Abstract
Aberrant activation of Notch signaling pathway has been correlated with high grade ovarian carcinoma and carcinogenesis. However, the predictive and prognostic values of Notch signaling pathway in ovarian cancer patients remains unclear. We utilize “The Kaplan-Meier plotter” (KM plotter) background database to access the prognostic values including overall survival (OS), progression-free survival (PFS), as well as post-progression survival (PPS) of four Notch receptor mRNA expression in ovarian cancer patients. Notch1 mRNA high expression was not correlated with OS, PFS and PPS for all ovarian cancer patients, but significantly correlated with poor PFS in TP53 wild type and favorite PFS in TP53 mutation type ovarian cancer patients. Notch2 mRNA high expression was significantly correlated with poor PFS for all ovarian cancer patients, especially in grade II patients. Notch3 mRNA high expression was significantly correlated with favorite PFS for all ovarian cancer patients. Notch4 mRNA high expression was significantly correlated with favorite OS, but not PFS and PPS for all ovarian cancer patients. The results strongly support that there are distinct prognostic values of four Notch receptor mRNA expression in ovarian cancer patients.
Keywords: Notch receptor, mRNA expression, KM plotter, hazard ratio, prognosis
INTRODUCTION
Cancer of the ovary is not common, but it ranks fourth as the cause of cancer deaths and causes more deaths than other female reproductive cancers in women [1–2]. Most ovarian tumors often initiated from ovarian surface epithelial (OSE) cells, thus have epithelial origins [3]. Although the improvement in early diagnosis, surgery, various operations for the radical cure, chemotherapy, targeted therapeutic treatment and the emerging immunotherapy, most of the cancer patients would experience recurrent disease following first-line therapy [4–5]. Therefore, the study on the molecular mechanisms of carcinogenesis and identification of differential diagnostic, prognostic marker is still needed.
The Notch signaling pathway that regulates the maintenance of stem cells and controls cell-fate decisions is an evolutionarily conserved system [6–7]. Deregulated expression of four Notch receptors and their ligands has been observed in several human malignancies including ovarian cancer [8–11]. The aberrant activation of Notch signaling pathway plays the imperative roles in ovarian cancer carcinogenesis and chemoresistance of ovarian cancer patients [12–17]. Recently, a number of studies also demonstrated that Notch signaling pathway, especially Notch1 is important for maintaining cancer stem cells in ovarian cancer [18–20]. DAPT, a γ-secretase inhibitor, which reduces gamma-secretase in Notch1 signaling pathway was reported as a highly promising novel therapeutic drug candidate for ovarian cancer patient [21]. LY900009, a first-in-human phase I study of the oral Notch inhibitor was also reported in patients with advanced cancer including ovarian cancer [22]. MK-0752 is another novel γ-secretase inhibitor, which is evaluated in clinical trial for treatment of several types of cancer including ovarian cancer [23]. However, at mRNA level, the predictive roles of individual Notch receptors in ovarian cancer patients remain unknown. In this study, we accessed the predictive roles of Notch receptor mRNA expression in human ovarian cancer patients.
The “Kaplan-Meier plotter” (KM plotter) was capable of assessing the effect of 54,675 genes on survival of 1,648 ovarian cancer patients (http://kmplot.com/analysis/) [24]. KM plotter [25] (http://kmplot.com/analysis/index.php?p=service&cancer=ovar), handled by a PostgreSQL server, which integrates gene expression and clinical data simultaneously. KM plotter was established using gene expression data and survival information [25]. Until now, several genes, such as ALDH1, ITIH5, CK2, GREB1 have been identified and validated by KM plotter in lung cancer [26–9], breast cancer [29–39], as well as in ovarian cancer [29, 40–41]. In this study, we took advantage of KM plotter and accessed the prognostic roles of four Notch receptors in 1,648 ovarian cancer patients.
RESULTS
Notch receptors include Notch1~4 family members. All Notch receptors can be found Kaplan-Meier OS, PFS, as well as PPS information in the KM plotter database.
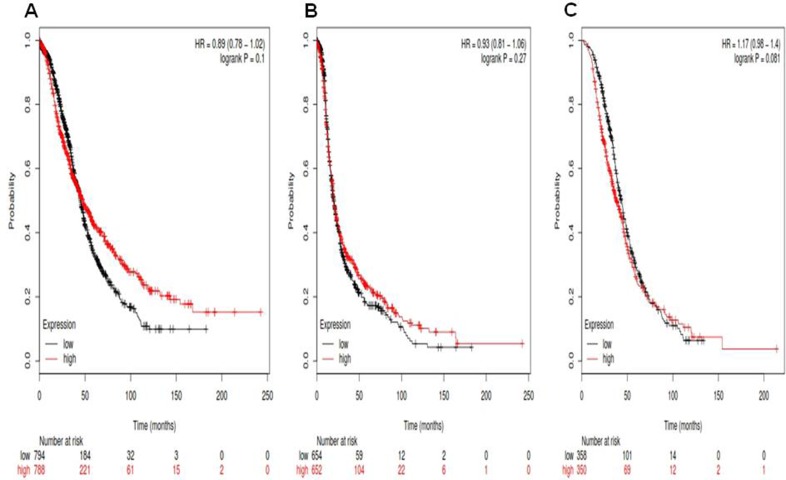
For Notch1, its Affymetrix ID is 218902_at. OS curves are plotted for ovarian cancer patients (n = 1,582) (Figure 1A), PFS curves are plotted for ovarian cancer patients (n = 1,306) (Figure 1B) and PPS curves are plotted for ovarian cancer patients (n = 708) (Figure 1C). Notch1 mRNA high expression was not correlated to OS for all ovarian cancer patients followed for 20 years, HR 0.89 (0.78–1.02), p = 0.1. Notch1 mRNA high expression was also not correlated to PFS ovarian cancer patients, 0.93 (0.81–1.06), p = 0.27, as well as PPS in ovarian cancer patients, HR 1.17 (0.98–1.4), p = 0.081.
Figure 1. For Notch1, its Affymetrix ID is 218902_at.
(A) OS curves are plotted for ovarian cancer patients (n = 1,582). (B) PFS curves are plotted for ovarian cancer patients (n = 1,306). (C) PPS curves are plotted for ovarian cancer patients (n = 708).
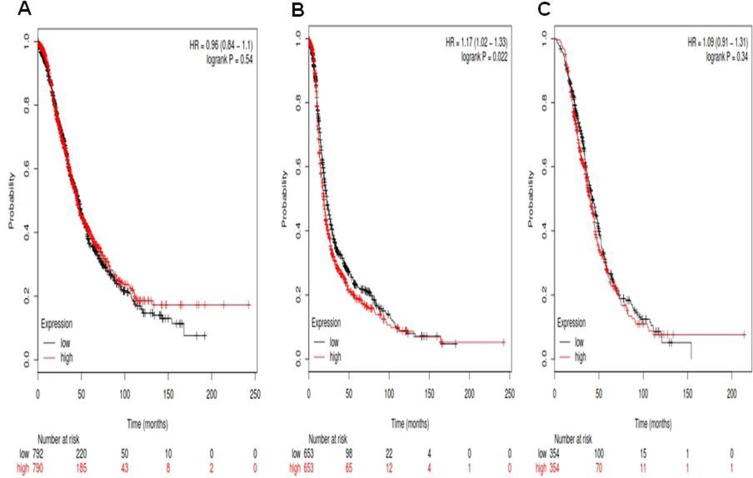
For Notch2, its Affymetrix ID is 210756_s_at. Notch2 mRNA high expression was not correlated to OS for all ovarian cancer patients HR, 0.96 (0.84–1.1), p = 0.54 (Figure 2A). However, Notch2 mRNA high expression was significantly correlated to poor PFS for all ovarian cancer patients, HR 1.17 (1.02–1.33), p = 0.022 (Figure 2B). Notch2 mRNA high expression was not correlated to PPS in ovarian cancer patients, HR 1.09 (0.91–1.31), p = 0.34 (Figure 2C).
Figure 2. For Notch2, its Affymetrix ID is 210756_s_at.
(A) OS curves are plotted for all ovarian cancer patients (n = 1,582). (B) PFS curves are plotted for ovarian cancer patients (n = 1,306). (C) PPS curves are plotted for ovarian cancer patients (n = 708).
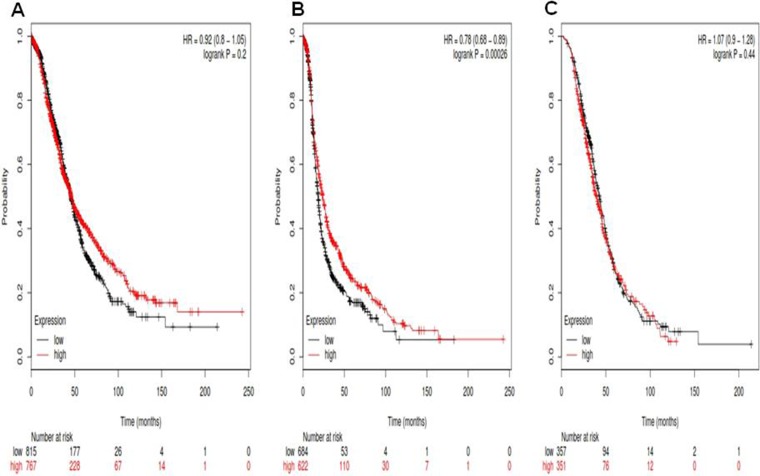
For Notch3, its Affymetrix ID is 203237_at. Notch3 mRNA high expression was not correlated to OS for all ovarian cancer patients HR, 0.92 (0.8–1.05), p = 0.2 (Figure 3A). However, Notch3 mRNA high expression was significantly correlated to favorite PFS for all ovarian cancer patients, HR 0.78 (0.68–0.89), p = 0.00026 (Figure 3B). Notch3 mRNA high expression was not correlated to PPS in ovarian cancer patients, HR 1.07 (0.9–1.28), p = 0.44 (Figure 3C).
Figure 3. For Notch3, its Affymetrix ID is 203237_at.
(A) OS curves are plotted for all ovarian cancer patients (n = 1,582). (B) PFS curves are plotted for ovarian cancer patients (n = 1,306). (C) PPS curves are plotted for ovarian cancer patients (n = 708).
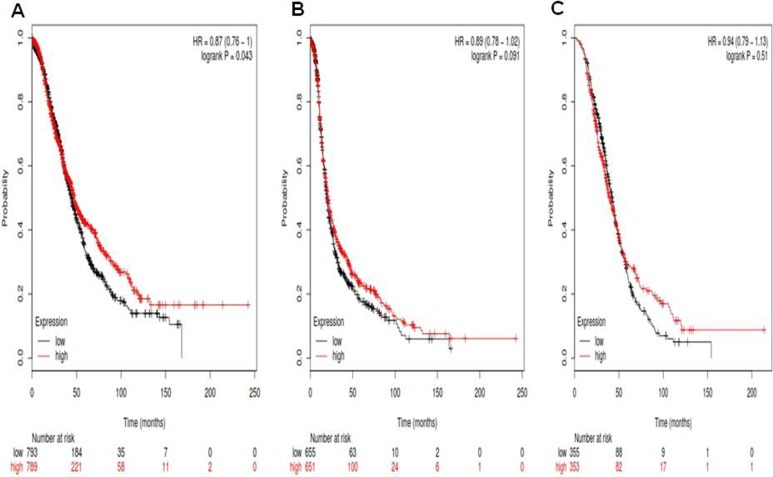
For Notch4, its Affymetrix ID is 205247_at. Notch4 mRNA high expression was significantly correlated to favorite OS for all ovarian cancer patients, HR 0.87 (0.76–1), p = 0.043 (Figure 4A). Notch4 mRNA high expression was not significantly correlated to PFS for all ovarian cancer patients, HR0.89 (0.78–1.02), p = 0.091 (Figure 4B). Notch4 mRNA high expression was not correlated to PPS in ovarian cancer patients, HR 0.94 (0.79–1.13), p = 0.51 (Figure 4C).
Figure 4. For Notch4, its Affymetrix ID is 205247_at.
(A) OS curves are plotted for all ovarian cancer patients (n = 1,582). (B) PFS curves are plotted for ovarian cancer patients (n = 1,306). (C) PPS curves are plotted for ovarian cancer patients (n = 708).
For further access the correlation of individual Notch receptor mRNA expression with other clinicopathological features, we examined the correlation of PFS with different histological types (Table 1), pathological grades (Table 2), clinical grades (Table 3) and TP53 mutation (Table 4) of ovarian cancer patients. As from Table 1, all the individual Notch receptors were not significantly associated with PFS in different histological types of ovarian cancer patients. In addition, all the individual Notch receptors were also not significantly associated with OS and PPS in different histological types of ovarian cancer patients (data not shown). From Table 2, all the individual Notch receptors except Notch 2 were not significantly associated with PFS in pathological grades of ovarian cancer patients. Notch 2 mRNA high expression was associated with poor PFS in grade II ovarian cancer patients, HR 1.45 (1.07–1.96), p = 0.016. In addition, Notch 4 mRNA high expression was associated with favorite OS in grade III ovarian cancer patients, HR 0.76 (0.64–0.9), p = 0.0018. From Table 3, all the individual Notch receptors were not significantly associated with PFS in clinical stages of ovarian cancer patients. However, Notch 1 mRNA high expression was associated with favorite OS in clinical stage III ovarian cancer patients, HR 0.83 (0.7–0.98), p = 0.03. Notch 3 mRNA high expression was also associated with favorite OS in clinical stage III ovarian cancer patients, HR 0.83 (0.7–0.99), p = 0.037. From Table 4, only Notch 1 mRNA high expression was significantly associated with poor PFS in TP53 wild type ovarian cancer patients, HR 1.86 (1.05–3.31), p = 0.031, but Notch 1 mRNA high expression was significantly associated with favorite PFS in TP53 mutation type ovarian cancer patients, HR 0.78 (0.6–0.99), p = 0.045.
Table 1. Correlation of Notch receptor mRNA high expression with PFS in different histology of ovarian cancer patients.
| Notch receptors | histology | Cases | HR 95% CI | P value |
|---|---|---|---|---|
| Notch 1 | serous | 1019 | 0.99 (0.85–1.15) | 0.9 |
| endometrioid | 36 | 0.48 (0.17–1.33) | 0.15 | |
| Notch 2 | serous | 1019 | 1.13 (0.97–1.32) | 0.11 |
| endometrioid | 36 | 1.35 (0.5–3.62) | 0.55 | |
| Notch 3 | serous | 1019 | 1.06 (0.91–1.23) | 0.45 |
| endometrioid | 36 | 1.59 (0.58–4.38) | 0.36 | |
| Notch 4 | serous | 1019 | 1.07 (0.92–1.25) | 0.37 |
| endometrioid | 36 | 0.69 (0.26–1.84) | 0.45 |
Table 2. Correlation of Notch receptor mRNA high expression with PFS in pathological grades of ovarian cancer patients.
| Notch receptors | Pathological grades | Cases | HR 95% CI | P value |
|---|---|---|---|---|
| Notch 1 | I | 37 | 0.68 (0.23–2.03) | 0.48 |
| II | 247 | 1.19 (0.89–1.61) | 0.25 | |
| III | 790 | 0.93 (0.78–1.11) | 0.44 | |
| Notch 2 | I | 37 | 0.93 (0.31–2.77) | 0.89 |
| II | 247 | 1.45 (1.07–1.96) | 0.016 | |
| III | 790 | 1.03 (0.87–1.23) | 0.71 | |
| Notch 3 | I | 37 | 1.66 (0.54–5.08) | 0.37 |
| II | 247 | 1.21 (0.89–1.63) | 0.22 | |
| III | 790 | 0.94 (0.79–1.12) | 0.48 | |
| Notch 4 | I | 37 | 0.38 (0.12 −1.22) | 0.091 |
| II | 247 | 1.03 (0.76–1.38) | 0.86 | |
| III | 790 | 1.04 (0.88–1.24) | 0.64 |
Note: three-tier grading scheme for pathological classification.
Table 3. Correlation of Notch receptor mRNA high expression with PFS in clinical stages of ovarian cancer patients.
| Notch receptors | Clinical stages | Cases | HR 95% CI | P value |
|---|---|---|---|---|
| Notch 1 | I + II III IV |
126 846 143 |
0.99 (0.55–1.79) 0.98 (0.83–1.15) 1.05 (0.7–1.57) |
0.97 0.80 0.81 |
| Notch 2 | I + II III IV |
126 846 143 |
0.79 (0.43–1.43) 1.07 (0.91–1.26) 1.48 (0.98–2.22) |
0.43 0.39 0.059 |
| Notch 3 | I + II III IV |
126 846 143 |
1.09 (0.6–1.98) 0.99 (0.84–1.16) 1.01 (0.67–1.5) |
0.77 0.86 0.98 |
| Notch 4 | I + II III IV |
126 846 143 |
1.12 (0.61–2.06) 1.09 (0.93–1.28) 1.37 (0.92–2.06) |
0.71 0.30 0.12 |
Note: four-tier grading scheme for clinical stages.
Table 4. Correlation of Notch receptor mRNA high expression with PFS in TP53 mutation status of ovarian cancer patients.
| Notch receptors | TP53 mutation | Cases | HR 95% CI | P value |
|---|---|---|---|---|
| Notch 1 | No Yes |
76 416 |
1.86 (1.05–3.31) 0.78 (0.6–0.99) |
0.031 0.045 |
| Notch 2 | No Yes |
76 416 |
0.84 (0.48–1.48) 1.05 (0.82–1.35) |
0.55 0.68 |
| Notch 3 | No Yes |
76 416 |
0.74 (0.42–1.31) 0.88 (0.69–1.13) |
0.31 0.33 |
| Notch 4 | No Yes |
76 416 |
1.15 (0.65–2.03) 0.94 (0.73–1.21) |
0.63 0.64 |
DISCUSSION
Notch1 was widely reported in ovarian carcinogenesis and was the best studied among Notch ligands and four Notch receptors [12–15]. The active form of Notch 1, the Notch 1 intracellular domain (NICD), was detected in ovarian cancer cell lines, ovarian cancer specimens and may led to growth inhibition of ovarian cancer cells upon depletion of Notch 1 by Notch 1 siRNA [42]. Down-regulation of Notch1 expression was significantly inhibit cell growth, induce G1 cell cycle arrest and induce cell apoptosis in A2780 ovarian cancer cells [43]. Notch1 NICD was reported to be an independently poor prognostic factor in ovarian cancer patients [44]. In this study, we found that Notch1 mRNA high expression was not correlated to PFS for all ovarian cancer patients. However, Notch 1 mRNA high expression is significantly associated with poor PFS in TP53 wild type, but favorite PFS in TP53 mutation type ovarian cancer patients. In addition, Notch1 mRNA high expression was also not correlated to PFS ovarian cancer patients, 0.93 (0.81–1.06), p = 0.27, as well as PPS in ovarian cancer patients, HR 1.17 (0.98–1.4), p = 0.081.
Notch2 was aberrant expressed ovarian cancer cells [44]. These results indicate that Notch2 seems to be a tumor suppressor in ovarian carcinogenesis. In this study, we found that Notch2 mRNA high expression was significantly correlated to poor PFS for all ovarian cancer patients, especially in grade II ovarian cancer patients. However, Notch2 mRNA high expression was not correlated to poor PFS in serous and endometrioid cancer patients. In addition, Notch2 mRNA high expression was not correlated to OS for all ovarian cancer patients HR, 0.96 (0.84–1.1), p = 0.54. Notch2 mRNA high expression was also not correlated to PPS in ovarian cancer patients, HR 1.09 (0.91–1.31), p = 0.34.
Notch3 high protein expression was detected in high-grade ovarian tumors [45]. Inactivation of Notch3 suppressed cell proliferation and induced apoptosis in the ovarian cancer cells [45]. Jagged-1/Notch3 interaction constitutes a juxtacrine loop promoting proliferation in ovarian cancer cells [46]. Notch 3 protein overexpression was associated with ovarian cancer metastasis, chemoresistance and poor overall survival in ovarian serous cancer patients [47]. Inhibition of Notch3 inhibited ovarian cancer growth and induced apoptosis [48]. In comparison with gamma-secretase inhibitor (GSI) in the treatment of paclitaxel in paclitaxel-resistant cancer cells, Notch3 siRNA specific inhibition showed more efficacy [49]. This approach of using more specific individual Notch member inhibitor would be likely to avoid the side effects of broad-spectrum GSI treatment and has more potential to use in clinical setting. Our results showed that Notch3 mRNA high expression was significantly correlated to favorite PFS for all ovarian cancer patients. However, Notch3 mRNA high expression was not correlated to OS for all ovarian cancer patients HR, 0.92 (0.8–1.05), p = 0.2; Notch3 mRNA high expression was also not correlated to PPS in ovarian cancer patients, HR 1.07 (0.9–1.28), p = 0.44.
Notch4 was reported as an oncogene in mammary carcinogenesis [50–51]. Notch4 significantly increased the tumorigenic potential [52–53]. Gao et al. [54] reported that Notch4 may be also an oncogene in ovarian carcinogenesis, since Notch4 was involved in modulating many functions of stem cells. We found that Notch4 mRNA high expression was not significantly correlated to PFS for all ovarian cancer patients. However, Notch4 mRNA high expression was significantly correlated to favorite OS for all ovarian cancer patients, HR 0.87 (0.76–1), p = 0.043. Notch4 mRNA high expression was not correlated to PPS in ovarian cancer patients, HR 0.94 (0.79–1.13), p = 0.51.
Notch members and TP53 are gene transcription regulators that are critically involved in various aspects of stem cell maintenance, cell differentiation, and tumor progression. Thus, extensive crosstalks between the Notch and TP53 pathways were reported about above processes [55]. TP53 and some of Notch members have also been identified as potential prognostic biomarkers in ovarian cancer patients [47, 56–57], however, there are no report about the association between TP53 and Notch members in ovarian cancer. Interestingly, there are strong evidences showing the correlation between TP53 and Notch members in breast cancer [58]. In our analysis, only Notch 1 mRNA high expression was significantly associated with poor PFS in TP53 wild type ovarian cancer patients, HR 1.86 (1.05–3.31), p = 0.031, but Notch 1 mRNA high expression was significantly associated with favorite PFS in TP53 mutation type ovarian cancer patients, HR 0.78 (0.6–0.99), p = 0.045. These results indicate that TP53 status significantly impact the prognostic value of Notch 1 in ovarian patients.
Previous results suggest that Notch signaling, especially Notch receptors may be essential drug target for ovarian cancer patients. However, so far, not many specific small molecular inhibitors or other antagonists of the different Notch members have been developed. γ-secretase inhibitor, DAPT was demonstrated to inhibit Notch activation and cell growth in ovarian cancer cells [21]. However, γ-secretase inhibitors are not able to distinguish individual Notch receptors and may cause intestinal toxicity [59] by inhibiting other signaling pathways [60]. Recently, highly specialized antibodies which can recognize each Notch receptor paralogue were developed by phage display technology in human patients and rodent models [61]. Based on our study that Notch1 mRNA high expression was significantly correlated with poor PFS in TP53 wild type ovarian cancer patients. Notch2 mRNA high expression was significantly correlated with poor PFS for all ovarian cancer patients, especially in grade II patients. Thus Notch1 and Notch2 might be potential drug targets for some types of ovarian cancer patients.
In summary, we demonstrated that Notch 1 mRNA high expression is significantly associated with poor PFS in TP53 wild type, but favorite PFS in TP53 mutation type ovarian cancer patients. Notch2 mRNA high expression was significantly correlated to poor PFS for all ovarian cancer patients, especially in grade II ovarian cancer patients. Notch3 mRNA high expression was significantly correlated to favorite PFS for all ovarian cancer patients. Notch4 mRNA high expression was not significantly correlated to PFS for all ovarian cancer patients. These results will be useful for favorite understand the heterogeneity and complexity in the molecular biology of ovarian cancer and to develop tools to more accurately predict their prognosis.
MATERIALS AND METHODS
KM plotter was used to analyze the correlation of individual Notch receptor mRNA expression to overall survival (OS), progression-free survival (PFS), as well as post-progression survival (PPS). The background database include lung cancer [26], breast cancer [24], gastric cancer, as well as ovarian cancer [25] database. Ovarian cancer patients in the database were identified from Cancer Biomedical Informatics Grid (caBIG, https://biospecimens.cancer.gov/relatedinitiatives/overview/caBig.asp), the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) ovarian cancer datasets [25]. They contain clinical data such as gender, age, histology, grade, stage, applied chemotherapy and TP53 mutation status for all patients in WinStat 2013. The ovarian cancer patients were followed up 20 years. The database collected survival information of 1,648 ovarian cancer patients downloaded from Gene Expression Omnibus (GEO). Four Notch sub-members (Notch1~4) were put into the database (http://kmplot.com/analysis/index.php?p=service&cancer=ovar) to obtain Kaplan-Meier various survival plots. In order to determine the prognostic value of a particular gene, the samples were split into two groups according to the median expression of the gene. The certain gene mRNA expression above or below the median separates the cases into high expression and low expression. KM plotter also provides options to split patients by lower quartile, lower tertile, upper tertile, upper quartile expression, but only median expression giving almost same numbers of two groups and less biasing. Hazard ratio (HR), 95% confidence intervals and log rank P were analyzed and presented on the main plots. P value of < 0.05 was considered to be statistically significant. HR is the ratio of the hazard rates corresponding to the conditions described by two levels of an explanatory variable in survival analysis.
Acknowledgments
This work was supported by Nature Science Foundation of China grant number 81472554, Third Military Medical University grant number SWH2016LCZD-03 and SWH2016JCYB-28.
Footnotes
Authors’ contributions
CC, XW, SY participated in the design of the study and drafted the manuscript. CC, XW, SH, LW, LH reviewed and extracted data. CC, XW and SY participated in the search the study and performed the statistical analysis. All authors reviewed the manuscript.
CONFLICTS OF INTEREST
The authors have no financial involvement in this work.
REFERENCES
- 1.Ovarian, cancer, national and alliance 2013 http://www.ovariancancer.org/about-ovarian-cancer/statistics/
- 2.Lee R, Yeung A, Hong S, Brose M, Michels D. Principles of medical oncology. Asian Pac J Surg Oncol. 2015;1:39–46. [Google Scholar]
- 3.Leung PC, Choi JH. Endocrine signaling in ovarian surface epithelium and cancer. Hum Reprod Update. 2007;13:143–162. doi: 10.1093/humupd/dml002. [DOI] [PubMed] [Google Scholar]
- 4.Foley OW, Rauh-Hain JA, del Carmen MG. Recurrent epithelial ovarian cancer: an update on treatment. Oncology (Williston Park) 2013;27:288–294. 298. [PubMed] [Google Scholar]
- 5.Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat Rev Clin Oncol. 2013;10:211–224. doi: 10.1038/nrclinonc.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis J. Notch signalling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol. 1998;9:583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- 7.Simpson P. Developmental genetics. The Notch connection. Nature. 1995;375:736–737. doi: 10.1038/375736a0. [DOI] [PubMed] [Google Scholar]
- 8.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miele L. Notch signaling. Clin Cancer Res. 2006;12:1074–1079. doi: 10.1158/1078-0432.CCR-05-2570. [DOI] [PubMed] [Google Scholar]
- 10.Miele L, Miao H, Nickoloff BJ. NOTCH signaling as a novel cancer therapeutic target. Curr Cancer Drug Targets. 2006;6:313–323. doi: 10.2174/156800906777441771. [DOI] [PubMed] [Google Scholar]
- 11.Alketbi A, Attoub S. Notch Signaling in Cancer: Rationale and Strategies for Targeting. Curr Cancer Drug Targets. 2015;15:364–374. doi: 10.2174/156800961505150710113353. [DOI] [PubMed] [Google Scholar]
- 12.Wang M, Ma X, Wang J, Wang L, Wang Y. Pretreatment with the gamma-secretase inhibitor DAPT sensitizes drug-resistant ovarian cancer cells to cisplatin by downregulation of Notch signaling. Int J Oncol. 2014;44:1401–1409. doi: 10.3892/ijo.2014.2301. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Huang X, Zhang J, Shao N, Chen LO, Ma D, Ji C. The expression of VEGF and Dll4/Notch pathway molecules in ovarian cancer. Clin Chim Acta. 2014;436:243–248. doi: 10.1016/j.cca.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Gou WF, Zhao Y, Lu H, Yang XF, Xiu YL, Zhao S, Liu JM, Zhu ZT, Sun HZ, Liu YP, Xu F, Takano Y, Zheng HC. The role of RhoC in epithelial-to-mesenchymal transition of ovarian carcinoma cells. BMC Cancer. 2014;14:477. doi: 10.1186/1471-2407-14-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young MJ, Wu YH, Chiu WT, Weng TY, Huang YF, Chou CY. All-trans retinoic acid downregulates ALDH1-mediated stemness and inhibits tumour formation in ovarian cancer cells. Carcinogenesis. 2015;36:498–507. doi: 10.1093/carcin/bgv018. [DOI] [PubMed] [Google Scholar]
- 16.Chiaramonte R, Colombo M, Bulfamante G, Falleni M, Tosi D, Garavelli S, De E, Vigolo SD, Todoerti K, Neri A, Platonova N. Notch pathway promotes ovarian cancer growth and migration via CXCR4/SDF1alpha chemokine system. Int J Biochem Cell Biol. 2015;66:134–140. doi: 10.1016/j.biocel.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Yen WC, Fischer MM, Axelrod F, Bond C, Cain J, Cancilla B, Henner WR, Meisner R, Sato A, Shah J, Tang T, Wallace B, Wang M, et al. Targeting Notch signaling with a Notch2/Notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor-initiating cell frequency. Clin Cancer Res. 2015;21:2084–2095. doi: 10.1158/1078-0432.CCR-14-2808. [DOI] [PubMed] [Google Scholar]
- 18.Seo EJ, Kim DK, Jang IH, Choi EJ, Shin SH, Lee SI, Kwon SM, Kim KH, Suh DS, Kim JH. Hypoxia-NOTCH1-SOX2 signaling is important for maintaining cancer stem cells in ovarian cancer. Oncotarget. 2016;7:55624–55638. doi: 10.18632/oncotarget.10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang HG, Kim DH, Kim SJ, Cho Y, Jung J, Jang W, Chun KH. Galectin-3 supports stemness in ovarian cancer stem cells by activation of the Notch1 intracellular domain. Oncotarget. 2016;7:68229–68241. doi: 10.18632/oncotarget.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakar SS, Worth CA, Wang Z, Carter K, Ratajczak M, Gunjal P. DOXIL when combined with Withaferin A (WFA) targets ALDH1 positive cancer stem cells in ovarian cancer. J Cancer Stem Cell Res. 2016;4:19. doi: 10.14343/JCSCR.2016.4e1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Z, Xu W, Zhang C, Liu M, Wen H. Inhibition of gamma-secretase in Notch1 signaling pathway as a novel treatment for ovarian cancer. Oncotarget. 2016;24:14152. doi: 10.18632/oncotarget.14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pant S, Jones SF, Kurkjian CD, Infante JR, Moore KN, Burris HA, McMeekin DS, Benhadji KA, Patel BK, Frenzel MJ, Kursar JD, Zamek-Gliszczynski MJ, Yuen ES, et al. A first-in-human phase I study of the oral Notch inhibitor, LY900009, in patients with advanced cancer. Eur J Cancer. 2016;56:1–9. doi: 10.1016/j.ejca.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Gong L, Ou R, Zheng Z, Chen J, Xie F, Huang X, Qiu J, Zhang W, Jiang Q, Yang Y, Zhu H, Shi Z, et al. Sequential combination therapy of ovarian cancer with cisplatin and gamma-secretase inhibitor MK-0752. Gynecol Oncol. 2016;140:537–544. doi: 10.1016/j.ygyno.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 24.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 25.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 26.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. Plos one. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You Q, Guo H, Xu D. Distinct prognostic values and potential drug targets of ALDH1 isoenzymes in non-small-cell lung cancer. Drug Des Devel Ther. 2015;9:5087–5097. doi: 10.2147/DDDT.S87197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dotsch MM, Kloten V, Schlensog M, Heide T, Braunschweig T, Veeck J, Petersen I, Knuchel R, Dahl E. Low expression of ITIH5 in adenocarcinoma of the lung is associated with unfavorable patients’ outcome. Epigenetics. 2015;10:903–912. doi: 10.1080/15592294.2015.1078049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortega CE, Seidner Y, Dominguez I. Mining CK2 in cancer. Plos one. 2014;9:e115609. doi: 10.1371/journal.pone.0115609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu M, Wang G, Gomez-Fernandez CR, Guo S. GREB1 Functions as a Growth Promoter and Is Modulated by IL6/STAT3 in Breast Cancer. Plos one. 2012;7:e46410. doi: 10.1371/journal.pone.0046410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Tilghman SL, Townley I, Zhong Q, Carriere PP, Zou J, Llopis SD, Preyan LC, Williams CC, Skripnikova E, Bratton MR, Zhang Q, Wang G. Proteomic signatures of acquired letrozole resistance in breast cancer: suppressed estrogen signaling and increased cell motility and invasiveness. Mol Cell Proteomics. 2013;12:2440–2455. doi: 10.1074/mcp.M112.023861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou C, Zhong Q, Rhodes LV, Townley I, Bratton MR, Zhang Q, Martin EC, Elliott S, Collins-Burow BM, Burow ME, Wang G. Proteomic analysis of acquired tamoxifen resistance in MCF-7 cells reveals expression signatures associated with enhanced migration. Breast Cancer Res. 2012;14:R45. doi: 10.1186/bcr3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maciejczyk A, Szelachowska J, Czapiga B, Matkowski R, Halon A, Gyorffy B, Surowiak P. Elevated BUBR1 expression is associated with poor survival in early breast cancer patients: 15-year follow-up analysis. J Histochem Cytochem. 2013;61:330–339. doi: 10.1369/0022155413480148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maciejczyk A, Lacko A, Ekiert M, Jagoda E, Wysocka T, Matkowski R, Halon A, Gyorffy B, Lage H, Surowiak P. Elevated nuclear S100P expression is associated with poor survival in early breast cancer patients. Histol Histopathol. 2013;28:513–524. doi: 10.14670/HH-28.513. [DOI] [PubMed] [Google Scholar]
- 35.Maciejczyk A, Jagoda E, Wysocka T, Matkowski R, Gyorffy B, Lage H, Surowiak P. ABCC2 (MRP2, cMOAT) localized in the nuclear envelope of breast carcinoma cells correlates with poor clinical outcome. Pathol Oncol Res. 2012;18:331–342. doi: 10.1007/s12253-011-9449-9. [DOI] [PubMed] [Google Scholar]
- 36.Adam MA. New prognostic factors in breast cancer. Adv Clin Exp Med. 2013;22:5–15. [PubMed] [Google Scholar]
- 37.Ivanova L, Zandberga E, Silina K, Kalnina Z, Abols A, Endzelins E, Vendina I, Romanchikova N, Hegmane A, Trapencieris P, Eglitis J, Line A. Prognostic relevance of carbonic anhydrase IX expression is distinct in various subtypes of breast cancer and its silencing suppresses self-renewal capacity of breast cancer cells. Cancer Chemother Pharmacol. 2015;75:235–246. doi: 10.1007/s00280-014-2635-1. [DOI] [PubMed] [Google Scholar]
- 38.Wu S, Xue W, Huang X, Yu X, Luo M, Huang Y, Liu Y, Bi Z, Qiu X, Bai S. Distinct prognostic values of ALDH1 isoenzymes in breast cancer. Tumour Biol. 2015;13:13. doi: 10.1007/s13277-014-2852-6. [DOI] [PubMed] [Google Scholar]
- 39.Hong CQ, Zhang F, You YJ, Qiu WL, Giuliano AE, Cui XJ, Zhang GJ, Cui YK. Elevated C1orf63 expression is correlated with CDK10 and predicts better outcome for advanced breast cancers: a retrospective study. BMC Cancer. 2015;15:548. doi: 10.1186/s12885-015-1569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamieniak MM, Rico D, Milne RL, Munoz-Repeto I, Ibanez K, Grillo MA, Domingo S, Borrego S, Cazorla A, Garcia-Bueno JM, Hernando S, Garcia-Donas J, Hernandez-Agudo E, et al. Deletion at 6q24.2-26 predicts longer survival of high-grade serous epithelial ovarian cancer patients. Mol Oncol. 2015;9:422–436. doi: 10.1016/j.molonc.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gayarre J, Kamieniak MM, Cazorla-Jimenez A, Munoz-Repeto I, Borrego S, Garcia-Donas J, Hernando S, Robles-Diaz L, Garcia-Bueno JM, Cajal TR, Hernandez-Agudo E, Soto VH, Marquez-Rodas I, et al. The NER-related gene GTF2H5 predicts survival in high-grade serous ovarian cancer patients. J Gynecol Oncol. 2015;12:12. doi: 10.3802/jgo.2016.27.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose SL, Kunnimalaiyaan M, Drenzek J, Seiler N. Notch 1 signaling is active in ovarian cancer. Gynecol Oncol. 2010;117:130–133. doi: 10.1016/j.ygyno.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Wang M, Wu L, Wang L, Xin X. Down-regulation of Notch1 by gamma-secretase inhibition contributes to cell growth inhibition and apoptosis in ovarian cancer cells A2780. Biochem Biophys Res Commun. 2010;393:144–149. doi: 10.1016/j.bbrc.2010.01.103. [DOI] [PubMed] [Google Scholar]
- 44.Alniaimi AN, Demorest-Hayes K, Alexander VM, Seo S, Yang D, Rose S. Increased Notch1 expression is associated with poor overall survival in patients with ovarian cancer. Int J Gynecol Cancer. 2015;25:208–213. doi: 10.1097/IGC.0000000000000359. [DOI] [PubMed] [Google Scholar]
- 45.Park JT, Li M, Nakayama K, Mao TL, Davidson B, Zhang Z, Kurman RJ, Eberhart CG, Shih I, Wang TL. Notch3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 46.Choi JH, Park JT, Davidson B, Morin PJ, Shih I, Wang TL. Jagged-1 and Notch3 juxtacrine loop regulates ovarian tumor growth and adhesion. Cancer Res. 2008;68:5716–5723. doi: 10.1158/0008-5472.CAN-08-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung SG, Kwon YD, Song JA, Back MJ, Lee SY, Lee C, Hwang YY, An HJ. Prognostic significance of Notch 3 gene expression in ovarian serous carcinoma. Cancer Sci. 2010;101:1977–1983. doi: 10.1111/j.1349-7006.2010.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu W, Liu T, Ivan C, Sun Y, Huang J, Mangala LS, Miyake T, Dalton HJ, Pradeep S, Rupaimoole R, Previs RA, Han HD, Bottsford-Miller J, et al. Notch3 pathway alterations in ovarian cancer. Cancer Res. 2014;74:3282–3293. doi: 10.1158/0008-5472.CAN-13-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang H, Jeong JY, Song JY, Kim TH, Kim G, Huh JH, Kwon AY, Jung SG, An HJ. Notch3-specific inhibition using siRNA knockdown or GSI sensitizes paclitaxel-resistant ovarian cancer cells. Mol Carcinog. 2016;55:1196–1209. doi: 10.1002/mc.22363. [DOI] [PubMed] [Google Scholar]
- 50.Gallahan D, Jhappan C, Robinson G, Hennighausen L, Sharp R, Kordon E, Callahan R, Merlino G, Smith GH. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 1996;56:1775–1785. [PubMed] [Google Scholar]
- 51.Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, Callahan R. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992;6:345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- 52.Soriano JV, Uyttendaele H, Kitajewski J, Montesano R. Expression of an activated Notch4(int-3) oncoprotein disrupts morphogenesis and induces an invasive phenotype in mammary epithelial cells in vitro. Int J Cancer. 2000;86:652–659. doi: 10.1002/(sici)1097-0215(20000601)86:5<652::aid-ijc8>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 53.Callahan R, Raafat A. Notch signaling in mammary gland tumorigenesis. J Mammary Gland Biol Neoplasia. 2001;6:23–36. doi: 10.1023/a:1009512414430. [DOI] [PubMed] [Google Scholar]
- 54.Gao MQ, Choi YP, Kang S, Youn JH, Cho NH. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene. 2010;29:2672–2680. doi: 10.1038/onc.2010.35. [DOI] [PubMed] [Google Scholar]
- 55.Roemer K. Notch and the p53 clan of transcription factors. Adv Exp Med Biol. 2012;727:223–240. doi: 10.1007/978-1-4614-0899-4_17. [DOI] [PubMed] [Google Scholar]
- 56.de Graeff P, Crijns AP, de Jong S, Boezen M, Post WJ, de Vries EG, van der Zee AG, de Bock GH. Modest effect of p53, EGFR and HER-2/neu on prognosis in epithelial ovarian cancer: a meta-analysis. Br J Cancer. 2009;101:149–159. doi: 10.1038/sj.bjc.6605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garziera M, Montico M, Bidoli E, Scalone S, Sorio R, Giorda G, Lucia E, Toffoli G. Prognostic Role of Serum Antibody Immunity to p53 Oncogenic Protein in Ovarian Cancer: A Systematic Review and a Meta-Analysis. Plos one. 2015;10:e0140351. doi: 10.1371/journal.pone.0140351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yun J, Espinoza I, Pannuti A, Romero D, Martinez L, Caskey M, Stanculescu A, Bocchetta M, Rizzo P, Band V, Band H, Kim HM, Park SK, et al. p53 Modulates Notch Signaling in MCF-7 Breast Cancer Cells by Associating With the Notch Transcriptional Complex Via MAML1. J Cell Physiol. 2015;230:3115–3127. doi: 10.1002/jcp.25052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 60.Beel AJ, Sanders CR. Substrate specificity of gamma-secretase and other intramembrane proteases. Cell Mol Life Sci. 2008;65:1311–1334. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]