Abstract
Cervical cancer is one of the most common types of cancer in women worldwide. Most deaths due to the disease occur in less developed areas of the world. In this work, we introduce a new image dataset along with expert annotated diagnoses for evaluating image-based cervical disease classification algorithms. A large number of Cervigram® images are selected from a database provided by the US National Cancer Institute. For each image, we extract three complementary pyramid features: Pyramid histogram in L*A*B* color space (PLAB), Pyramid Histogram of Oriented Gradients (PHOG), and Pyramid histogram of Local Binary Patterns (PLBP). Other than hand-crafted pyramid features, we investigate the performance of convolutional neural network (CNN) features for cervical disease classification. Our experimental results demonstrate the effectiveness of both our hand-crafted and our deep features. We intend to release this multi-feature dataset and our extensive evaluations using seven classic classifiers can serve as the baseline.
Keywords: Cervical cancer screening, computer aided diagnosis, image classification, pyramid histogram, local binary patterns, convolutional neural network
1. Introduction
Cervical cancer ranks as the second most common type of cancer in women aged 15 to 44 years worldwide [1]. Over 80% of deaths due to the disease occur in less developed regions of the world [1]. Therefore, there is a need for lower cost and more automated screening methods for early detection of cervical cancer, especially those applicable in low-resource regions. Screening procedures can help prevent cervical cancer by detecting cervical intraepithelial neoplasia (CIN), which is the potentially precancerous change and abnormal growth of squamous cells on the surface of the cervix. According to the World Health Organization (WHO) [1], CIN is divided into three grades: CIN1 (mild), CIN2 (moderate), and CIN3 (severe). Lesions in CIN2/3+ require treatment, whereas mild dysplasia in CIN1 only needs conservative observation because it will typically be cleared by an immune response in one year. Thus, in clinical practice one important goal of screening is to differentiate CIN1 from CIN2/3 or cancer (denoted as CIN2/3+ [2]).
Widely used cervical cancer screening methods today include Pap tests, HPV tests, and visual examination. Pap tests involve collecting a small sample of cells from the cervix and need a laboratory and trained personnel to examine these samples under a microscope for squamous and glandular intraepithelial lesions (SIL). Also Pap tests suffer from low sensitivity in detecting CIN 2/3+ [3]. HPV tests are DNA tests which detect human papillomavirus (HPV) strains associated with cervical cancer. The sensitivity of HPV tests in detecting CIN 2/3+ lesions varies greatly [3]. Colposcopy is a visual diagnostic procedure that often involves taking a biopsy. Digital cervicography, a non-invasive visual examination method that takes a photograph of the cervix (called a Cervigram®) after the application of 5% acetic acid to the cervix epithelium, has great potential to be a primary or adjunctive screening tool in developing countries because of its low cost and accessibility in resource-poor regions. However, one concern with cervicography is that its overall effectiveness has been questioned by reports of poor correlation between visual lesion recognition and high-grade disease, as well as disagreement among experts when grading visual findings. To address this concern and investigate the feasibility of using images as a screening method for cervical cancer, we conjecture that computer algorithms can be developed to improve the accuracy in grading lesions using visual (and image) information. This conjecture is inspired and encouraged by recent successes in computer-assisted Pap tests such as the ThinPrep Imaging System (TIS) [4], FocalPoint [5], and the work by Zhang et al. [6]; these computer-assisted Pap tests apply multi-feature Pap smear image classification using support vector machines (SVM) and other machine learning algorithms, and they have been shown to be statistically more sensitive than manual methods with equivalent specificity.
In this work, we describe our efforts in building a dataset of multiple features extracted from Cervigram images along with patient diagnosis ground truth for evaluating image-based cervical disease classification algorithms. First, we design a new type of pyramid features. From each image, we extract three complementary pyramid features: Pyramid histogram in L*A*B* color space (PLAB), Pyramid Histogram of Oriented Gradients (PHOG), and Pyramid histogram of Local Binary Patterns (PLBP). Second, besides hand-crafted pyramid features, we investigate the performance of convolutional neuron network (CNN) features for cervical disease classification. Third, on this multi-feature dataset, we also present some baseline results of applying different classic machine-learning algorithms (e.g., SVM, random forest) to differentiate patient visits that are high-risk from those visits that are low-risk. We train binary classifiers to separate CIN1/Normal and CIN2/3+ images. All the classifiers are trained and tested on the same dataset, with a uniform parameter optimization strategy. They are then compared by ROC curves and other evaluation measures. On the same dataset, our lower-cost image-based classifiers can perform comparably or better than human interpretation on other traditional screening results, such as Pap tests and HPV tests.
2. Related Work
Several computer-assisted Pap tests have been approved by United States Food and Drug Administration (USFDA), such as ThinPrep Imaging System (TIS) [4] and FocalPoint [5]. These methods were shown to be statistically more sensitive than manual methods with equivalent specificity. Encouraged by these developments, a data-driven algorithm [2] was developed for automated cancer diagnosis via analyzing Cervigram images. In contrast to Pap tests [4, 5], Cervigrams are images captured by the non-invasive and low cost digital cervicography. To further improve the classification performance, Song et al. [7] combined the Cervigram information with other clinical test results such as Pap and HPV; however, these other clinical tests require additional resources that may not be available in resource-poor areas of the world.
The choice of feature descriptors is one of the most important factors for image segmentation and classification. Several types of features [2, 7, 8, 9, 10] have been proposed to encode Cervigram information. Li et al. [8] identified acetowhite regions by analyzing local color features. Zimmerman et al. [9] detected specularities in Cervigrams by utilizing image intensity, saturation, and gradient information. In the work by Ji et al. [10], texture features were used to recognize important vascular patterns in Cervigrams. In [2, 7], the authors combined the pyramid histogram of oriented gradients (PHOG) and the pyramid color histogram in L*A*B space (PLAB) features to perform region of interest (ROI) segmentation and CIN classification.
In addition to feature descriptors, classifiers also have great influence on the performance of a machine-learning based classification method. Neural networks, support vector machines (SVM), k-Nearest Neighbors (KNN), linear discriminant analysis (LDA), and decision trees are commonly used for studying cervical cancer [11]. Kim et al. [2] applied a linear SVM to classify Cervigrams into CIN1/normal or CIN2/3+, while Song et al. [7] utilized KNN coupled with a majority voting algorithm to perform the CIN classification. Zhang et al. [12] proposed a discriminative sparse representation for tissue classification in Cervigrams. In the work by Lee et al. [13], the authors developed a system which integrates multiple classifiers including neural network classifiers, statistical binary decision tree classifiers, and a hybrid classifier.
3. An Image Data Set with Multiple Features for CIN Classification
Here we introduce a dataset for image-based CIN classification, built from a large medical data archive collected by the National Cancer Institute (NCI) in the Guanacaste project [14]. The archive consists of data from 10,000 anonymized women, and the data is stored in the Multimedia Database Tool (MDT) developed by the National Library of Medicine [15]. In the archive, each patient typically had multiple visits at different ages. During each visit, multiple cervical screening tests including cervicography were performed. The cervicography test produced two Cervigram images for a patient during her visit and the images were later sent to an expert for interpretation.
In our dataset, we used 1112 patient visits, 345 positive (CIN2/3/cancer) and 767 negative (CIN1/Normal). For each patient, the ground truth diagnosis is based on the Worst Histology result of that patient visit: multiple expert histology interpretations were done on each biopsy; the most severe interpretation is labeled the Worst Histology for that visit in the database. Note that our dataset is unbalanced, i.e. it contains more negative cases than positive cases. Since many classification methods assume a balanced distribution of classes and require additional strategies to handle unbalanced data, we apply under-sampling to the negative visits and randomly choose 345 negative visits from each dataset. In this paper, we will use this balanced sub-dataset, including all 345 positive visits and the randomly selected 345 negative visits.
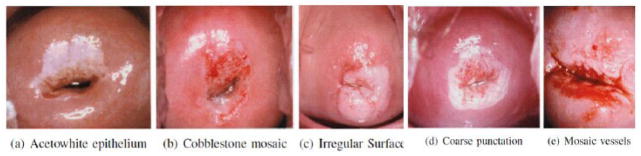
Interpretations based on Cervigram images have been shown to be an effective way to detect CIN2/3+ [2]. Some of the most important visual observations in Cervigrams include the acetowhite region, and features within that region, such as mosaicism, punctation, and atypical vessels; it is important to distinguish these possibly disease-related features from benign features such as polyps or cysts. Fig. 1 shows some example images of those observations [7]. To robustly identify these characteristics which are helpful for diagnosis, we propose a type of hand-crafted pyramid features. We also investigate the performance of deep features for cervical disease classification, which have achieved superior performance in many other domains [16].
Figure 1.
Illustration of visual observations in Cervigrams
3.1. Hand-crafted Pyramid Features
Previous works [2, 7, 8, 9, 10] have shown that the local color, gradient and texture features are good at encoding Cervigram information. For example, color plays a key role to detect the presence of acetowhitened regions in Cervigrams; gradient plays important role in detecting specularities; texture is important for the identification of mosaicism and vessel pattern. We convert the pixel colors in a Cervigram into the perceptually uniform L*A*B color space because of its property: a small change in the color value corresponds to about the same small change in visual appearance. We utilize LBP for texture encoding and HOG for gradient encoding because of their great success in various classification tasks.
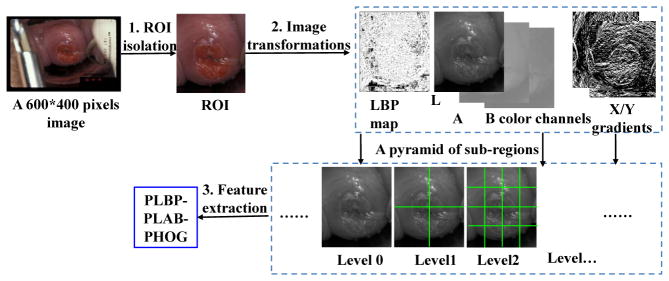
We extract multi-scale pyramid histogram features to encode the statistical appearance information in Cervigrams, as shown in Fig. 2. First, we isolate the cervix region of interest (ROI) from the input image and resize it to 300*250 pixels. We use the method proposed in [2] to segment the ROI. Second, we transform the ROI image patch into different types of feature maps, including the local binary pattern (LBP) map, L*A*B color channels, and the image gradient maps. Third, we construct a spatial pyramid of sub-regions for each feature map. We then extract and concatenate pyramid LBP (PLBP), pyramid LAB (PLAB) and pyramid Histogram of Oriented Gradients (PHOG) features to be a multi-feature descriptor.
Figure 2.
Image features extraction
Color and Image Gradient
Color plays an important role in cervical lesion classification, because one of the most important visual features on the cervix that have relevant diagnostic properties is the presence of acetowhitened regions. Thus, the color feature is widely used in Cervigram analysis [2, 7, 12]. We calculate the L*A*B color channels as our color feature maps. To capture edge and shape information, we calculate the gradient map, which is shown to be complementary to the color feature [2, 7].
Texture
In addition to the color and gradient features, we introduce a new local binary pattern (LBP) feature that extracts local texture characteristics for cervical lesion classification. Ojala et al. [17] first introduced LBP and showed its powerful ability for texture classification. In a local neighborhood of an input image, given a pixel (xc, yc) which is surrounded by 8 neighbors, we can calculate its LBP value by Eq. (1),
| (1) |
Where ic indicates the gray-scale value of the center pixel (xc, yc); ip corresponds to the gray-scale value of the pth neighbor. s(x) is a sign function where s(x) = 1, if x ≥ 0; else, s(x) = 0.
Later, several extensions of the original LBP operator were presented [18]. First, the LBP was extended to a circular neighborhood of different radii, denoted as LBPP,R which refers to P equally spaced pixels on a circle of radius R. Furthermore, the rotation invariant local binary pattern is defined in Eq. (2),
| (2) |
Where ROR(LBPP,R, i) performs a circular bit-wise right shift on the P-bit LBPP,R, for i number of times.
To obtain the LBP map, we compute the value for each pixel in the input image. Because of the neighborhood constraints when capturing LBP features, pixels on the boundary of the input image within the R range do not have any LBP values. We set those pixels’ values to be zeros or to be their closest neighbors’ LBP values.
In this paper, we use . There is no need to use LBP with other radii because our pyramid histogram LBP feature (PLBP) can encode a multi-scale local binary pattern.
Pyramid Feature Extraction
As Fig. 2 shows, we construct a spatial pyramid for each feature map. A pyramid is constructed by splitting the image into rectangular sub-regions, increasing the number of regions at each level, i.e., level 0 has 1 sub-region; level 1 has 4 sub-regions; level 2 has 16 sub-regions, and so forth. Histogram features are extracted from each of these pyramid sub-regions. The extracted pyramid histograms encode the statistical distribution of feature values at different positions and scales in a cervigram.
For the PLBP feature, the total number of bins is 10 for the histogram of a sub-region. A 4-level pyramid is constructed resulting in a PLBP histogram feature that has 850 dimensions. For the PLAB feature, we use 3 pyramid levels with a 16-bin histogram for each channel in L*A*B color space in each sub-region. Thus, the PLAB color feature has 1,008 dimensions. In the gradient map, we calculate the histogram of oriented gradients (PHOG) for each subregion in the pyramid. An 8-bin orientation histogram over a 4-level pyramid is used. Hence, the total vector size of our PHOG feature is 680. Finally, we construct a multi-feature descriptor by concatenating the three different types of features, PLBP-PLAB-PHOG. Thus, this handcrafted multi-feature descriptor has a vector size of 2,538.
3.2. CNN Deep Features
The work by Razavian et al. [16] indicates that the deep features extracted from convolutional neural networks (CNN) are very powerful for many recognition tasks. In this work, we investigate the performance of CNN deep features for cervical disease classification. In contrast to hand-crafted features, CNN features are automatically learned from a large number of images. We extract CNN features using the open-source package Caffe [19] with its ImageNet pre-trained CaffeNet. CaffeNet is a variant of AlexNet [20]. It consists of five convolutional layers and two fully connected layers (fc6 and fc7) and a final 1000-way softmax (fc8). Besides those main layers, there are some other layers, such as max-pooling layers and normalization layers. As in the published work [16], we extract the 4096 dimensional CNN features from the fully connected layer (fc6 or fc7).
To make the CNN features more discriminative for our CIN classification task, we also fine-tune the pre-trained CaffeNet from ROIs extracted in Cervigram images. We replace the original 1000-way fc8 layer in CaffeNet with our new 2-way fc8 layer with randomly initialized weights drawn from a Gaussian distribution with σ = 0.01 and μ = 0. Based on the loss curve on the training dataset, we find the appropriate base learning rate and weight decay. We set 0.0001 as the learning rate of all pre-trained convolutional layers and fully connected layers and increase the learning rate by a factor of 10 (i.e., to 0.001) for our new fc8 layer. The weight decay is set to be 0.5. The ROI of each training image is resized to 256*256 pixels and then cropped to the 227*227 network input size. Flipped training images are also used in the fine-tuning process. For testing and for feature extraction, each ROI is directly resized to 227*227 and no cropping and flipping are used.
4. Seven Classifiers for Comparison
On the Cervigram image dataset introduced above, we compare seven classic machine learning methods, including random forest (RF), gradient boosting decision tree (GBDT), AdaBoost, support vector machines (SVM), logistic regression (LR), multilayer perceptron (MLP), and k-Nearest Neighbors (kNN). Some of these, such as SVM, have been widely used in the field of medical image analysis [21, 22, 23, 24], while others, like random forest and GBDT, have been used only in the recent few years [25]. There are additional published works that aim to compare classifier performances on benchmark datasets. For example, Morra et al. [21] compared AdaBoost with SVM while Osareh et al. [22] compared SVM with neural networks. In both papers, the comparisons were done between two classifiers. In the work by Wei et al. [23], more classifiers were studied, but ensemble methods like RF and GBDT were not included. In this paper, we conduct a comprehensive comparison of seven popular classifiers. Next, we briefly introduce each of them.
Random Forest (RF) is an increasingly popular machine learning method [26]. It builds an ensemble of many decision trees trained separately on a bootstrapped sample set of the original data. Each decision tree grows by randomly selecting a subset of candidate attributes for splitting at each node. We optimize parameters for RF by searching the number of trees in {10, 100, 200, 500, 1000, 2000} and searching the subset size of features for node splitting among {’sqrt’, 100, 200, 500, 1000, 2000} where ’sqrt’ is the square root of the whole feature size.
Gradient boosting decision tree (GBDT) is a kind of additive boosting model which, in general, can be expressed as Eq. (3)
| (3) |
where β is called the expansion coefficient, and serves as the weight of the tree in each iteration, and b(x; γ) are usually simple basic functions, e.g. decision tree, characterized by parameters γ. Details for the training process of GBDT can be found in [26]. We optimize the parameters for GBDT by searching the number of trees among {10, 100, 200, 500, 1000, 2000} and the learning rate in {1, 0.1, 0.01, 0.001, 0.0001}.
Adaboost is a classic boosting tree model [27]. It has the form H(x) = Σtαtht(x), which can be trained by minimizing the loss function in a greedy fashion. An optimal weak classifier ht is selected for each training iteration t. We use shallow decision trees (i.e. stumps) as the weak learners. In the final strong classifier H(x), the weight of the weak classifier ht(x) is αt, which is inversely proportional to the classification error of ht(x). To optimize parameters for AdaBoost, we search the depth (d) of each decision tree in {1, 2, 3, 4} and the number of weak classifiers from 10 to the whole feature size with an increment of 120/d.
Multilayer perceptron (MLP) is a feed-forward neural network. MLP uses layer-wise connected nodes to build the architecture of the model. Each node (except for the input nodes) can be viewed as a neuron with a nonlinear activation function. In this paper, we use the sigmoid Eq. (4) as the activation function,
| (4) |
where the weight vector w and bias vector b in each layer pair are trained by the Back Propagation algorithm. We also introduce L2 regularization weight decay to prevent over-fitting. We optimize hyper-parameters for MLP by searching the hidden layer size in {2, 3}, the hidden unit size in {0.0625*m, 0.125*m, 0.25*m} where m is the feature size 2538, and searching the weight decay strength among {0.0005, 0.0001, 0.00001, 0.0}.
Logistic regression is a type of probabilistic statistical classification model. For the binary classification problem, with labeled sample set , it computes the positive probability by Eq. 5 and the model parameter is trained to minimize the cost Eq. (6).
| (5) |
| (6) |
In our experiments, we use the batch gradient descent algorithm with L2 regularization to train the model. The strength of regularization is searched from 10−5 to 105, with an increment of 1 for the exponent.
Support vector machines (SVM) is one of the most widely used classifiers in medical image analysis [2, 6, 21, 22]. It performs classification by constructing a hyperplane in a high-dimensional feature space. It can use either linear or non-linear kernels, and its effectiveness depends on the selection of kernel, the kernel’s parameters, and the soft margin parameter C. Linear SVM is widely used because it has good performance and fast speed in many tasks. In this paper, we also choose to use the linear SVM; we did try nonlinear kernels such as the radial basis functions (RBF) but they are time consuming and did not improve performance in our task. For linear SVM, we need to optimize the parameter C. Let C = 2m, we search m in the range [−8, 9] with a step increment of 1.
k-Nearest Neighbors (kNN) is one of the simplest classifiers, which classifies a new instance by a majority vote of its k nearest neighbors. In this paper, we use the Euclidean distance metric to find the k nearest neighbors. We search the optimal k value for our task in the range [1, 50] with a step increment of 1.
5. Experiments
On the image dataset with multiple types of features introduced in Section 3, we use the same ten-round ten-fold cross validation to evaluate our features and compare different classifiers. We randomly divide the samples (Cervigrams) into ten folds. In the ten rounds, we rotationally use one fold for testing and nine folds for training (or fine-tuning the CaffeNet). On the training set, we use a uniform strategy, Exhaustive Grid Search [28], to search for the optimal parameters of each classifier. The exact parameters and search ranges for each classifier are discussed in the Section 4. Note that there are two images from each patient visit, which are visually similar but not identical. We have to avoid using one image for training while the other image is being used for testing. Thus we construct two separate image datasets, D1 and D2, and randomly assign one image of a visit to D1 and assign the other image from the same visit to D2. We compute the average results on D1 and D2 to represent the visit-level performance. By default, we show the visit-level performance in all our experiments.
The results of our ten rounds are used to draw ROC curves. We compare different features or classifiers by analyzing their ROC curves, areas under ROC curves (AUC), and accuracy, sensitivity and specificity values at the point with the default probability threshold of 0.5. We also compare the results of our image-based classifiers with several other screening tests results, obtained for the same visits that are used to construct our dataset.
All our experiments are conducted on the computer with 3.0Ghz Intel Xeon E5-2623 CPU and 64 GB memory. The GPU card used in training the deep CNN network is Nvidia TITAN X. Since we use the ImageNet pre-trained model as the weight initialization for our deep CNN network, the training converges fast for our dataset and the total training time is about half an hour. For testing, the proposed CNN framework can achieve real-time speed (18ms per image), which demonstrates promising efficiency for future applications.
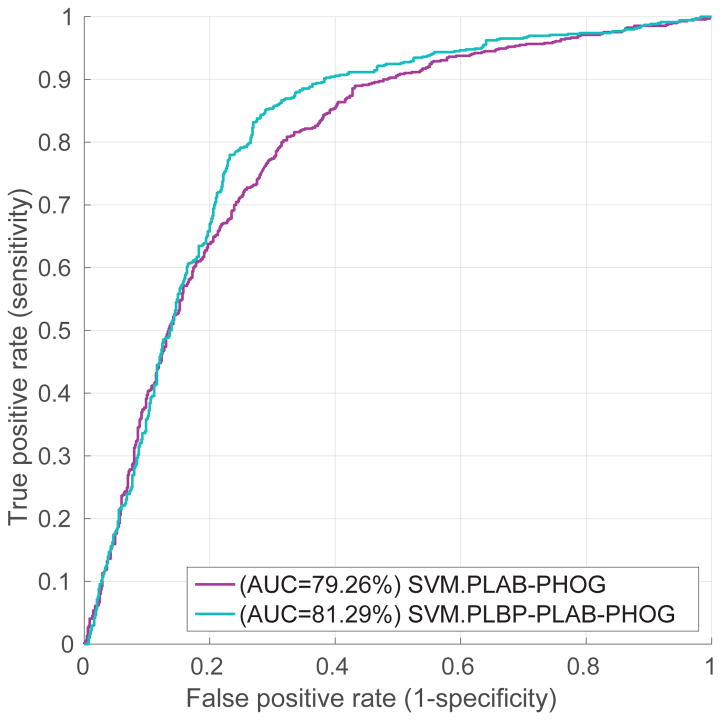
5.1. PLBP-PLAB-PHOG Feature vs. PLAB-PHOG
We evaluate the performance of our PLBP-PLAB-PHOG feature descriptor by comparing it with the baseline feature PLAB-PHOG [2, 7]. In Fig. 3, we compare their visit-level performance in ROC curves produced by linear SVM classifier trained on different features. It shows that the PLBP-PLAB-PHOG feature outperforms PLAB-PHOG. For example, our PLBP-PLAB-PHOG increases the accuracy from 73.70% to 77.17% at the probability threshold of 0.5. The best accuracy of PLBP-PLAB-PHOG feature is 78.12% achieved at 83.19% sensitivity and 73.04% specificity, while the best accuracy of PLAB-PHOG is 74.28%. Consequently, adding PLBP makes a better feature descriptor for Cervigram images.
Figure 3.
Comparison of PLBP-PLAB-PHOG and PLAB-PHOG feature descriptors
5.2. Evaluation of Seven Classic Classifiers with PLBP-PLAB-PHOG Feature
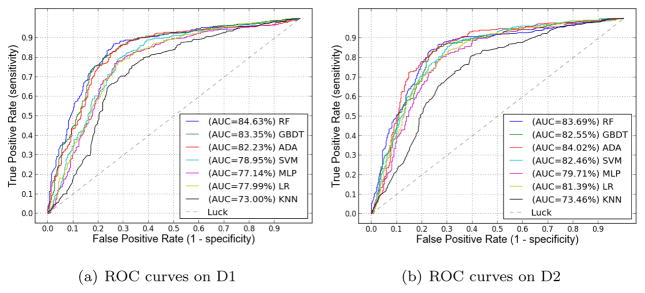
In this set of experiments, we compare seven classifiers described in Section 4 based on our handcrafted PLBP-PLAB-PHOG feature. The implementations for the seven classic classifiers are from well known open source libraries. The Random Forest, GBDT, and LR classifiers are implemented with scikit-learn [29]; the MLP classifier is provided by pylearn2 [30]; the SVM is from Libsvm [28]; the AdaBoost is provided by Appel et. al. [27]; and the kNN classifier is provided by the implementation in MATLAB.
Our comparison results on D1 and D2 are shown in Fig. 4 as ROC curves, which illustrate that the three ensemble-tree models— RandomForest (RF), GBDT, and AdaBoost—outperform other classifiers. At the 5% significance level, there is no difference between RandomForest, GBDT and AdaBoost. For instance, on D1 the p value is 0.0708 by paired t-test between RF (1st rank) and AdaBoost (3rd rank). However, these three ensemble-tree classifiers are significantly better than all other classifiers. The p value is 0.0062 and 1.7191 * 10−4, by paired t-test between RF (1st rank) and SVM (4th rank), and between RF and kNN (lowest rank), respectively. We conjecture that the ensemble-tree models perform best because they are more robust to over-fitting than other models such as SVM and MLP when dealing with scalar data sets that are not too large.
Figure 4.
Comparison of seven classifiers based on PLBP-PLAB-PHOG feature.
5.3. Evaluation of CNN Deep Features
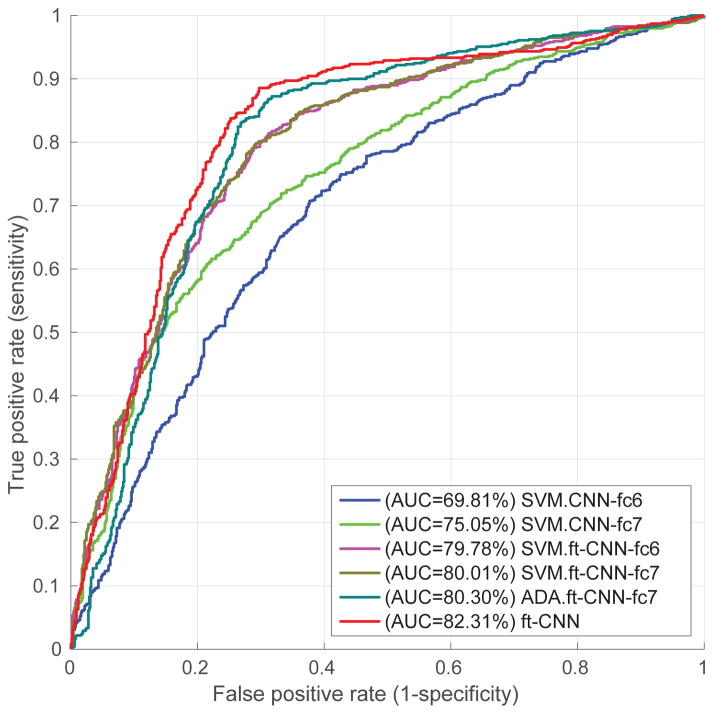
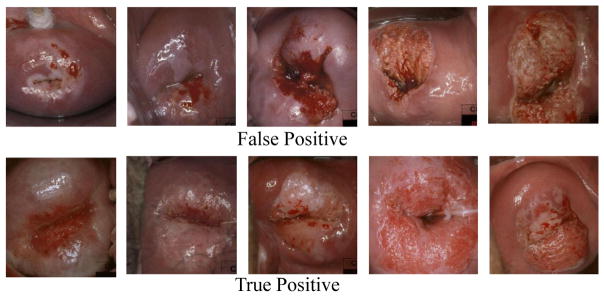
In this subsection, we evaluate the CNN deep features extracted from different layers and trained with different classifiers. Based on the results shown in Fig. 5 and Table 1, we have several observations. (1) CNN features extracted directly from pre-trained CaffeNet perform much worse than our hand-crafted PLBP-PLAB-PHOG feature descriptor. We believe the reason is that our task (i.e. cervical disease classification) is far too different from the original task of CaffeNet (i.e. object recognition in natural image scenes). (2) CNN features extracted from fc7 greatly outperform those from fc6 in the pre-trained model. The work in [16] also indicates that later layers in the CNN network can improve performance. Fine-tuning, however, did not improve the performance of fc7 as much as that of fc6, thus fine-tuned fc6 and fine-tuned fc7 achieved similar performance; one of the reasons for this could be that our dataset is too small to fine tune the large number of parameters in fully connected layers so that there is no big difference between fc6 and fc7 after fine-tuning. (3) Compared with AdaBoost and SVM classifiers trained on fine-tuned CNN features, the end-to-end CNN architecture achieves better performance. Fig. 6 shows some false positive and true positive examples according to the diagnosis given by the end-to-end CNN classifier. As one can see, some of the false positive examples are difficult to distinguish from true positive examples, based on image information alone. Multi-modal interpretation, using multiple sources of information, may be able to improve classification performance further.
Figure 5.
Results of CNN features
Table 1.
Overall performance of CNN features at the default probability threshold 0.5. ft indicates fine-tuned model. The ft-CNN model utilizes the fine-tuned CNN architecture as an end-to-end classifier; while all other models use either handcrafted or CNN features to train classifiers. This table lists the means ± standard deviations of our ten-fold ten-cross validation results.
| Model | AUC(%) | accu(%) | sensi(%) | speci(%) |
|---|---|---|---|---|
| SVM.PLBP-PLAB-PHOG | 80.71±6.15 | 77.17±6.62 | 78.55±6.17 | 75.80±8.39 |
|
| ||||
| SVM.CNN-fc6 | 69.81±5.02 | 66.01±3.10 | 65.07±5.52 | 66.96±6.79 |
| SVM.CNN-fc7 | 75.05±5.50 | 69.13±5.16 | 69.57±8.08 | 68.70±7.29 |
|
| ||||
| SVM.ft-CNN-fc6 | 79.78±4.60 | 74.20±4.65 | 75.36±7.48 | 73.04±6.55 |
| SVM.ft-CNN-fc7 | 80.01±4.99 | 74.64±5.71 | 76.52±9.11 | 72.75±6.09 |
| ADA.ft-CNN-fc7 | 80.30±4.07 | 77.39±3.89 | 80.87±6.69 | 73.91±9.23 |
|
| ||||
| ft-CNN | 82.31±4.63 | 78.41±5.01 | 80.87±7.43 | 75.94±7.46 |
Figure 6.
False positive (1st row) and true positive (2nd row) examples as diagnosed by the CNN classifier.
5.4. Image-based CIN classification vs. Pap and HPV tests
In clinical practice, screening methods should have high specificity (e.g., higher than 90%) because it is important to have low risk for unnecessary treatment for women that do not have disease. In Table 2, we compare our image-based CIN classification methods with several conventional screening methods (Pap tests and HPV tests), which are available for the same visits that are used to construct our dataset. As discussed in Section 1, Pap tests involve collecting a small sample of cells from the cervix and need a laboratory and trained personnel to examine these samples under a microscope. Based on the degree of the disease, the examination result can be classified to be low-risk (negative) or high-risk (positive). HPV tests are DNA tests which detect human papillomavirus strains associated with cervical cancer. The detection result can be negative or positive. The performance for Pap or HPV tests is computed based on their examination results and the ground truth. It is clear that those conventional methods are designed to have high specificity. For fair comparison, we constrain our methods to have 90% specificity in Table 2
Table 2.
Comparison of visit-level performance: our image-based classifiers vs. Pap tests and HPV tests.
| Method | accu(%) | sensi(%) | speci(%) |
|---|---|---|---|
| Alfaro ThinPrep | 51.26±10.02 | 20.69±19 | 81.82±5.07 |
| Cytyc ThinPrep | 69.01±4.77 | 49.55±8.14 | 88.46±3.33 |
| Costa Rica Pap | 63.77±4.18 | 39.42±7.65 | 88.12±3.18 |
| Hopkins Pap | 66.56±9.54 | 36.00±20.67 | 97.11±2.51 |
| HPV16 | 64.01±4.66 | 33.82±7.41 | 94.19±3.69 |
| HPV18 | 53.07±1.95 | 08.16±3.98 | 97.97±0.91 |
|
| |||
| our RF.PLBP-PLAB-PHOG | 70.50±6.02 | 51.00±6.07 | 90.00±0 |
| our ft-CNN | 65.00±5.11 | 40.00±7.34 | 90.00±0 |
As illustrated in Table 2, with respect to accuracy and sensitivity, our handcrafted PLBP-PLAB-PHOG feature descriptor with random forest classifier (RF.PLBP-PLAB-PHOG) outperforms every single Pap test or HPV test, when achieving a specificity of 90%. When not constrained by the 90% specificity requirement, our image-based classifier can achieve even better overall accuracy. For example, our fine-tuned CNN features with Softmax classifier can achieve an accuracy of 78.41% with 80.87% sensitivity and 75.94% specificity at the default probability threshold 0.5. Consequently, on this dataset, our lower-cost image-based classifiers can perform comparably or better than human interpretation based on widely-used Pap and HPV tests; in particular, the image-based classifiers can achieve higher sensitivity in detecting CIN2/3+.
5.5. Discussion
Besides testing the performance of using hand-crafted or deep features separately, we have evaluated the performance of combining hand-crafted and deep features. Unfortunately, the performance is not improved. For example, the SVM classifier trained on the combined features achieves 79.99% AUC; but the SVM classifier using hand-crafted features only or using deep features alone gives 80.71% and 80.01% AUC, respectively. At the 5% significance level, they are proven to have no significant difference. The p value is 0.5963 and 0.3572 by paired t-test between the combined features and hand-crafted features, and between combined features and deep features, respectively. This result shows that the deep features and hand-crafted features are not complementary for our task.
6. Conclusions
In this paper, we present a new benchmark dataset with multiple types of features for evaluating cervical dysplasia classification or grading algorithms. Both image features and ground truth diagnoses are included in the dataset. We will publish 1 the original dataset, sample images, fine-tuned CNN model and the source code for extracting the multiple image features. We will also add information from other screening tests such as Pap and HPV and expand the size of the dataset in the future.
Our experimental results indicate that our hand-crafted PLBP-PLAB-PHOG descriptor and fine-tuned CNN features outperform the baseline feature descriptor [2, 7]. Based on those features, our lower-cost image-based classifiers perform comparably or better than human interpretation on traditional Pap and HPV test, on our test dataset. Further, we adopt a uniform experimentation and parameter optimization framework to compare seven classic machine learning algorithms in terms of their performance in classifying an image into either CIN1/Normal (i.e. low-grade lesion/healthy) or CIN2/3+ (i.e. high-grade lesion/cancer). The reported results can serve as a baseline for future comparisons of automated cervical dysplasia classification methods. From the results, we find that ensemble-tree models—Random Forest, Gradient Boosting Decision Tree, and AdaBoost—outperform other classifiers such as multi-layer perceptron, SVM, logistic regression and kNN, on this task. This finding is consistent with the conclusion in other works [31].
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM), and Lister Hill National Center for Biomedical Communications (LHNCBC), under Contract HHSN276201000693P. We would also like to acknowledge the expert advice and support from Dr. Mark Schiffman and Dr. Nicolas Wentzensen of the National Cancer Institute (NCI) in the use and interpretation of data from the NCI Guanacaste Project.
Footnotes
Project webpage at http://www.cse.lehigh.edu/~idealab/cervitor
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.WHO. Summary report, ICO Information Centre on HPV and Cancer. Aug, 2014. Human papillomavirus and related cancers in the world. [Google Scholar]
- 2.Kim E, Huang X. A data driven approach to cervigram image analysis and classification. Color Medical Image analysis, Lecture Notes in Computational Vision and Biomechanics. 2013;6:1–13. [Google Scholar]
- 3.Sankaranarayanan R, Gaffikin L, Jacob M, et al. A critical assessment of screening methods for cervical neoplasia. International Journal of Gynecology and Obstetrics. 2005;89:4–12. doi: 10.1016/j.ijgo.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Biscotti CV, Dawson AE, et al. Assisted primary screening using the automated thinprep imaging system. American Journal of Clinical Pathology. 2005;123(2):281–287. [PubMed] [Google Scholar]
- 5.Wilbur DC, Black-Schaffer WS, Luff RD, et al. The becton dickinson focalpoint gs imaging system: Clinical trials demonstrate significantly improved sensitivity for the detection of important cervical lesions. American Journal of Clinical Pathology. 2009;132(5):767–775. doi: 10.1309/AJCP8VE7AWBZCVQT. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Liu Y. Cervical cancer detection using svm based feature screening. Medical Image Computing and Computer-Assisted Intervention, MICCAI. 2004;3217:873–880. [Google Scholar]
- 7.Song D, Kim E, Huang X, et al. Multi-modal entity coreference for cervical dysplasia diagnosis. IEEE Trans on Medical Imaging, TMI. 2015;34(1):229–245. doi: 10.1109/TMI.2014.2352311. [DOI] [PubMed] [Google Scholar]
- 8.Li W, Gu J, Ferris D, Poirson A. Automated image analysis of uterine cervical images. SPIE Medical Imaging. 2007 [Google Scholar]
- 9.Zimmerman-Moreno G, Greenspan H. Automatic detection of specular reflections in uterine cervix images. SPIE Medical Imaging. 2006 [Google Scholar]
- 10.Ji Q, Engel J, Craine E. Classifying cervix tissue patterns with texture analysis. Pattern Recognition. 2000;33(9):1561–1574. [Google Scholar]
- 11.Jusman Y, Ng S, Osman N. Intelligent screening systems for cervical cancer. The Scientific World Journal. 2014;2014:15. doi: 10.1155/2014/810368. Article ID 810368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, Huang J, et al. Discriminative sparse representations for cervigram image segmentation. International Symposium on Biomedical Imaging, ISBI. 2010:133–136. [Google Scholar]
- 13.Lee J-J, Hwang J, et al. Integration of neural networks and decision tree classifiers for automated cytology screening. International Joint Conference on Neural Networks, IJCNN. 1991;1:257–262. [Google Scholar]
- 14.Herrero R, Schiffman M, Bratti C, et al. Design and methods of a population-based natural history study of cervical neoplasia in a rural province of costa rica: the guanacaste project. Rev Panam Salud Publica. 1997;1:362–375. doi: 10.1590/s1020-49891997000500005. [DOI] [PubMed] [Google Scholar]
- 15.Jeronimo J, Long LR, Neve L, et al. Digital tools for collecting data from cervigrams for research and training in colposcopy. Journal of Lower Genital Tract Disease. 2006;10(1):16–25. doi: 10.1097/01.lgt.0000194057.20485.5a. [DOI] [PubMed] [Google Scholar]
- 16.Razavian AS, Azizpour H, Sullivan J, Carlsson S. CNN features off-the-shelf: An astounding baseline for recognition. IEEE Conference on Computer Vision and Pattern Recognition, CVPR Workshops. 2014:512–519. [Google Scholar]
- 17.Ojala T, Pietikinen M, Harwood D. A comparative study of texture measures with classification based on feature distributions. Pattern Recognition. 1996;29:51–59. [Google Scholar]
- 18.Ojala T, Pietikinen M, Menp T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans on Pattern Analysis and Machine Intelligence, PAMI. 2002;24:971–987. [Google Scholar]
- 19.Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S, Darrell T. Caffe: Convolutional architecture for fast feature embedding. arXiv preprint arXiv:1408.5093. [Google Scholar]
- 20.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. 26th Annual Conference on Neural Information Processing Systems, NIPS; 2012. [Google Scholar]
- 21.Morra JH, Tu Z, Apostolova LG, et al. Comparison of adaboost and support vector machines for detecting alzheimer’s disease through automated hippocampal segmentation. IEEE Trans on Medical Imaging, TMI. 2010;29(1):30–43. doi: 10.1109/TMI.2009.2021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osareh A, Mirmehdi M, et al. Comparative exudate classification using support vector machines and neural networks. Medical Image Computing and Computer-Assisted Intervention, MICCAI. 2002:413–420. [Google Scholar]
- 23.Wei L, Yang Y, Nishikawa RM, Jiang Y. A study on several machine-learning methods for classification of malignant and benign clustered micro-calcifications. IEEE Trans on Medical Imaging, TMI. 2005;24(3):371–380. doi: 10.1109/tmi.2004.842457. [DOI] [PubMed] [Google Scholar]
- 24.Timoner SJ, Golland P, Kikinis R, et al. Performance issues in shape classification. Medical Image Computing and Computer-Assisted Intervention, MICCAI. 2002:355–362. [Google Scholar]
- 25.Alexander D, Zikic D, Zhang J, et al. Image quality transfer via random forest regression: Applications in diffusion mri. Medical Image Computing and Computer-Assisted Intervention, MICCAI. 2014:225–232. doi: 10.1007/978-3-319-10443-0_29. [DOI] [PubMed] [Google Scholar]
- 26.Hastie T, Tibshirani R, Friedman J, et al. The elements of statistical learning. 2009;2 [Google Scholar]
- 27.Appel R, Fuchs T, Dollr P, Perona P. Quickly boosting decision trees pruning underachieving features early. International Conference on Machine Learning, ICML. 2013;28:594–602. [Google Scholar]
- 28.Chang C, Lin C. LIBSVM: a library for support vector machines. 2001. [Google Scholar]
- 29.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 30.Goodfellow IJ, Warde-Farley D, Lamblin P, et al. Pylearn2: a machine learning research library. arXiv:1308.4214. [Google Scholar]
- 31.Fernández-Delgado M, Cernadas E, Barro S, Amorim D. Do we need hundreds of classifiers to solve real world classification problems? The Journal of Machine Learning Research. 2014;15(1):3133–3181. [Google Scholar]