Abstract
The Global Invertebrate Genomics Alliance (GIGA), a collaborative network of diverse scientists, marked its second anniversary with a workshop in Munich, Germany, where international attendees focused on discussing current progress, milestones and bioinformatics resources. The community determined the recruitment and training talented researchers as one of the most pressing future needs and identified opportunities for network funding. GIGA also promotes future research efforts to prioritize taxonomic diversity and create new synergies. Here, we announce the generation of a central and simple data repository portal with a wide coverage of available sequence data, via the compagen platform, in parallel with more focused and specialized organism databases to globally advance invertebrate genomics. Therefore this article serves the objectives of GIGA by disseminating current progress and future prospects in the science of invertebrate genomics with the aim of promotion and facilitation of interdisciplinary and international research.
Additional keywords: Metazoa, genetics, Biodiversity, evolution
Introduction
Genomic research will likely exhibit exponential growth for years to come (Stephens et al. 2015). In anticipation of this, the Global Invertebrate Genomics Alliance (GIGA) formed to address the multiple needs and opportunities of the expanding invertebrate genomics community (GIGA COS 2014). GIGA complements taxonomically similar but differently focused efforts, such as the arthropod-focused Genome 10K (Genome 10K COS 2009; Koepfli et al. 2015) and the Arthropod Genomics Consortium (i5k) (Robinson et al. 2011), and interacts with other initiatives, such as the Genomic Observatories Network (Davies et al. 2014), the Global Genome Initiative (GGI, http://naturalhistory.si.edu/ggi/), and the Ocean Genome Legacy (OGL, http://www.northeastern.edu/cos/marinescience/ogl/). These latter initiatives were created to archive diverse arrays of specimens, tissues, nucleic acid samples, voucher material, and supporting data in a global effort to help coordinate and disseminate these data and materials across the broader scientific community.
GIGA has adopted a “bottom up” philosophy to advance its community goals with voluntary and vigorous contributions of members to build a consensus approach to avoiding duplication of research efforts through effective coordination within the community (Supplementary Figure 1). GIGA was launched with an initial workshop in 2013 and an inaugural white paper (GIGA COS 2014). The second GIGA workshop (March 2015 at the Ludwig-Maximilians-Universität, Munich, Germany) reviewed the progress of this growing initiative. More than 80 scientists from 23 countries participated in the three-day event, which explored aspects of project implementation, comparative analyses across invertebrates, and training and funding efforts and opportunities. We briefly report here on the major outcomes of these discussions.
Current status of invertebrate genome/transcriptome sequencing efforts
Genomes, transcriptomes, and ‘priority species’
The first GIGA white paper (GIGA COS 2014) and two workshops identified many major hurdles hindering large-scale genome research, which have started to be addressed by concerted efforts from the community. For example, GIGA is tallying the growing number of completed invertebrate genomes from individual laboratories (Dunn and Ryan 2015), tracking new advances in sequencing and bioinformatics, and monitoring the investment of specific research consortia. To keep abreast of these ongoing efforts, the updated GIGA homepage (www.giga-cos.org) features improved access to member projects and relevant publications, and now provides phylogenetically sorted links to organismal genomes and curated transcriptome resources (see Box 1). In addition, the GIGA listserver (https://lists.lrz.de/mailman/listinfo/giga) was set up to facilitate information exchange within the community.
Box 1. The GIGA homepage as a community hub for invertebrate genomics data.
The GIGA homepage (http://giga-cos.org) provides an informative, interactive and mobile-friendly portal to relevant invertebrate genomic research, data, and news. A sign-up button welcomes new members to register, while another link invites members to upload current publications. Importantly, GIGA provides a landing page to relevant transcriptome and genome “links” with the opportunity for users to add their own assembled sequence data. The GIGA website is not meant to be a database in itself. Rather, the community nature of this homepage is emphasized by its open access to data. GIGA strongly supports contributions from the community (bottom-up philosophy) to share their new publications, links to data sets, and news items via web forms that can be filled without any prior knowledge on coding languages. Our anticipation is that the usefulness of the webpage will grow with the community.
While GIGA supports the focused efforts of single labs, the consortium acknowledges the need to collectively target priority species for sequencing due to their economic, evolutionary, and/or conservation significance (see Box 2). These priority species (not exhaustive) should occupy a distinct space in the GIGA framework, hopefully becoming the focus of future concerted community efforts (Table 1).
Box 2. Criteria for sequencing candidacy - Priority species for sequencing.
Several detailed biological criteria for species selection were discussed previously (see GIGA COS 2014), including:
keystone, endangered or threatened and invasive species status;
species representing deep, isolated phylogenetic branches;
species with contrasting life spans (particularly long or very short);
species with specific physiological abilities such as regeneration or anhydrobiosis; and
species inhabiting ‘extreme’ environments.
Practical constraints include technological, technical, financial, and cultural obstacles. Sample availability, individual researcher motivations, collaborations, and availability of genome size estimates must also be considered.
Major technical criteria for taxon selection includes:
the availability of appropriate sampling permits (following CITES, Rio Biodiversity and Nagoya Protocol guidelines (UNEP/CBD/COP 27 May 2002); https://www.cbd.int/abs/);
availability of metadata;
proper vouchering;
the species being model and/or popular;
availability of transcriptome data (or plans to generate them);
agreements for data sharing and access.
Table 1.
Proposed priority species for whole genome sequencing.
| Species | Common Name | Phylum/Class | Genome size (Gb) | Significance |
|---|---|---|---|---|
| Alitta (Nereis) virens | King ragworm or sandworm | Annelida/Polychaeta | TBD | Ecological model |
| Magelona pitelkai | N/A | Annelida/Polychaeta | TBD | early branching annelid |
| Owenia fusiformis | Spindle-shaped tubeworm | Annelida/Polychaeta | TBD | early branching annelid |
| Platynereis massiliensis | N/A | Annelida/Polychaeta | 0.3 | Ecological/developmental model |
| Spirobranchus lamarcki | Keel worm | Annelida/Polychaeta | 1.0 | Ecological/developmental model |
| Streblospio benedicti | N/A | Annelida/Polychaeta | 1.4 | developmental/ecological model |
| Artemia franciscana | Brine shrimp | Arthropoda/Branchiopoda | 0.9 | commercial, model |
| Acanthephyra purpurea | Mid-water shrimp | Arthropoda/Malacostraca | TBD | ecological importance, diurnal migrator |
| Asellus aquaticus | Waterlouse | Arthropoda/Malacostraca | 1.9 | research (ecology, evolution) |
| Barbouria cubensis | Cave shrimp | Arthropoda/Malacostraca | TBD | cave conservation importance |
| Glyptonotus spp. | Antarctic isopod | Arthropoda/Malacostraca | 0.5 | model for adaptation (cold) |
| Scylla olivacea | Orange mud crab | Arthropoda/Malacostraca | TBD | commercial |
| Sergia robusta | White shrimp | Arthropoda/Malacostraca | 3 | ecological, commercial |
| Systellaspis debilis | Mid-water shrimp | Arthropoda/Malacostraca | TBD | bioluminescent model species |
| Petrolisthes cinctipes | Porcelain crab | Arthropoda/Maxillopoda | TBD | Ecophysiology model intertidal species |
| Amphibalanus amphitrite | Striped barnacle | Arthropoda/Maxillopoda | TBD | commercial, pest |
| Pollicipes polymerus | Gooseneck Barnacle | Arthropoda/Maxillopoda | TBD | commercial, adaptation (intertidal) |
| Bugula neritina | Brown bryozoan | Bryozoa/Gymnolaemata | 0.2 | pharmaceutical |
| Cerianthus membranaceus | Cylinder anemone | Cnidaria/Anthozoa | 0.5 | sister group to Aiptasia and corals |
| Eguchipsammia fistula | Deep sea coral | Cnidaria/Anthozoa | 0.5 | deep sea ecology, adaptation (cold, depth) |
| Eunicella cavolini | Yellow sea whip | Cnidaria/Anthozoa | 0.5 | foundation species for temperature marine ecosystems |
| Heliopora coerulea | Blue coral | Cnidaria/Anthozoa | TBD | research |
| Montipora digitata | Finger coral | Cnidaria/Anthozoa | TBD | research |
| Orbicella annularis | Boulder star coral | Cnidaria/Anthozoa | TBD | research, taxonomy |
| Ricordea florida | Florida false coral | Cnidaria/Anthozoa | TBD | |
| Tubipora musica | Organ pipe coral | Cnidaria/Anthozoa | TBD | research |
| Millepora dichotoma | Net fire coral | Cnidaria/Hydrozoa | TBD | research |
| Cassiopea frondosa | Upside-down jellyfish | Cnidaria/Scyphozoa | 0.3 | research |
| Gephyrocrinus messingi | Sea lily | Echinodermata/Crinoidea | TBD | sister to remaining echinoderms |
| Himerometra robustipinna | Feather star | Echinodermata/Crinoidea | TBD | research |
| Pontiometra andersoni | Feather star | Echinodermata/Crinoidea | TBD | research, taxonomy |
| Bankia setacea | Feathery shipworm | Mollusca/Bivalvia | 1.5 | physiology, invasive |
| Bathymodiolus azoricus. | N/A | Mollusca/Bivalvia | deep sea ecology | |
| Corbicula fluminea | Asian clam or golden clam | Mollusca/Bivalvia | 1.5 | physiology, invasive |
| Dreissena polymorpha | Zebra mussel | Mollusca/Bivalvia | 1.7 | invasive, pest |
| Limnoperna fortunei | Golden mussel | Mollusca/Bivalvia | TBD | invasive |
| Mya arenaria | Soft-shell clam | Mollusca/Bivalvia | 1.4 | invasive, commercial |
| Ruditapes decussatus | Grooved carpet shell | Mollusca/Bivalvia | 1.8 | commercial, ecological |
| Ruditapes philippinarum | Manila clam | Mollusca/Bivalvia | 1.9 | commercial, invasive, model for mitochondrial biology |
| Cranchia scabra | Glass squid | Mollusca/Cephalopoda | TBD | deep sea ecology |
| Graneledone verrucosa | Atlantic longarm octopus | Mollusca/Cephalopoda | TBD | benthic deep sea species |
| Japetella diaphana | Pelagic bolitaenid octopod | Mollusca/Cephalopoda | TBD | pelagic deep sea species |
| Loligo pealeii | Longfin inshore squid | Mollusca/Cephalopoda | 2.7 | cellular neurobiology, fishery |
| Macrotritopus defillipi | N/A | Mollusca/Cephalopoda | TBD | research |
| Conus bullatus | Bubble cone | Mollusca/Gastropoda | 2.7 | natural products |
| Pomacea maculata | Apple snail | Mollusca/Gastropoda | 0.6 | invasive species, biomedical research |
| Cellana sandwicensis | Hawaiian limpet | Mollusca/Gastropoda | TBD | Model for sympatric diversification |
| Aplysina cauliformis | Row pore rope sponge | Porifera/Demospongiae | TBD | research |
| Cinachyrella alloclada | Golfball sponge | Porifera/Demospongiae | TBD | research |
| Haliclona oculata | N/A | Porifera/Demospongiae | 0.1–0.2 | research |
| Tedania ignis | Fire sponge | Porifera/Demospongiae | ||
| Themiste lageniformis | Peanut worm | Sipuncula/Sipunculidea | 0.5 | taxonomy, developmental biology |
| Milnesium tardigradum | Limno-terrestrial tardigrade | Tartigrada/Eutardigrada | TBD | taxonomy |
| Xenoturbella profundus | N/A | Xenacoelomorpha/Xenoturbellida | 0.6 | early diverging bilaterian clade |
(N/A = not available, TBD = to be determined).
We are limited in the number of taxa we can list, and therefore Table 1 only shows a small cross section of priority taxa.
Distributed effort model and ambassador framework
The GIGA II workshop identified a “divide-and-conquer” as the best strategy for the distribution of efforts concerning the coordination of genome and transcriptome sequencing and knowledge exchange in different taxonomic and phylogenetic groups and scientific communities. For example, clade-specific “ambassadors” (see http://giga-cos.org/index.php/members) were chosen to recruit within their own smaller communities and discuss candidate taxa for future genomics. The ambassador framework will also effectively distribute the efforts to provide a complete and comprehensive overview of current and future invertebrate genomic and transcriptomic resources, and GIGA members will have direct “go-to” experts for the organisms of interest.
Developing Online Resources for Sharing GIGA Data
Sequence data archives
High-throughput sequencing technologies have boosted the field of comparative genomics (Green et al. 2015; Koepfli et al. 2015). The volume of sequence data available through public databases such as the International Nucleotide Sequence Database Collaboration (INSDC) portal and its members, GenBank (at NCBI), European Nucleotide Archive (ENA) (at European Bioinformatics Institute), and DDBJ, has increased exponentially (Baxevanis 2011; Stephens et al. 2015). As overall sequencing costs continue to decrease, individual laboratories and community efforts can generate ever larger amounts of novel genomic and transcriptomic data from non-model organisms. Aggregative databases such as Compagen (http://compagen.org) (Hemmrich & Bosch 2008) or Reefgenomics (http://reefgenomics.org) (Liew et al. 2016) coordinate data presentation and analyses for particular species or ecosystem groups. While developing a single database covering data from a wide variety of invertebrate species would likely have wide appeal, the programming, curatorial, and funding burden of developing and maintaining a centralized resource goes beyond the reach of GIGA and similar consortia at present. Many researchers only need simple queries (e.g. BLAST-based sequence similarity searches), while a minority needs more complex, multivariate queries (e.g. synteny-based orthology inference). We propose an approach that will cater to both needs by providing: (1) a central sequence database that focuses on simple queries accessing a large range of data, in combination with (2) more focused, de-centralized species databases that satisfy complex queries and more specific requirements (Parkhill et al. 2010). The GIGA homepage will serve as a hub from which to access these platforms.
Compagen.org as a general data portal for GIGA transcriptomes and genomes
Compagen.org, originally designed to cater to the early branching (non-bilaterian) animal community, will be extended to serve as our central sequence database providing access to processed data sets such as assembled transcriptomes/genomes and predicted peptides, thereby complementing sequence datasets available from public domains (e.g., NCBI SRA, NCBI dbEST, and ENA trace archives). Compagen.org provides a critical extension to public databases due to the disconnect between what is submitted in the form of primary sequence data and what is available in the form of assembled and annotated data (Figure 1). Having compagen.org as a central repository will clearly facilitate provision of assembled invertebrate sequence data as well as retrieval of data for analyses. This present choice does not preclude the additional development of other (taxon or feature specific) portals to GIGA-related sequences in the future.
Figure 1.
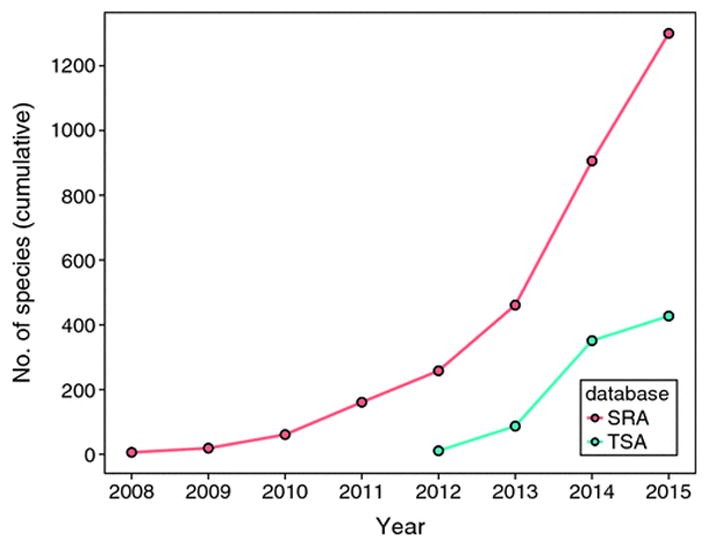
Disconnect between primary (Sequence Read Archive, SRA) and assembled (Transcriptome Shotgun Assembly, TSA) high throughput sequence data available for invertebrates at NCBI. The plot shows a strong difference between the number of species for which primary transcriptomic sequence data are available (SRA) in comparison to the number of species for which assembled transcriptomes are available (TSA). One aim of GIGA is to fill this gap by providing a hub for assembled transcriptome and genome data currently not accessible from public databases.
Databases for GIGA-relevant taxa with a larger research community
For taxon groups studied by larger research communities, it is realistic (and tractable) to develop or coordinate independent, researcher/lab-curated, taxon-specific databases, such as www.spongebase.net for sponges or http://reefgenomics.org for coral reef invertebrates. These databases can accommodate the needs of the specific research groups focusing on specific taxa or groups of taxa, emphasizing genomics, transcriptomics, variation or multi-species interaction as required. In connection with those, several single-species databases already exist, including the Aiptasia genome platform (http://aiptasia.reefgenomics.org/) (Baumgarten et al. 2015) and the Hypsibius dujardini genome server (http://www.tardigrades.org) (Koutsovoulos et al. 2016). From these efforts, cross-species databases can arise, such as http://comparative.reefgenomics.org, which provides a comparative platform for coral transcriptomes focusing on orthologous genes that can be used to interrogate coral species-specific genes (Bhattacharya et al. 2016). Community-managed data repositories should be extended to link genomic efforts to physical specimens deposited in natural history collections or to genetic resources deposited in genetic repositories in natural history museums or in other institutions. Overall, the accessibility of novel sequence data enhanced with metadata will open multiple doors to new hypotheses and crossover collaborations, thereby propelling broad scientific, technological and societal advances (i5K Consortium 2013; Dubilier et al. 2015; Green et al. 2015; Koepfli et al. 2015).
Community Opportunities
Collaboration
The GIGA II workshop highlighted the common goals shared among i5k and GIGA initiatives, primarily with regards to help in structuring and advising the community for handling large genome project initiatives. Other key shared goals include funding, expert genome annotation, and dissemination of results. There will be additional opportunities for collaboration between research groups involved in these and other large invertebrate genomics/transcriptomics consortia (such as 1Kite or the nematode.net) due to the shared methodological and bioinformatic approaches and training opportunities.
Training
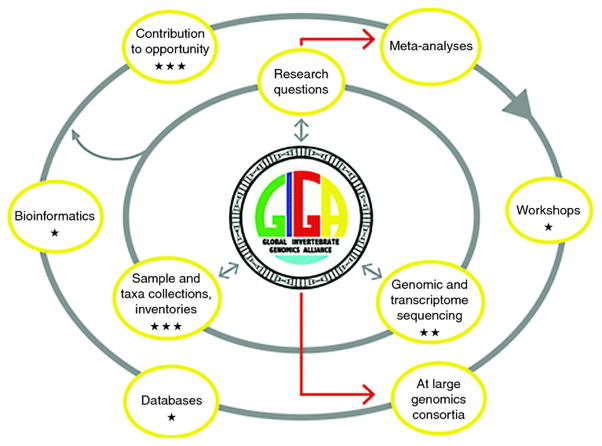
Achieving the broad goals of GIGA will require new commitment and investment in training and educating the next generation of genome scientists (Figure 2). Computational biology skills are needed to allow students to navigate, customize, and populate increasingly complex databases, but on the other hand training in aspects of organismal and evolutionary biology, ecology, and biodiversity legislation that regulates the exchange of genetic resources across national boundaries needs to be provided. Besides a sweeping view and comparison of various Big Data issues, recent reviews provide a compelling argument for the genomics community to prepare an educational foundation to handle the deluge of genomic- and its associated metadata (Stephens et al. 2015; Dougherty et al. 2016). In general, current genomics and bioinformatics curricula appear relatively staid, while independent bioinformatics workshops (e.g., evomics.org, and those listed at http://meetings.cshl.edu/courseshome.aspx) can provide cutting edge approaches and showcase innovation; however, scheduling may be more uncertain and irregular for the latter and those specialist courses do not cover all aspects needed for the holistic training needed to cater for future job markets in (invertebrate/biomedical) genomics.
Figure 2.
Proposed GIGA deliverables and funding opportunities - Funding opportunities intersect with GIGA activities. Various activities have cumulative and synergistic effect. Specific areas for funding opportunities and requests are indicated by stars, with the number proportional to estimated relative costs (e.g., workshops and conferences cost less than current sample collection or bioinformatics training – see Stephens et al. 2015)). “At Large” genomics consortia can represent new and old groups with similar or complementary goals (Genome 10K 2009; Koepfli et al. 2015).
Funding Agencies for invertebrate genomes
Currently, targeted funding for invertebrate genomes pales in comparison to the investments being made in vertebrate genomics (Couzin-Frankel 2012; AAAS 2015), so we compiled a limited selection of some of the agencies that we thought might contribute funding to further GIGA efforts (Table 2). Realizing these will eventually expire, individual scientists, small collaborative cohorts, or larger groups will have to forge imagination, fortitude, and the scientific foundations and infrastructure discussed herein to persuade funding entities on the values of each particular project. However, during GIGA II the current dearth of international, multilateral funding programs that allow researchers in different continents (e.g., EU, Asia, and Americas) to collaborate became strikingly obvious.
Table 2.
Potential funding agencies/opportunities.
| Program | URL |
|---|---|
| NSF Genealogy of Life (GoLife) | http://www.nsf.gov/funding/pgm_summ.jsp?pims_id=5129 |
| EU Innovative Training Networks (ITN) | http://ec.europa.eu/research/mariecurieactions/about-msca/actions/itn/index_en.htm |
| JGI Community Science Program (CSP) | http://jgi.doe.gov/collaborate-with-jgi/community-science-program/ |
| Earth Cube | http://earthcube.org/home |
| Alfred P. Sloan Foundation | http://www.sloan.org/apply-for-grants/the-grant-application-process/ |
| Gordon and Betty Moore Foundation | https://www.moore.org/grants/list |
Concluding Statement
Many benefits derived from single or multi-species genome sequencing projects have been described, realized, and projected in recent years (GIGA COS 2014). Discoveries will continue to accrue with further innovations. Efforts will be further enhanced through the GIGA framework, which coordinates and promotes the concerted contributions of diverse experts. In the words of Immanuel Kant (Durant 1938a) (by way of William Durant (Durant 1938b)), “Science is organized knowledge; wisdom is organized life”. In a similar spirit, GIGA will facilitate scientific organization, and consequently the collective progress (wisdom) of humanity.
Supplementary Material
Supplementary Figure 1. Current GIGA membership by country (n = 255). Numbers combine both registrations on the GIGA-cos.org website and in the GIGA listserver.
Acknowledgments
The GIGA Community of Scientists gives special thanks to the German Research Foundation (DFG, project Wo896/16-1 to G. Wörheide) for substantially supporting the GIGAII workshop in Munich (Germany) in March 2015. This work was funded in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
Appendix
GIGA community of Scientists (COS) – Marcin Adamski (University of Bergen, Bergen, Norway), Shobhit Agrawal (King Abdullah University of Science and Technology, Thuwal, Saudi Arabia), Agostinho Antunes (CIIMAR, University of Porto, Portugal), Manuel Aranda (King Abdullah University of Science and Technology, Thuwal, Saudi Arabia), Andreas D. Baxevanis (National Institutes of Health, Bethesda MD, USA), Mark Blaxter (University of Edinburgh, Edinburgh, Scotland), Thomas Bosch (University of Kiel, Kiel, Germany), Heather Bracken-Grissom (Florida International University, Miami FL, USA), Jonathan Coddington (Smithsonian Institution, Washington DC, USA), Timothy Collins (Florida International University, Miami FL, USA), Keith Crandall (George Washington University, Washington DC, USA), Mikael Dahl (The Arctic University of Tromsø, Norway), Daniel Distel (Ocean Genome Legacy, Nahant, MA, USA), Casey Dunn (Brown University, Providence RI, USA), Michael Eitel (Ludwig-Maximilians-Universität München, München, Germany), Jean-Francois Flot (Université Libre de Bruxelles, Belgium), Gonzalo Giribet (Museum of Comparative Zoology, Harvard University, Cambridge MA, USA), Fabrizio Ghiselli (University of Bologna, Bologna, Italy), Steven Haddock (MBARI, Monterey CA, USA), Kevin Kocot (University of Alabama, Tuscaloosa AL, USA), Yi Jin Liew (King Abdullah University of Science and Technology, Thuwal, Saudi Arabia), Audrey Majeske (University of Puerto Rico Mayagüez, Puerto Rico), Bernhard Misof (Zoological Research Museum Alexander Koenig, Leibniz, Germany), Christopher Mungal (Lawrence Berkeley National Laboratory, Berkeley CA, USA), Stephen J. O’Brien (Nova Southeastern University, Dania Beach FL, USA), Timothy Ravasi (King Abdullah University of Science and Technology, Thuwal, Saudi Arabia), Mauro de Freitas Rebelo (Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil), Ana Riesgo (Natural History Museum, London, United Kingdom), Christine Schnitzler (National Institutes of Health, Bethesda MD, USA), Anja Schulze (Texas A&M University at Galveston, TX, USA), Billie Swalla (Friday Harbor Laboratory WA, USA), Denis Tagu (French Institute for Agricultural Research, Paris, France), Robert Toonen (University of Hawaii at Manoa HI, USA), Marcela Uliano da Silva (Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil), Sergio Vargas (Ludwig-Maximilians-Universität München, München, Germany), Xin Zhou (Beijing Genomics Institute, Shenzhen, China).
References
- AAAS. Report: Research & Development series and analysis of FY 2016 budget request (AAAS) 2015. [Google Scholar]
- Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, Li Y, Hambleton EA, Guse A, Oates ME, Gough J, Weis VM, Aranda M, Pringle JR, Voolstra CR. The genome of Aiptasia a sea anemone model for coral symbiosis. Proceedings of the National Academy of Sciences of the USA. 2015;112:11893–11898. doi: 10.1073/pnas.1513318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis AD. The importance of biological databases in biological discovery. Current Protocols in Bioinformatics. 2011;Chapter 1(Unit 1 1) doi: 10.1002/0471250953.bi0101s34. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Agrawal S, Aranda M, Baumgarten S, Belcaid M, Drake JL, Erwin D, Foret S, Gates RD, Gruber DF, Kamel B, Lesser MP, Levy O, Liew YJ, MacManes M, Mass T, Medina M, Mehr S, Meyer E, Price DC, Putnam HM, Qiu H, Shinzato C, Shoguchi E, Stokes AJ, Tambutté S, Tchernov D, Voolstra CR, Wagner N, Walker CW, Weber APM, Weis V, Zelzion E, Zoccola D, Falkowski PG. Comparative genomics explains the evolutionary success of reef-forming corals. eLife. 2016;5:e13288. doi: 10.7554/eLife.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzin-Frankel J. Research philanthropy. Billionaire restores funding for novel biomedical network. Science. 2012;338:873–874. doi: 10.1126/science.338.6109.873. [DOI] [PubMed] [Google Scholar]
- Davies N, Field D, Amaral-Zettler L, Clark MS, Deck J, Drummond A, Faith DP, Geller J, Gilbert J, Glockner FO, Hirsch PR, Leong JA, Meyer C, Obst M, Planes S, Scholin C, Vogler AP, Gates RD, Toonen R, Berteaux-Lecellier V, Barbier M, Barker K, Bertilsson S, Bicak M, Bietz MJ, Bobe J, Bodrossy L, Borja A, Coddington J, Fuhrman J, Gerdts G, Gillespie R, Goodwin K, Hanson PC, Hero JM, Hoekman D, Jansson J, Jeanthon C, Kao R, Klindworth A, Knight R, Kottmann R, Koo MS, Kotoulas G, Lowe AJ, Marteinsson VT, Meyer F, Morrison N, Myrold DD, Pafilis E, Parker S, Parnell JJ, Polymenakou PN, Ratnasingham S, Roderick GK, Rodriguez-Ezpeleta N, Schonrogge K, Simon N, Valette-Silver NJ, Springer YP, Stone GN, Stones-Havas S, Sansone SA, Thibault KM, Wecker P, Wichels A, Wooley JC, Yahara T, Zingone A, COS GO. The founding charter of the Genomic Observatories Network. Gigascience. 2014;3:2. doi: 10.1186/2047-217X-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty MJ, Wicklund C, Johansen Taber KA. Challenges and Opportunities for Genomics Education: Insights from an Institute of Medicine Roundtable Activity. The Journal of continuing education in the health professions. 2016;36:82–85. doi: 10.1097/CEH.0000000000000019. [DOI] [PubMed] [Google Scholar]
- Dubilier N, McFall-Ngai M, Zhao L. Microbiology: Create a global microbiome effort. Nature. 2015;526:631–634. doi: 10.1038/526631a. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Ryan JF. The evolution of animal genomes. Current Opinion in Genetics & Development. 2015;35:25–32. doi: 10.1016/j.gde.2015.08.006. http://dx.doi.org/10.1016/j.gde.2015.08.006. [DOI] [PubMed] [Google Scholar]
- Durant W, editor. The Story of Philosophy. Garden City Publishing Company; Garden City, New York: 1938a. Kant and German Idealism: Transcental Analytic. [Google Scholar]
- Durant W. The Story of Philosophy. Simon and Schuster; New York: 1938b. [Google Scholar]
- Genome 10K Community of Scientists. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. Journal of Heredity. 2009;100:659–674. doi: 10.1093/jhered/esp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIGA Community of Scientists. The Global Invertebrate Genomics Alliance (GIGA): developing community resources to study diverse invertebrate genomes. Journal of Heredity. 2014;105:1–18. doi: 10.1093/jhered/est084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ED, Watson JD, Collins FS. Human Genome Project: Twenty-five years of big biology. Nature. 2015;526:29–31. doi: 10.1038/526029a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmrich G, Bosch TC. Compagen, a comparative genomics platform for early branching metazoan animals, reveals early origins of genes regulating stem-cell differentiation. Bioessays. 2008;30:1010–1018. doi: 10.1002/bies.20813. [DOI] [PubMed] [Google Scholar]
- i5K Consortium. The i5K Initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. Journal of Heredity. 2013;104:595–600. doi: 10.1093/jhered/est050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli KP, Paten B, Genome KCOS, O’Brien SJ. The Genome 10K Project: a way forward. Annual Review of Animal Biosciences. 2015;3:57–111. doi: 10.1146/annurev-animal-090414-014900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsovoulos G, Kumar S, Laetsch DR, Stevens L, Daub J, Conlon C, Maroon H, Thomas F, Aboobaker AA, Blaxter M. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proceedings of the National Academy of Sciences of the USA. 2016;113:5053–5058. doi: 10.1073/pnas.1600338113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew YJ, Aranda M, Voolstra CR. reefgenomics.org-a repository for marine genomics data. Database. 2016 doi: 10.1093/database/baw152. in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Birney E, Kersey P. Genomic information infrastructure after the deluge. Genome Biology. 2010;11:402. doi: 10.1186/gb-2010-11-7-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GE, Hackett KJ, Purcell-Miramontes M, Brown SJ, Evans JD, Goldsmith MR, Lawson D, Okamuro J, Robertson HM, Schneider DJ. Creating a buzz about insect genomes. Science. 2011;331:1386. doi: 10.1126/science.331.6023.1386. [DOI] [PubMed] [Google Scholar]
- Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, Iyer R, Schatz MC, Sinha S, Robinson GE. Big Data: Astronomical or Genomical? PLoS Biology. 2015;13:e1002195. doi: 10.1371/journal.pbio.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNEP/CBD/COP. Report of the sixth meeting of the conference of the parties to the convention on biological diversity: Decision VI/7: Identification, monitoring, indicators and assessments. The Hague; Netherlands: May 27, 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Current GIGA membership by country (n = 255). Numbers combine both registrations on the GIGA-cos.org website and in the GIGA listserver.