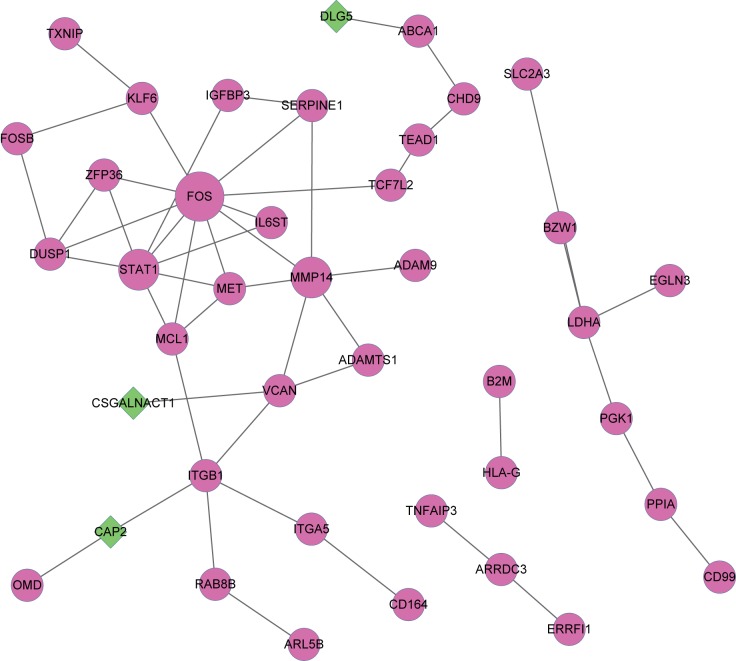
PPI network of the DEGs
The PPI network of the DEGs was constructed using String. In the PPI network, there were 41 nodes, including 38 up-regulated DEGs and 3 down-regulated DEGs, and 50 interactions (Fig 3). The hub nodes were FOS, STAT1, MMP14, ITGB1, VCAN, DUSP1, LDHA, MCL1, MET and ZFP36. Among all of the proteins in the PPI network, only CAP2, CSGALNACT1 and DLG5 were down-regulated.
Fig 3. Protein-protein interaction network of DEGs.
Purple cycles represent up-regulated genes, green diamonds represent down-regulated genes. The size indicates the interaction degrees.
Discussion
PE is a multisystem disorder specific to pregnancy, and deficiency in our knowledge of the exact aetiology and pathogenesis of PE restricts the ability to treat this disease. Therefore, understanding the molecular mechanism involved in PE is extremely important to develop more effective diagnostic and therapeutic strategies. In the present study, a total of 65 DEMIs and 91 DEGs were identified. The up-regulated DEMIs were enriched in the Hippo signalling pathway and MAPK signalling pathway, and the down-regulated DEMIs were enriched in HTLV-I infection and miRNAs in cancers. FOS, STAT1, MMP14, ITGB1, VCAN, DUSP1, LDHA, MCL1, MET and ZFP36 were defined as key proteins that might provide new ideas for further studies on PE.
MiRNAs have been increasingly recognized to have a vital association with disease including PE through post-transcriptional regulation of gene expression. In the present study, miRNA expression profiles showed that miRNAs in placentas were quite different between the PE and normal group. Eight up-regulated miRNAs, including miR-19a-3p, miR-877-3p, miR-148a-3p, miR-212-5p, miR-1825, miR-210-3p, miR-940, and miR-134-5p, and two down-regulated miRNAs, miR-3609 and miR-145-5p, were identified as statistically significant different miRNAs. MiR-148a and miR-19a have been reported to influence the +3142 C/G polymorphism of HLA-G, resulting in the down-regulation of HLA-G in PE[14]. It is widely accepted that the PE syndrome consists of two successive processes, including poor placentation in early pregnancy and the following placental oxidative stress[4]. Hypoxia of the placenta is a crucial factor leading to poor biological functions of trophoblast cells. MiR-210 is a hypoxia-inducible miRNA[15] and inhibits invasion of trophoblast cells[16]. Down-regulation of miR-145 has been identified in the placenta of PE women[17]. No studies on the relationship between the other miRNAs, miR-877-3p, miR-212-5p, miR-1825, miR-940, miR-134-5p, and miR-3609, and PE have been reported. However, they are all related to the occurrence and development of carcinomas. For example, miR-212 was down-regulated in ovarian cancer, potentially due to the significant enrichment of EZH2 and H3K27me3 in the promoter region[18]. Decreased miR-940 in hepatocellular carcinoma acted as an adaptor of CXCR2 and suppressed the invasion and migration of HCC cells[19]. Considering that the conversion of the biological functions of normal cells is fundamental to the pathology of PE and carcinoma, we infer that miR-877-3p, miR-212-5p, miR-1825, miR-940, miR-134-5p, and miR-3609 might take part in the progression of PE.
KEGG pathway analysis of the DEMIs revealed that the development of PE was associated with the Hippo signalling pathway and MAPK signalling pathway. The Hippo signalling pathway could provide novel anti-cancer drug targets. Components of the Hippo signalling pathway such as Yes-associated protein 1 (YAP) and transcription regulator protein 1 (TAZ) are synergistically associated with other signalling pathways such as G protein-coupled receptor, epidermal growth factor and Wnt pathways, which play a crucial role in cell proliferation, differentiation, apoptosis, and development[20]. Recent evidence indicates that the p38 MAPK signalling pathway is one of the key pathways in vascular endothelial cell dysfunction in PE. Activated p38 MAPK in the placenta of PE could significantly increase the levels of sEng and sFlt-l in maternal serum[21]. Gadd45α(DNA damage-inducible 45 alpha) is an oxidative stress-induced factor with high levels in PE. Gadd45αinhibits trophoblast invasion and regulates anti-angiogenesis factor secretion via the p38 MAPK signalling pathway[22].
GO and KEGG pathway analyses were performed to better understand the interactions of the DEGs. The GO analyses showed that up-regulated DEGs were intensively involved in the BP of the response to cAMP, response to hydrogen peroxide and cell-cell adhesion mediated by integrin. Furthermore, the KEGG pathways of the up-regulated DEGs included the Hippo signalling pathway and pathways in cancer. The hub genes with top degrees in the PPI network were FOS, STAT1, MMP14, ITGB1, VCAN, DUSP1, LDHA, MCL1, MET, and ZFP36. FOS was identified as up-regulated in PE, which was consist with that reported by Song[23]. FOS is involved in the regulation of angiogenesis by encoding the transcription factor c-fos proto-oncogene[24]. The second hub gene, STAT1 (signal transducers and activators of transcription 1), is phosphorylated, forming a dimer that activates Janus tyrosine kinases (JAKs) when initiated by IFN-γ[25]. Endothelial activation and excessive inflammation are the characteristics of PE, which can be induced by the IFN-γ/STAT1 signalling pathway[26]. It has been proposed that the mouse systolic arterial pressure and plasma levels of sEng were increased compared to those exposed to doxycycline, a compound that could block the transcription of MMP-14. sEng and sFlt-1, contributing to the maternal vascular dysfunction, while up-regulated MMP14 released by endothelial cells induced the release of sEng and sFlt-1[27]. Previous studies have reported that LDHA was up-regulated in PE[28–29]. Activated by hypoxia, the LDH isozyme in trophoblasts can induce higher lactate production[30], which might inhibit germ cell death dose-dependently in the human testis[31]. The MKP/DUSPs family acts as negative feedback regulators of MAPK activity by dephosphorylating phosphorylated tyrosine or serine/threonine[32]. Christe et al. reported that DUSP9/MKP-4 was essential for placental function [33]. DUSP5 may mediate the H19 down-regulation-induced suppression of proliferation and apoptosis of JAR cells [34]. The other five hub genes are ITGB1, MCL1, VCAN, MET and ZFP36. Two previous studies have reported that ITGB1 is related to PE through encoding the beta subunit of integrin. Additionally, up-regulated miR-29b might contribute to PE via its target genes ITGB1 and MCL1[35–36]. The Mtd/Mcl-1 system plays a crucial role in regulating trophoblast cell functions in both physiological and pathological conditions; Mcl-1 induces apoptosis and reduced proliferation, while Mtd likely shows different properties[37]. In preeclampsia, the Mtd/Mcl-1 system is altered towards the production of killer isoforms, meaning that both Mtd-L and Mtd-P were increased, and the expression of Mcl-1 was down-regulated in PE[38–39]. Further studies are needed to identify the functions of ITGB1 and MCL1 in PE. No studies on VCAN in PE have been reported. Met, an anti-angiogenic factor was significantly elevated in both the second and third trimesters of PE[40], but Zeng found that the plasma sMet concentration was significantly lower in women with severe PE than in control groups [41]. ZFP36 is a zinc-finger protein and can regulate the production of growth factors and cytokines by destabilizing mRNAs. Recently, one study found that ZFP36 might be a potential regulator of VEGF to control reepithelialization and angiogenesis in the skin[42].
In conclusion, we identified several abnormally expressed miRNAs and genes in PE that may participate in the pathogenesis of PE. Our study provides a comprehensive bioinformatic analysis of DEMIs and DEGs in PE, helps to understand the underlying molecular mechanisms of PE, and may provide potential biomarkers and therapeutic targets for PE. Further experiments are required to confirm the expression and potential functions of the identified miRNAs and genes in PE.
Supporting information
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Natural Science Foundation of China (Grant No. 81471470 to W-RG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Duley L. The Global Impact of Pre-eclampsia and Eclampsia. SEMIN PERINATOL. 2009;33(3):130–7. doi: 10.1053/j.semperi.2009.02.010 [DOI] [PubMed] [Google Scholar]
- 2.Chelbi ST, Vaiman D. Genetic and epigenetic factors contribute to the onset of preeclampsia. MOL CELL ENDOCRINOL. [Journal Article; Review]. 2008. 2008-01-30;282(1–2):120–9. doi: 10.1016/j.mce.2007.11.022 [DOI] [PubMed] [Google Scholar]
- 3.Nikuei P, Davoodian N, Tahamtan I, Keshtkar AA. Predictive value of miR-210 as a novel biomarker for pre-eclampsia: a systematic review protocol. BMJ OPEN. [Journal Article]. 2016. 2016-09-28;6(9):e11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ura B, Feriotto G, Monasta L, Bilel S, Zweyer M, Celeghini C. Potential role of circulating microRNAs as early markers of preeclampsia. Taiwan J Obstet Gynecol. [Journal Article]. 2014. 2014-06-01;53(2):232–4. doi: 10.1016/j.tjog.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 5.Zhu X, Yang Y, Han T, Yin G, Gao P, Ni Y, et al. Suppression of microRNA-18a expression inhibits invasion and promotes apoptosis of human trophoblast cells by targeting the estrogen receptor alpha gene. MOL MED REP. [Journal Article; Research Support, Non-U.S. Gov't]. 2015. 2015-08-01;12(2):2701–6. doi: 10.3892/mmr.2015.3724 [DOI] [PubMed] [Google Scholar]
- 6.Zhang M, Muralimanoharan S, Wortman AC, Mendelson CR. Primate-specific miR-515 family members inhibit key genes in human trophoblast differentiation and are upregulated in preeclampsia. Proceedings of the National Academy of Sciences. 2016. 2016-11-08;113(45):E7069–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. NUCLEIC ACIDS RES. [Comparative Study; Journal Article; Research Support, Non-U.S. Gov't; Validation Studies]. 2008. 2008-02-01;36(2):e11 doi: 10.1093/nar/gkm1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. BIOINFORMATICS. [Journal Article; Research Support, N.I.H., Extramural; Research Support, Non-U.S. Gov't]. 2010. 2010-10-01;26(19):2363–7. doi: 10.1093/bioinformatics/btq431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. GENOME BIOL. [Journal Article]. 2004. 2004-01-20;5(10):R80 doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. GENOME RES. [Journal Article; Research Support, Non-U.S. Gov't; Research Support, U.S. Gov't, Non-P.H.S.; Research Support, U.S. Gov't, P.H.S.]. 2003. 2003-11-01;13(11):2498–504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Gene Ontology (GO) project in 2006. NUCLEIC ACIDS RES. 2006; 34:D322–6. doi: 10.1093/nar/gkj021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. NUCLEIC ACIDS RES. [Journal Article; Research Support, Non-U.S. Gov't]. 2000 2000-01-01;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. NUCLEIC ACIDS RES. [Journal Article; Research Support, Non-U.S. Gov't]. 2011. 2011-01-01;39(Database issue):D561–8. doi: 10.1093/nar/gkq973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castelli EC, Moreau P, Oya ECA, Mendes-Junior CT, Veiga-Castelli LC, Yaghi L, et al. In silico analysis of microRNAS targeting the HLA-G 3' untranslated region alleles and haplotypes. HUM IMMUNOL. [Journal Article; Research Support, Non-U.S. Gov't]. 2009. 2009-12-01;70(12):1020–5. doi: 10.1016/j.humimm.2009.07.028 [DOI] [PubMed] [Google Scholar]
- 15.Luo R, Wang Y, Xu P, Cao G, Zhao Y, Shao X, et al. Hypoxia-inducible miR-210 contributes to preeclampsia via targeting thrombospondin type I domain containing 7A. Sci Rep. [Journal Article; Research Support, Non-U.S. Gov't]. 2016. 2016-01-22;6:19588 doi: 10.1038/srep19588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anton L, Olarerin-George AO, Schwartz N, Srinivas S, Bastek J, Hogenesch JB, et al. miR-210 inhibits trophoblast invasion and is a serum biomarker for preeclampsia. AM J PATHOL. [Journal Article; Research Support, N.I.H., Extramural; Research Support, U.S. Gov't, Non-P.H.S.]. 2013. 2013-11-01;183(5):1437–45. doi: 10.1016/j.ajpath.2013.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hromadnikova I, Kotlabova K, Hympanova L, Krofta L. Cardiovascular and Cerebrovascular Disease Associated microRNAs Are Dysregulated in Placental Tissues Affected with Gestational Hypertension, Preeclampsia and Intrauterine Growth Restriction. PLOS ONE. 2015. 2015-09-22;10(9):e138383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin L, Wang Z, Jin H, Shi H, Lu Z, Qi Z. MiR-212/132 is epigenetically downregulated by SOX4/EZH2-H3K27me3 feedback loop in ovarian cancer cells. Tumour Biol. [Journal Article]. 2016. 2016-11-03. [DOI] [PubMed] [Google Scholar]
- 19.Ding D, Zhang Y, Yang R, Wang X, Ji G, Huo L, et al. miR-940 Suppresses Tumor Cell Invasion and Migration via Regulation of CXCR2 in Hepatocellular Carcinoma. BIOMED RES INT. [Journal Article]. 2016. 2016-01-20;2016:7618342 doi: 10.1155/2016/7618342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae JS, Kim SM, Lee H. The Hippo signaling pathway provides novel anti-cancer drug targets. ONCOTARGET. [Review; Journal Article]. 2016. 2016-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo X, Yao ZW, Qi HB, Liu DD, Chen GQ, Huang S, et al. Gadd45alpha as an upstream signaling molecule of p38 MAPK triggers oxidative stress-induced sFlt-1 and sEng upregulation in preeclampsia. CELL TISSUE RES. [Journal Article; Research Support, Non-U.S. Gov't]. 2011. 2011-06-01;344(3):551–65. doi: 10.1007/s00441-011-1164-z [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Deng Q, Luo X, Chen Y, Shan N, Qi H. Oxidative stress-induced Gadd45alpha inhibits trophoblast invasion and increases sFlt1/sEng secretions via p38 MAPK involving in the pathology of pre-eclampsia. J Matern Fetal Neonatal Med. [Journal Article]. 2016. 2016-12-01;29(23):3776–85. doi: 10.3109/14767058.2016.1144744 [DOI] [PubMed] [Google Scholar]
- 23.Song J, Li Y, An RF. Identification of Early-Onset Preeclampsia-Related Genes and MicroRNAs by Bioinformatics Approaches. REPROD SCI. 2015. 2015-08-01;22(8):954–63. doi: 10.1177/1933719115570898 [DOI] [PubMed] [Google Scholar]
- 24.Dony C, Gruss P. Proto-oncogene c-fos expression in growth regions of fetal bone and mesodermal web tissue. NATURE. [Journal Article; Research Support, Non-U.S. Gov't]. 1987. 1987-08-20;328(6132):711–4. doi: 10.1038/328711a0 [DOI] [PubMed] [Google Scholar]
- 25.Aaronson DS, Horvath CM. A road map for those who don't know JAK-STAT. SCIENCE. [Journal Article; Research Support, Non-U.S. Gov't; Research Support, U.S. Gov't, P.H.S.; Review]. 2002. 2002-05-31;296(5573):1653–5. doi: 10.1126/science.1071545 [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Hu Y, Liu X, Zheng Y, Luo M, Liu W, et al. EPHB4, a down stream target of IFN-γ/STAT1 signal pathway, regulates endothelial activation possibly contributing to the development of preeclampsia. AM J REPROD IMMUNOL. 2016;76(4):307–17. doi: 10.1111/aji.12555 [DOI] [PubMed] [Google Scholar]
- 27.Valbuena-Diez AC, Blanco FJ, Oujo B, Langa C, Gonzalez-Nunez M, Llano E, et al. Oxysterol-induced soluble endoglin release and its involvement in hypertension. CIRCULATION. [Journal Article; Research Support, Non-U.S. Gov't]. 2012. 2012-11-27;126(22):2612–24. doi: 10.1161/CIRCULATIONAHA.112.101261 [DOI] [PubMed] [Google Scholar]
- 28.Kay HH, Zhu S, Tsoi S. Hypoxia and Lactate Production in Trophoblast Cells. PLACENTA. 2007;28(8–9):854–60. doi: 10.1016/j.placenta.2006.11.011 [DOI] [PubMed] [Google Scholar]
- 29.Lee GSR, Joe YS, Kim SJ, Shin JC. Cytokine-related genes and oxidation-related genes detected in preeclamptic placentas. ARCH GYNECOL OBSTET. 2010;282(4):363–9. doi: 10.1007/s00404-009-1222-x [DOI] [PubMed] [Google Scholar]
- 30.Tsoi SCM, Zheng J, Xu F, Kay HH. Differential Expression of Lactate Dehydrogenase Isozymes (LDH) in Human Placenta with High Expression of LDH-A4Isozyme in the Endothelial Cells of Pre-eclampsia Villi. PLACENTA. 2001;22(4):317–22. doi: 10.1053/plac.2000.0620 [DOI] [PubMed] [Google Scholar]
- 31.Erkkila K, Aito H, Aalto K, Pentikainen V, Dunkel L. Lactate inhibits germ cell apoptosis in the human testis. MOL HUM REPROD. [Journal Article; Research Support, Non-U.S. Gov't]. 2002. 2002-02-01;8(2):109–17. [DOI] [PubMed] [Google Scholar]
- 32.Kidger AM, Rushworth LK, Stellzig J, Davidson J, Bryant CJ, Bayley C, et al. Dual-specificity phosphatase 5 controls the localized inhibition, propagation, and transforming potential of ERK signaling. Proceedings of the National Academy of Sciences. 2017. 2017-01-04:201614684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christie GR, Williams DJ, Macisaac F, Dickinson RJ, Rosewell I, Keyse SM. The dual-specificity protein phosphatase DUSP9/MKP-4 is essential for placental function but is not required for normal embryonic development. MOL CELL BIOL. [Journal Article; Research Support, Non-U.S. Gov't]. 2005. 2005-09-01;25(18):8323–33. doi: 10.1128/MCB.25.18.8323-8333.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu LL, Chang K, Lu LS, Zhao D, Han J, Zheng YR, et al. Lentivirus-mediated RNA interference targeting the H19 gene inhibits cell proliferation and apoptosis in human choriocarcinoma cell line JAR. BMC CELL BIOL. [Journal Article; Research Support, Non-U.S. Gov't]. 2013. 2013-05-27;14:26 doi: 10.1186/1471-2121-14-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li P, Guo W, Du L, Zhao J, Wang Y, Liu L, et al. microRNA-29b contributes to pre-eclampsia through its effects on apoptosis, invasion and angiogenesis of trophoblast cells. Clin Sci (Lond). [Journal Article; Research Support, Non-U.S. Gov't]. 2013. 2013-01-01;124(1):27–40. [DOI] [PubMed] [Google Scholar]
- 36.Jiang F, Yang Y, Li J, Li W, Luo Y, Li Y, et al. Partial least squares-based gene expression analysis in preeclampsia. GENET MOL RES. [Journal Article; Research Support, Non-U.S. Gov't]. 2015. 2015-06-18;14(2):6598–604. doi: 10.4238/2015.June.18.2 [DOI] [PubMed] [Google Scholar]
- 37.Ray J, Jurisicova A, Caniggia I. IFPA Trophoblast Research Award Lecture: the dynamic role of Bcl-2 family members in trophoblast cell fate. PLACENTA. [Lectures; Research Support, Non-U.S. Gov't]. 2009. 2009-03-01;30 Suppl A:S96–100. [DOI] [PubMed] [Google Scholar]
- 38.Soleymanlou N, Wu Y, Wang JX, Todros T, Ietta F, Jurisicova A, et al. A novel Mtd splice isoform is responsible for trophoblast cell death in pre-eclampsia. CELL DEATH DIFFER. [Comparative Study; Journal Article; Research Support, Non-U.S. Gov't]. 2005. 2005-05-01;12(5):441–52. doi: 10.1038/sj.cdd.4401593 [DOI] [PubMed] [Google Scholar]
- 39.Soleymanlou N, Jurisicova A, Wu Y, Chijiiwa M, Ray JE, Detmar J, et al. Hypoxic switch in mitochondrial myeloid cell leukemia factor-1/Mtd apoptotic rheostat contributes to human trophoblast cell death in preeclampsia. AM J PATHOL. [Journal Article; Research Support, N.I.H., Extramural; Research Support, Non-U.S. Gov't]. 2007. 2007-08-01;171(2):496–506. doi: 10.2353/ajpath.2007.070094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim SY, Park SY, Kim MJ, Lee BY, Han JY, Ryu HM. Preeclampsia is associated with an elevation of plasma sMet concentrations in the second trimester. J Matern Fetal Neonatal Med. [Journal Article; Research Support, Non-U.S. Gov't]. 2013. 2013-06-01;26(9):860–5. doi: 10.3109/14767058.2013.769952 [DOI] [PubMed] [Google Scholar]
- 41.Zeng X, Sun Y, Yang HX, Li D, Li YX, Liao QP, et al. Plasma level of soluble c-Met is tightly associated with the clinical risk of preeclampsia. AM J OBSTET GYNECOL. [Journal Article; Research Support, Non-U.S. Gov't]. 2009. 2009-12-01;201(6):611–8. [DOI] [PubMed] [Google Scholar]
- 42.Prenzler F, Fragasso A, Schmitt A, Munz B. Functional analysis of ZFP36 proteins in keratinocytes. EUR J CELL BIOL. [Journal Article]. 2016. 2016-08-01;95(8):277–84. doi: 10.1016/j.ejcb.2016.04.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.